Abstract
Sucrose-nonfermentation1-related protein kinase1 (SnRK1) is an evolutionarily conserved energy sensor protein that regulates gene expression in response to energy depletion in plants. Efforts to elucidate the functions and mechanisms of this protein kinase are hampered, however, by inherent growth defects of snrk1-null mutant plants. To overcome these limitations and study SnRK1 functions in vivo, we applied a method combining transient expression in leaf mesophyll protoplasts and stable expression in transgenic plants. We found that both rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) SnRK1 activities critically influence stress-inducible gene expression and the induction of stress tolerance. Genetic, molecular, and chromatin immunoprecipitation analyses further revealed that the nuclear SnRK1 modulated target gene transcription in a submergence-dependent manner. From early seedling development through late senescence, SnRK1 activities appeared to modulate developmental processes in the plants. Our findings offer insight into the regulatory functions of plant SnRK1 in stress-responsive gene regulation and in plant growth and development throughout the life cycle.
Living organisms sense and respond to cellular sugar starvation and/or energy depletion to survive and sustain function. Limited availability of sugar or energy in eukaryotic cells activates an evolutionarily conserved energy-sensing protein kinase (PK) known as Sucrose-nonfermentation1 (Snf1) in yeast, AMPK in mammals, and Snf1-related protein kinase1 (SnRK1) in plants (Polge and Thomas, 2007; Hardie, 2011). As sessile organisms, land plants endure continual environmental fluctuations that may abruptly deplete their energy stores. To survive such challenges, plant SnRK1s modulate transcriptional, metabolic, and developmental processes (Hey et al., 2010; Smeekens et al., 2010). In Arabidopsis (Arabidopsis thaliana), SnRK1s KIN10 and KIN11 manage energy-depleting conditions through the control of stress-responsive gene expression and stress hormone abscisic acid signaling, which also modulates plant susceptibility to pathogens (Baena-González et al., 2007; Jossier et al., 2009). Rice (Oryza sativa) OsSnRK1, which is under the control of calcineurin B-like interacting protein kinase 15 (Lee et al., 2009), derepresses Glc-repressed gene expression in the embryo to modulate seed germination and early seedling growth (Lu et al., 2007).
AtSnRK1 activity is repressed by inorganic phosphate starvation and trehalose-6-phosphate application (Fragoso et al., 2009; Zhang et al., 2009) and binds to myoinositol polyphosphate-5-phosphatase, which negatively regulates the kinase activity (Ananieva et al., 2008). AtSnRK1-activating kinases, AtSnAK1 and AtSnAK2, and AtSnRK1 establish a dynamic and reciprocal regulatory process though feedback phosphorylation (Shen et al., 2009; Crozet et al., 2010). Although the function and regulation of SnRK1 have been studied in various plants, the molecular and cellular mechanisms for the multiple functions of this signaling kinase in plant growth and development are not well understood.
In this study, we investigated the mechanisms of SnRK1 signaling through an integration of plant cell- and transgenic plant-based assays. This PK has a central role in flooding-responsive gene expression through its direct association with target gene chromatins in the nucleus, and by this route the PK contributes to the induction of plant resistance to flooding stress. SnRK1 signaling also plays a central role in modulating early seedling development after seed germination and in controlling age-dependent leaf senescence.
RESULTS
SnRK1 Activates Specific Stress-Responsive Gene Expression in the Nucleus
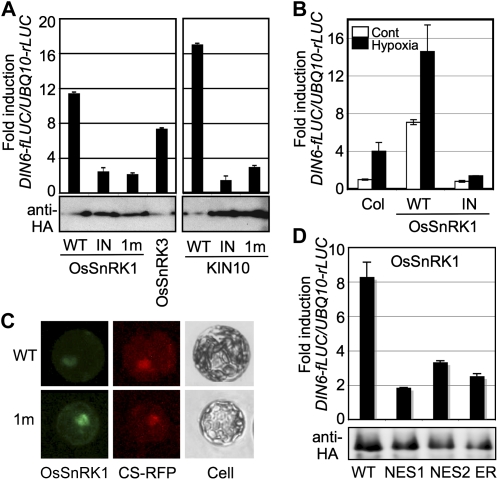
To deal with energy-depleting stress, eukaryotic organisms express orthologous signaling kinases, notably Snf1 in yeast, AMPK in mammals, and SnRK1 in plants (Polge and Thomas, 2007). Arabidopsis SnRK1s (KIN10 and KIN11) have high structural similarity to the catalytic α-subunits of yeast Snf1 and mammalian AMPK (Amodeo et al., 2007; Chen et al., 2009; Xiao et al., 2011) and play similar regulatory roles in stress-inducible gene expression (Baena-González et al., 2007; Jossier et al., 2009; Hardie, 2011). In this study, we used a transient expression system based on Arabidopsis leaf mesophyll protoplasts (Yoo et al., 2007) to demonstrate the evolutionarily conserved gene regulatory functions of rice OsSnRK1. When an individual effector construct of OsSnRK1, OsSnRK3, or KIN10 was transfected to leaf mesophyll protoplasts, with either a DARK-INDUCED6 (DIN6; At3g47340) or GLUTATHIONE S-TRANSFERASE6 (GST6; At2g47730) promoter-driven firefly luciferase (fLUC) construct as a reporter gene, each effector could activate the DIN6 promoter (Fig. 1A) but not the GST6 promoter (Supplemental Fig. S1A). The inability to activate the promoter of GST6, which is categorized as a general stress-responsive gene (Branco-Price et al., 2008), showed that plant SnRK1s mediate a specific pathway of stress signaling.
Figure 1.
SnRK1 activates specific stress-responsive gene expression. A, The DIN6-fLUC reporter was activated by wild-type (WT) SnRK1s but not by inactive ones. Inactive forms were ATP-binding site (IN)- and catalytic site (1m)-mutated SnRK1s. Protoplasts were cotransfected with a designated effector construct with DIN6-fLUC and UBQ10-rLUC constructs as reporters. UBQ10 promoter activity served as a transfection control. HA-tagged SnRK1 expression is shown by protein blot analysis using anti-HA antibody. B, Wild-type OsSnRK1 activated DIN6-fLUC under the hypoxia condition but not the inactive one. All experiments were repeated three times with consistent results. The means of triplicate measurements are shown with se bars. C, Transiently expressed GFP fusion proteins of the wild type and the inactive form (1m) of OsSnRK1 are shown in Arabidopsis leaf mesophyll protoplasts under fluorescence microscopy (200× magnification). CS-RFP (for red fluorescent protein) was used as a nuclear marker. Cell images were also taken under bright field as a control. D, OsSnRK1-NES1, -NES2, and -ER compromised the PK capacity to induce DIN6 promoter activity in leaf mesophyll cells. HA-tagged SnRK1 expression is shown by protein-blot analysis using anti-HA antibody. [See online article for color version of this figure.]
To validate the PK function in gene regulation, inactive forms of SnRK1s were prepared (Fig. 1A) as ATP-binding site-mutated PKs (OsSnRK1K43M [OsSnRK1_IN] and KIN10K48M [KIN10_IN]) and as catalytically inactive PKs (OsSnRK1K139R [SnRK1_1m] and KIN10K144R [KIN10_1m]). The phosphorylation of OsSnRK1Thr-170 and KIN10Thr-175 was detected in all types of SnRK1s by anti-p-AMPKα antibody (Supplemental Fig. S2A). Even so, none of the mutant kinases was able to activate the DIN6-fLUC reporter (Fig. 1A), suggesting that intact PK activities of OsSnRK1 and KIN10 are essential for gene regulation.
It is reported that KIN10 participates in response to an energy-depleting hypoxic condition (Baena-González et al., 2007). When we applied the same condition in the presence of rice SnRK1, hypoxia-inducible DIN6-fLUC reporter activity was further enhanced (Fig. 1B) in a manner similar to that of KIN10. Conversely, the inactive form of OsSnRK1_IN suppressed the hypoxia-induced DIN6-fLUC activity, confirming that the PK activity of OsSnRK1 is essential in activating specific stress (hypoxia)-responsive gene expression. These results indicated that rice OsSnRK1 shares not only protein sequence and structure but also stress-responsive regulatory functions with Arabidopsis KIN10 (Fig. 1, A and B). Since OsSnRK1 exhibited highly conserved signaling functions, we decided to further investigate the molecular and cellular mechanisms of SnRK1 from both Arabidopsis and rice in Arabidopsis leaf mesophyll protoplasts and transgenic plant systems.
The nuclear function of plant SnRK1s was expected from the orthologous gene functions in yeast and mammals (Ahuatzi et al., 2007; Bungard et al., 2010); hence, we carefully examined the subcellular localization of a GFP fusion protein of the wild type and an inactive form of OsSnRK1 in leaf mesophyll cells. To exclude artifacts of overexpression, the transfected protoplasts were incubated for less than 6 h. Both the wild type and the catalytically inactive form (1m) of OsSnRK1 were observed in multiple locations including the nucleus, implying that the nuclear localization of OsSnRK1 is independent of its kinase activity (Fig. 1C). The nuclear localization of OsSnRK1 was further confirmed through its colocalization with a nuclear marker protein fused with red fluorescent protein. This finding is consistent with a previous report of Arabidopsis SnRK1 localization (Bitrián et al., 2011).
Since OsSnRK1-GFP is approximately 84.5 kD, this fusion protein cannot move into the nucleus by diffusion. The pattern of OsSnRK1-GFP localization implied that SnRK1 is actively shuttled into the nucleus. Cellular trafficking between the nucleus and the cytoplasm occurs through nuclear pore complexes in the nuclear membrane. Most proteins actively transported into the nucleus possess a nuclear localization signal, whereas proteins destined to leave the nucleus are typically tagged with a nuclear export signal (NES; Poon and Jans, 2005). Although SnRK1s do not possess an obvious nuclear localization signal, they must be able to leave the cytoplasm and enter the nucleus under certain conditions.
Although the transport mechanism of plant SnRK1s is unclear, its nuclear localization (Bitrián et al., 2011; Fig. 1C) provided a rationale to further explore the roles of SnRK1s in gene regulation. We first excluded OsSnRK1 from the nucleus by fusing the C terminus of OsSnRK1 to the two different types of NES or to the endoplasmic reticulum (ER) retention signal peptide. Fusion of OsSnRK1 with the NES1, NES2, or ER retention signal peptide significantly reduced the capacity of the PK to induce DIN6 promoter activity in leaf mesophyll cells, suggesting that the nuclear localization of SnRK1 is essential for the PK gene regulatory function (Fig. 1D).
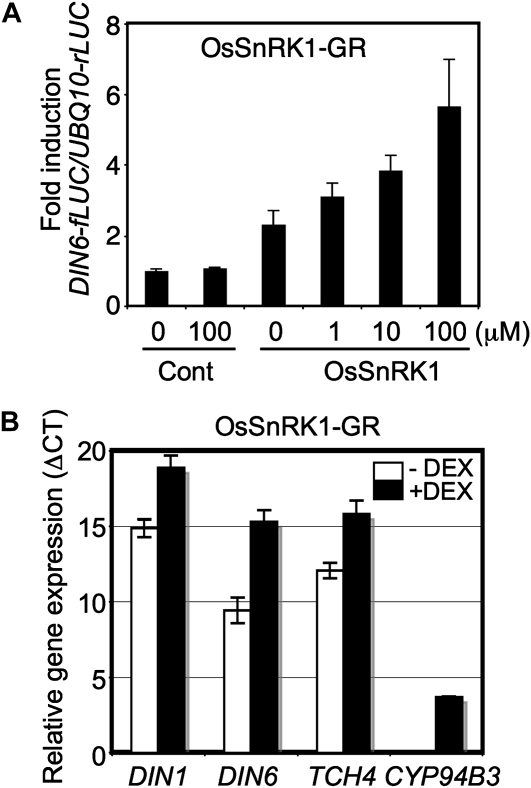
We further tested the importance of nuclear localization by fusing the C-terminal end of OsSnRK1 to the ligand-binding domain of rat glucocorticoid receptor (GR), followed by a hemagglutinin (HA) epitope (Supplemental Fig. S2A). On binding to a glucocorticoid hormone (i.e. dexamethasone [DEX]), the GR conveys the associated protein into the nucleus (Aoyama and Chua, 1997). DEX application did not alter the protein abundance or phosphorylation status of OsSnRK1, as shown by protein blotting using anti-HA or anti-p-AMPKα antibody, respectively (Supplemental Fig. S2B). Even so, OsSnRK1-GR activated the DIN6-fLUC reporter in a DEX dosage-dependent manner (Fig. 2A), showing that SnRK1 retains activity in the nucleus. We noted, however, that activation of the reporter was relatively weak, which may be attributed to the perturbation of OsSnRK1 function by the fused GR domain. The relatively large size of the GR presents a limitation in maintaining intact full activity of the fused protein. For instance, OsSnRK1 with GR fused to its N-terminal end (Supplemental Fig. S2A) could not induce DIN6 promoter activity at all, even in the presence of DEX (Supplemental Fig. S2C).
Figure 2.
SnRK1 needs to be localized in the nucleus to affect gene expression. A, The DIN6-fLUC reporter was activated by OsSnRK1-GR-HA upon treatment with DEX. The experiments were repeated twice with consistent results. The means of triplicate measurements are shown with se bars. B, DIN1, DIN6, TCH4, and CYP94B3 expression was measured in OsSnRK1-GR-expressing transgenic plants using real-time RT-PCR after DEX application (10 μm for 1 h). All experiments were repeated three times with consistent results. The means of triplicate measurements are shown with se bars.
To validate the cell-based findings in whole plants, we generated transgenic Arabidopsis plants expressing OsSnRK1-GR. After the selection of T2 plants, we searched for transgenic lines expressing OsSnRK1-GR at a level comparable to that of endogenous KIN10 using quantitative reverse transcription (RT)-PCR (Supplemental Fig. S2D). Gene expression was monitored in the selected transgenic lines in 1 h after DEX application. As markers for SnRK1 signaling, DIN1 (SEN1; At4g35770) and DIN6 (Baena-González et al., 2007) expression was measured, and CYP94B3 (At3g48520) and TCH4 (At5g57560) served as markers for spray-induced touch stimulus (Reymond et al., 2000) in the transgenic plants (Fig. 2B). DEX spraying led to significant induction of the marker gene expression, but the same DEX spraying protocol did not yield a similar degree of induction in Columbia (Col) plants (data not shown). These results gave further evidence that OsSnRK1-GR at a nuclear location induced the expression of marker genes. More generally, evidence from both the cellular and plant-based systems showed that plant SnRK1 activity regulated specific gene expression from the nucleus.
Nuclear SnRK1 Activity Regulates Gene Expression under Flooding Stress
To examine SnRK1 function in stress-responsive gene expression in plants, we generated transgenic Arabidopsis expressing wild-type OsSnRK1 and KIN10 as well as inactive OsSnRK1 (IN and 1m) and KIN10 (IN). The IN and 1m genes were mutated as described in Figure 1A. Transgene expression was measured using quantitative RT-PCR, and lines with similar levels of expression and normal plant growth were selected for further analyses (Supplemental Fig. S3).
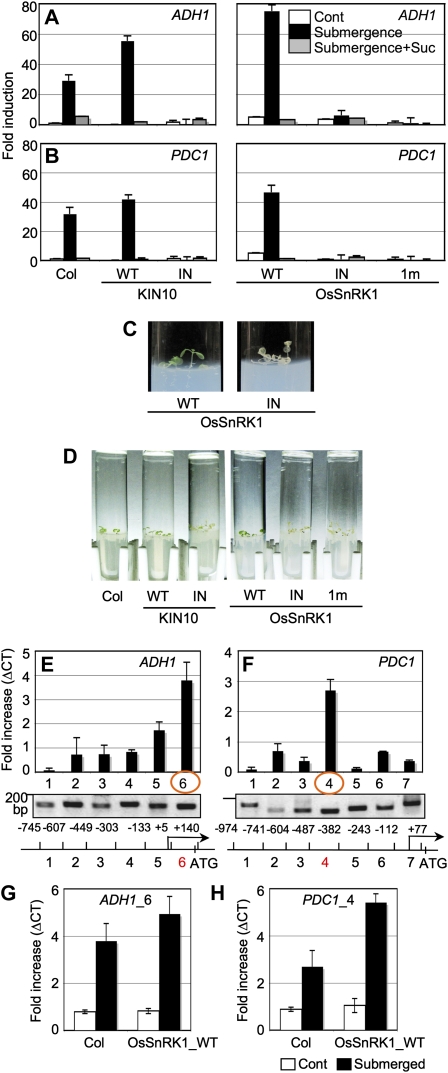
Hypoxia (i.e. oxygen deprivation) activated OsSnRK1, which led to DIN6 promoter activation (Fig. 1B). To test whether SnRK1 activity is essential for gene regulation during oxygen deprivation, total RNA was extracted from Col plants and transgenic plants expressing SnRK1s grown on Murashige and Skoog (MS) agar medium containing 0.5% Suc for 8 d followed by complete submergence for 24 h. Real-time RT-PCR was used to measure the expression of two marker genes for the flooding stress response, ALCOHOL DEHYDROGENASE1 (ADH1; At1g77120) and PYRUVATE DECARBOXYLASE1 (PDC1; At4g33070). ADH1 and PDC1 were up-regulated in Col by submergence and were further up-regulated in transgenic plants expressing wild-type KIN10 and OsSnRK1 (Fig. 3, A and B). Strikingly, the gene induction by submergence was completely absent in transgenic plants expressing inactive forms of KIN10 and OsSnRK1. These inactive SnRK1s expressed the strong dominant-negative function in whole plants (Fig. 3) as in leaf mesophyll cells (Fig. 1A), although no SnRK1 activity was measured in this study. Expression of a general stress-responsive GST6 was not induced by the treatment (Supplemental Fig. S4), confirming that SnRK1 activity mediates a specific stress-signaling pathway. To further investigate the relationship between cellular energy depletion and oxygen deprivation imposed by submergence, we examined the gene responses to Suc supplements. Interestingly, the induction of ADH1 and PDC1 expression caused by submergence was abolished in the presence of 90 mm Suc (Fig. 3, A and B). Thus, Suc application appeared to repress SnRK1 activity and its downstream pathway, which are important to induce ADH1 and PDC1 by submergence.
Figure 3.
SnRK1 regulates gene expression as well as binds directly to specific target genes in response to oxygen deprivation under flooding. A and B, ADH1 (A) and PDC1 (B) expression was induced by submergence. This induction was abolished in the presence of inactive forms of SnRK1s or 90 mm Suc. RNA was extracted from Col as well as transgenic plants expressing wild-type (WT) or inactive forms of SnRK1 under the submergence condition (24 h), and marker gene expression was measured using real-time RT-PCR. All experiments were repeated three times with consistent results. The means of triplicate measurements are shown with se bars. C, Wild-type OsSnRK1-expressing transgenic plants distinctively displayed green shoots, but OsSnRK1_IN-expressing transgenic plants turned albino under a prolonged flooding condition in the dark. D, Transgenic plants expressing inactive forms of SnRK1 resulted in the accelerated loss of chlorophyll during flooding conditions in the dark. Experiments were repeated three times with triplicates and showed consistent results. E and F, SnRK1 associated with ADH1 (E) and PDC1 (F) promoters under flooding. All gene-specific primer sets generated PCR products of expected sizes with Arabidopsis genomic DNA isolated from Col as controls (gels at bottom). Structures of promoters were analyzed and depicted as schematic diagrams. G and H, Chromatin association of SnRK1s at ADH1_6 (G) and PDC1_4 (H) regions was further increased in transgenic plants expressing wild-type OsSnRK1 under flooding. The experiments were repeated twice with consistent results. The means of triplicate measurements are shown with se bars. [See online article for color version of this figure.]
To further support the role of SnRK1 in stress-responsive gene expression, we designed a phenotypic assay to monitor SnRK1 activity in planta under the conditions of flooding stress. For the initial 3 d, Col and transgenic plants were grown on MS agar medium containing 0.5% Suc prepared in a test tube under a photoperiod of 16 h of light/8 h of dark (Supplemental Fig. S5A). Seedlings were then completely submerged and incubated in the dark for another 37 d to observe seedling phenotype in response to oxygen deprivation. The seedling growth was arrested more or less completely by flooding, but transgenic seedlings expressing wild-type OsSnRK1 maintained green shoots, quite in contrast to the pale transparent shoots of transgenic plants expressing OsSnRK1_IN (Fig. 3C). Surprisingly, Col also preserved relatively green shoots during the prolonged submergence in the dark, similar to transgenic seedlings expressing wild-type SnRK1s (Fig. 3D; Supplemental Fig. S6). This indicated that endogenous SnRK1 activity may be sufficient to induce the adaptive responses. Since all the transgenic plants expressing inactive forms of SnRK1s failed to preserve any green shoots under these conditions, SnRK1 activity must be essential to establish tolerance in plants against flooding stress. Alternatively, when plants were continually kept under a photoperiod of 16 h of light/8 h of dark without any treatment, all the transgenic seedlings grew more or less similarly to Col (Supplemental Fig. S5B).
Signaling kinases that control gene regulation are often directly associated with target gene chromatins in the nucleus (Chow and Davis, 2006). Bungard et al. (2010) reported that metabolic stress-activated mammalian AMPK accumulates at target gene chromatins in the nucleus and phosphorylates histone H2B Ser-36 at target gene promoters and transcribed regions. Based on our own data indicating that plant SnRK1s regulate gene function through nuclear activities (Figs. 1 and 2; Supplemental Figs. S1 and S2), we tested whether SnRK1s associate directly with target gene promoters (ADH1 and PDC1) under submergence. We performed chromatin immunoprecipitation (ChIP) using anti-phospho-AMPKα antibody to pull down the phosphorylated SnRK1s cross-linked to the endogenous genomic DNA in vivo. An equal level of PCR products of a control gene was analyzed concurrently as an input control using DNA de-cross-linked before immunoprecipitation (Supplemental Fig. S7A). To detect SnRK1 association, the genomic DNA was isolated from the chromatins and analyzed by real-time PCR using a set of primers spanning promoter regions of about 0.8 and 1 kb in ADH1 and PDC1, respectively (Fig. 3, E and F). As a positive control, all primer sets generated PCR products of the expected size using Col genomic DNA (Fig. 3, E and F, gels at bottom). When gene expression levels were presented as differences of cycle threshold values (ΔCT) normalized to those of Col plants not submerged, OsSnRK1 and KIN10 showed high binding specificity at the ADH1_6 and PDC1_4 regions but not at neighboring regions in submerged Col and wild-type OsSnRK1-expressing transgenic plants. These results clearly showed that the PK associated with specific target gene chromatins in a signaling-dependent manner (Fig. 3, E and F). Moreover, these DNA regions were further enriched in DNA pools collected from submerged transgenic plants expressing wild-type OsSnRK1 compared with submerged Col plants (Fig. 3, G and H). All results indicated that SnRK1s bound directly to target chromatins and modulated stress-responsive gene expression in the nucleus. The combined molecular, cellular, and phenotypic analyses of wild-type SnRK1 and inactive forms of SnRK1 expressed in transgenic plants provide evidence for specific roles of SnRK1 activity in response to oxygen deprivation by submergence.
SnRK1 Modulates Early Seedling Development
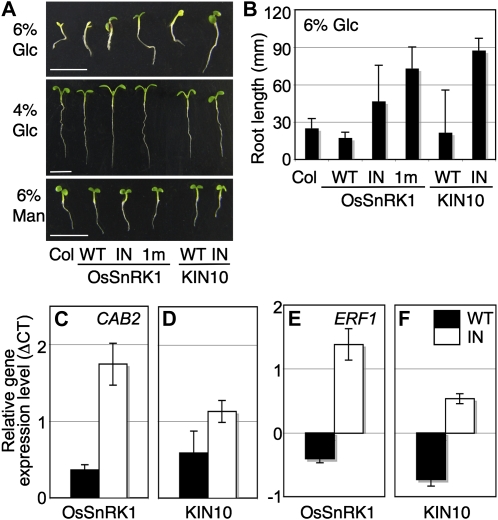
Plant seedlings undergo a metabolic transition from auxotrophic to autotrophic nutrition after seed germination (Moore et al., 2003; Cho et al., 2010). A complete shift in trophism at the early seedling stage is critical for plants to secure a healthy life span. When we examined seedling growth on 6% (w/v) Glc agar medium with full-strength MS salts, Col seedlings exhibited a typical early developmental arrest characterized by the inhibition of hypocotyl and root growth and the repression of cotyledon expansion and chlorophyll accumulation (Fig. 4A). These responses depend largely on the plant Glc sensor Hexokinase1 (HXK1; Moore et al., 2003). Mannitol at 6% did not evoke a similar developmental arrest phenotype, thereby excluding a nonspecific osmotic effect in this plant response (Fig. 4A). Transgenic plants expressing wild-type OsSnRK1 and KIN10 were sensitive to high (6%) Glc (Fig. 4A), which is consistent with the hypersensitivity shown in the presence of high Glc by Arabidopsis seedlings that overexpress KIN10 (Jossier et al., 2009). The effect seemed to be marginal, however, because these transgenic lines did not show a similar developmental arrest phenotype at 4% Glc.
Figure 4.
SnRK1 modulates early seedling development. A, Transgenic plants expressing wild-type (WT) and inactive forms of OsSnRK1 or KIN10 showed different sensitivities to 6% Glc. The similar seedling repression was not observed under 6% mannitol (Man), which was used as an osmotic control, and 4% Glc as a subpotent condition. All experiments were repeated three times with consistent results. Bars = 5 mm. B, Quantitative analysis of the Glc repression of root elongation of Col and transgenic plants. Each measurement represents the mean of primary root length with an error bar indicating sd of 50 samples. C and D, Inactive forms of SnRK1-expressing transgenic plants were insensitive to Glc repression of CAB2 (At1g29920). E and F, ERF1 (At3g23240) expression was further enhanced in inactive forms of SnRK1-expressing transgenic seedlings by Glc signaling. Values were normalized based on those obtained from seedlings treated with Man, and means of triplicate measurements with se bars are shown. The experiments were repeated twice with similar results. [See online article for color version of this figure.]
More interestingly, all transgenic lines expressing inactive SnRK1s (OsSnRK1_IN, 1m, and KIN10_IN) were insensitive to high Glc repression (Fig. 4A). To quantify seedling sensitivity to Glc, we measured the root lengths of Col and transgenic seedlings (Fig. 4B). The root lengths of transgenic seedlings expressing inactive SnRK1s were longer than Col and transgenic seedlings expressing wild-type SnRK1s, further supporting the involvement of PK activity in seedling Glc sensitivity. It appears that early seedling establishment may require the coordination of SnRK1 activity with the well-studied HXK1 signaling.
To gain insight into the relationships between Glc-inducible responses and SnRK1 activity, we examined the expression of two well-known Glc (CHLOROPHYLL A/B-BINDING PROTEIN2 [CAB2]; At1g29920) and ethylene (ETHYLENE RESPONSE FACTOR1 [ERF1]; At3g23240) signaling marker genes using real-time RT-PCR. Total RNA was extracted from 7-d-old Col and transgenic seedlings expressing wild-type OsSnRK1, wild-type KIN10, OsSnRK1_IN, and KIN10_IN after 6% Glc treatment for 6 h. The gene responses were normalized to those of seedlings treated with 6% mannitol that served as an osmotic control. CAB2 expression was reduced in transgenic seedlings expressing wild-type OsSnRK1 and KIN10, but not in OsSnRK1_IN and KIN10_IN-expressing seedlings, which suggested that SnRK1 activity can positively modulate Glc signaling (Fig. 4, C and D). ERF1 expression was suppressed in transgenic seedlings expressing wild-type OsSnRK1 and KIN10 but clearly enhanced in transgenic seedlings expressing inactive forms of OsSnRK1 and KIN10 (Fig. 4, E and F), providing evidence that Glc and ethylene signaling antagonize each other (León and Sheen, 2003).
SnRK1 Activity Extends Leaf Longevity during Senescence
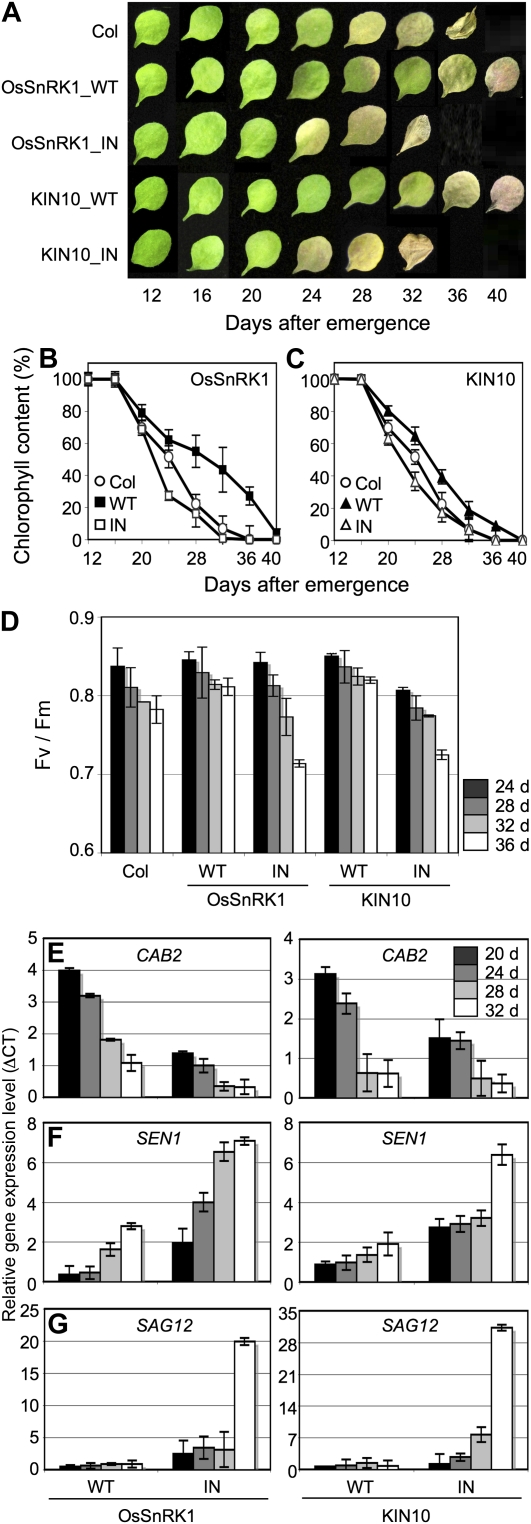
Since KIN10 and KIN11 double loss-of-function mutants are inherently predisposed to growth defects (Baena-González et al., 2007), transgenic lines expressing inactive forms of SnRK1s are especially useful to study PK functions in plants as they mature and senesce. Here, we observed leaf senescence in transgenic plants by monitoring leaf phenotype and several parameters of senescence to evaluate the regulatory roles of SnRK1 during the later stages of plant development. Since each individual leaf of a plant ages differently, only the third and the fourth leaves of a given plant were used for further analysis. Col leaves started to turn yellow at the leaf age of 28 d, counting from its emergence, and yellowing gradually spread through the whole leaf, as senescence proceeded (Fig. 5A). Col leaves started to show signs of necrosis at the age of 36 d. On the contrary, the leaves of transgenic plants expressing wild-type OsSnRK1 and KIN10 senesced more slowly; even the mutant leaves maintained their shapes until the age of 40 d. Consistently, transgenic plants expressing inactive forms of SnRK1s (OsSnRK1_IN and KIN10_IN) displayed accelerated yellowing of the leaves when compared with Col plants (Fig. 5A), suggesting that SnRK1 activity may influence leaf longevity. We observed a similar phenotype in transgenic plants expressing OsSnRK1_1m (data not shown).
Figure 5.
Age-dependent senescence. A, Phenotypes of the third or fourth rosette leaves of Col and transgenic plants expressing wild-type (WT) and inactive forms of OsSnRK1 or KIN10 at different ages. Wild-type SnRK1-expressing transgenic plants showed extended leaf longevity. The photographs show representative leaves at each time point. B and C, Chlorophyll content was measured from the third and fourth rosette leaves after leaves were fully grown at the indicated ages. Means of triplicate measurements presented with error bars indicating sd of triplicate measurements are shown. D, The photochemical efficiency of PSII (maximum photochemical efficiency of PSII in the dark-adapted state, [Fv/Fm]) was measured from the mature rosette leaves. Error bars indicate sd. E to G, Age-dependent changes in gene expression of CAB2 (E), SEN1 (F), and SAG12 (G). Values were normalized based on those obtained from Col, and means of triplicate measurements with se bars are shown. The experiments were repeated twice with similar results. [See online article for color version of this figure.]
The symptoms of age-dependent senescence were quantified by measuring changes in chlorophyll content (Fig. 5, B and C), photochemical efficiency (Fig. 5D), and the expression of senescence-associated genes (Fig. 5, E–G). Consistently, the chlorophyll loss was delayed in transgenic plants expressing wild-type OsSnRK1 and KIN10 when compared with that of Col plants during natural age-dependent senescence (Fig. 5, B and C). Analysis of chlorophyll fluorescence has been used to determine photosynthetic yields of PSII and for measuring photosynthetic activity (Akimoto and Mimuro, 2007). Measurements of photosynthetic yields of PSII in green leaf tissues using pulsed amplitude-modulated fluorescence showed that photochemical efficiency in Col leaves declined to 11% by the age of 28 d, whereas values in transgenic plants expressing wild-type OsSnRK1 and KIN10 leaves remained relatively unchanged even at the age of 36 d (Fig. 5D). This suggested that the functional integrity of PSII was maintained much longer in transgenic plant leaves expressing wild-type SnRK1s. Consistently, the photosynthetic yields of transgenic plants expressing inactive forms of SnRK1s diminished rapidly from the age of 36 d, suggesting that PSII lost integrity more rapidly than in plants with active SnRK1s. Moreover, in wild-type OsSnRK1 and KIN10-expressing transgenic plants, a photosynthesis-related gene, CAB2, was expressed at higher levels at the later stages compared with transgenic plants expressing inactive forms of SnRK1s (Fig. 5E). In addition, the induction of two senescence-associated genes, SENESCENCE1 (SEN1; At4g35770) and SENESCENCE-ASSOCIATED GENE12 (SAG12; At5g45890), was delayed (Fig. 5, F and G). These results revealed the involvement of SnRK1 in extending leaf longevity, delaying leaf senescence, and controlling various senescence-associated physiological and molecular symptoms in plants. In wild-type OsSnRK1 and KIN10-expressing transgenic plants, symptoms of senescence were also delayed under conditions imposed to accelerate senescence, specifically by treatment of detached leaves with 1-aminocyclopropane-1-carboxylic acid, in a manner similar to age-dependent senescence (data not shown). As a whole, these findings showed that SnRK1s are involved in diverse physiological processes throughout the life cycle of plants, from the time of seedling establishment to senescence.
DISCUSSION
This study provides molecular, cellular, and genetic evidence that SnRK1 activity plays multiple essential roles in plant growth and development and that, from a nuclear location, SnRK1 regulates specific stress-inducible genes involved in physiological adaptation to stress.
The nuclear SnRK1 function in plants was much anticipated from orthologous gene functions in yeast and mammals (Ahuatzi et al., 2007; Bungard et al., 2010). Yeast Hxk2 functions in the nucleus through protein-protein interactions with another nuclear protein, Mig1, which is a protein substrate of Snf1 (Ahuatzi et al., 2007). Under low-Glc conditions, Snf1 phosphorylates Mig1 and the phosphorylated Mig1 exits the nucleus. This exclusion of the phosphorylated Mig1 derepresses Glc-repressed gene expression. Furthermore, using a fluorescent reporter, AMPK activity can be directly measured in the nucleus (Tsou et al., 2011). In this study, we found evidence that not only Arabidopsis SnRK1 (Bitrián et al., 2011) but also rice SnRK1 regulates gene activity from a nuclear location. We demonstrated that rice OsSnRK1 and OsSnRK3, which share sequence and structure similarities with Arabidopsis KIN10, yeast Snf1, and mammalian AMPK (Amodeo et al., 2007; Chen et al., 2009; Xiao et al., 2011), induced the activity of the DIN6 promoter and responded to hypoxia in a manner similar to Arabidopsis SnRK1 (Fig. 1, A and B).
The observation of OsSnRK1 function with respect to subcellular location showed that the PK may regulate gene expression in the nucleus (Figs. 1, 2, and 3, E–H). First, OsSnRK1-GFP was observed in the nucleus (Fig. 1C). Nuclear SnRK1 function in gene regulation was also tested using OsSnRK1 fused with NES or the ER retention signal peptide, and none of these fusion proteins could activate DIN6 promoter activity as wild-type OsSnRK1 did (Fig. 1D). Furthermore, OsSnRK1-GR fusion proteins that shuttle from the cytosol into the nucleus upon DEX application clearly showed the induction of DIN6-fLUC reporter activity in leaf mesophyll cells after treatment of the cells with the ligand (Fig. 2A). In OsSnRK1-GR-expressing transgenic plants, DEX treatment appeared to enhance DIN1, DIN6, TCH4, and CYP94B3 gene induction (Fig. 2B). OsSnRK1-GR is a large protein, approaching 90 kD, and therefore it cannot move in and out of the nucleus by diffusion. Consequently, any activation of the reporter in this situation would reflect the active translocation of OsSnRK1-GR to the nucleus. In accordance with the mode of action of the glucocorticoid receptor, the activity of a GR fusion protein will require translocation of the ligand-bound receptor-fusion protein into the nucleus (Aoyama and Chua, 1997).
We also showed that SnRK1 induces stress-responsive gene expression (Figs. 1 and 2) through direct association with target gene chromatins and that this response increases tolerance to stress in plants under submergence (Fig. 3). Specifically, submergence induced ADH1 and PDC1 (Fig. 3, A and B), which are involved in establishing stress tolerance in plants (Fig. 3, C and D), and this induction was correlated with the direct association of SnRK1s with target gene chromatins (Fig. 3, E–H). The specific promoter regions of ADH1 and PDC1 were enriched in the chromatin-DNA immunoprecipitated from Arabidopsis seedlings subjected to submergence (Fig. 3, E–H).
Recent reviews present evidence supporting ours that signaling proteins involved in gene expression associate with target gene chromatins in the nucleus (Chow and Davis, 2006; Baek, 2011). Plant SnRK1s have no typical DNA-binding motif; hence, we cautiously speculate that, in the conditions of submergence we observed, the PKs associated with target gene chromatins by way of protein complexes that recruited specific DNA-binding partners. Accordingly, a computational analysis of the PDC1 promoter sequence using AGRIS (http://arabidopsis.med.ohio-state.edu/) revealed a highly conserved cis-element for binding of ATB2/AtbZIP53/AtbZIP44/GBF5 transcription factors (Supplemental Fig. S7B). Consistent with the location of a functional regulatory cis-element upstream of the PDC1 coding region, SnRK1s bound specifically to the PDC1_4 region of the PDC1 promoter (Fig. 3, F and H) containing the ATB2/AtbZIP53/AtbZIP44/GBF5-binding motif. Since AtGBF5 is known to mediate hypoxia-inducible gene regulation (Baena-González et al., 2007), GBF5 and/or this class of bZIP transcription factors (Dietrich et al., 2011) is expected to play a key role in PDC1 activation through SnRK1 in conditions of submergence. The identification of target gene chromatins and protein substrates of the chromatin-associated SnRK1 complex will help to clarify the gene-regulatory mechanisms of SnRK1.
The submergence of Arabidopsis seedlings for 24 h induced the expression of ADH1 and PDC1, genes encoding two important enzymes in alternative respiration (Fig. 3, A and B). The gene induction patterns were largely consistent with the transcriptome analysis under submergence (Lee et al., 2011). Loreti et al. (2005) reported a similar pattern of gene regulation in anoxia, a condition of complete oxygen deprivation, but these genes were up-regulated at 1 h and down-regulated at 24 h after the treatment. The difference in gene expression patterns between submergence and anoxia may reflect a difference in the degree of stress, since submergence may induce only slow and incomplete deprivation of oxygen, in contrast to anoxia. A recent study supports this view, demonstrating a slow decrease in oxygen content in Arabidopsis petioles under submergence by measurement of oxygen partial pressure (Lee et al., 2011). ADH1 and PDC1 expression was further enhanced by submergence in transgenic plants expressing wild-type OsSnRK1 or KIN10, but gene induction was abolished in transgenic plants expressing inactive forms of OsSnRK1 and KIN10 (Fig. 3, A and B). Moreover, Suc supplementation also repressed the stress-induced gene activation. Consistently, Col and transgenic plants expressing wild-type SnRK1 and KIN10 preserved green shoots after a prolonged submergence, while plants expressing inactive forms of SnRK1s did not (Fig. 3, C and D). All these results indicated that plant SnRK1 activity is important in gene regulation under oxygen deprivation during submergence and that cellular energy depletion may mediate such a stress response.
From the time that the seedling is established, SnRK1 regulates plant developmental physiology. Transgenic plants expressing wild-type OsSnRK1 and KIN10 were highly sensitive to HXK1-dependent Glc signaling during seedling establishment (Fig. 4, A and B). It remains to be explained how the seemingly unrelated sugar starvation and signaling sensors SnRK1 and HXK1 contribute to modulate early seedling growth and development.
Plant SnRK1s may also influence leaf senescence (Fig. 5). Transgenic plants expressing wild-type OsSnRK1 and KIN10 showed the delay of senescence, as reflected by several senescence-associated parameters, including chlorophyll content, the photochemical efficiency of PSII, and the expression of senescence-related genes. Conversely, plants expressing inactive forms of SnRK1s showed accelerated senescence. Tissue senescence induced by either energy depletion or developmental programming can activate SnRK1, a gene that controls a broad array of activities, including autophagy (Baena-González et al., 2007). Autophagy is an essential regulator in senescence-associated Rubisco degradation (Ishida et al., 2008). However, its physiological function appears to sustain rather than decrease cell viability. For example, Arabidopsis mutants missing one of the autophagy genes display hypersensitivity to nitrogen deficiency and carbon starvation, which enhance senescence (Thompson et al., 2005). Recently, the kinase component of autophagy was shown to be phosphorylated by AMPK in mammalian cells and to modulate autophagic activity (Egan et al., 2011). It would be interesting to test whether autophagy mediates the effects of SnRK1 on leaf senescence.
Plant SnRK1s orchestrate transcriptional networks to promote catabolism and suppress anabolism and to maintain cellular energy homeostasis in stressful conditions. SnRK1 appears to control the expression of stress-related genes by binding directly to target gene chromatins. In Arabidopsis, this PK modulates physiologic processes from early seedling development through leaf senescence and also acts to maintain tolerance to stress under flooding conditions. Further studies on SnRK1 activities inside and outside of the nucleus may lead to the development of new crop varieties with tolerance to stresses imposed by a changing climate (Coello et al., 2011).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in soil for 23 to 25 d under a photoperiod of 12 h of light/12 h of dark (60 μmol m−2 s−1) at 23°C. Col plants were used as the wild-type Arabidopsis in this study. Plasmid constructs for transgenic plants were generated by inserting cDNA of wild-type SnRK1 (OsSnRK1_WT and KIN10_WT), the ATP-binding site mutant PK (OsSnRK1K43M [OsSnRK1_IN] and KIN10K48M [KIN10_IN]), or the catalytically inactive PKs (OsSnRK1K139R [SnRK1_1m]) between the 35SC4PPDK promoter and the NOS terminator in a minibinary vector, pCB302. These constructs were then expressed in Col plants. The transgenic lines expressing transgenes at levels similar to the levels in Col were selected and used for further analyses. Multiple independent transgenic lines were generated and analyzed for consistency. We analyzed the phenotypes of transgenic plants and seedlings from at least two independent lines of the T2 or T3 generation. Protein expression was analyzed using anti-phospho-AMPKα antibody (Cell Signaling Tech) and anti-HA antibody (Roche).
For flooding experiments, seedlings were grown on MS (Caisson Laboratories) agar medium containing 0.5% Suc for 8 d with a photoperiod of 16 h of light/8 h of dark (50 μmol m−2 s−1) at 23°C. The plates were placed in a plastic box (13 cm × 11 cm × 8 cm) into which distilled and autoclaved water was poured with care to avoid air bubbles. The lid was closed for another 24 h. To simulate the condition of prolonged flooding, seeds were sown inside of 30-mL test tubes holding 5 mL of MS agar medium containing 0.5% Suc for 3 d with a photoperiod of 16 h of light/8 h of dark (50 μmol m−2 s−1) at 23°C. Then, 10 mL of distilled and autoclaved water was carefully pipetted into each test tube, and the tubes were placed in the dark for another 37 d. For each experiment, seeds were stratified at 4°C for 4 d before plating. The results were confirmed through several replications.
For Glc response experiments, seedlings were grown on one-tenth-strength MS medium containing 0.2% Suc for 7 d under constant light (50 μmol m−2 s−1) at 23°C and then treated with 6% Glc for 6 h. As an osmotic control, seedlings were treated with 6% mannitol. For each experiment, seeds were stratified at 4°C for 4 d before plating. The results were confirmed through replications.
Arabidopsis Mesophyll Protoplast Transient Expression Assay
Protoplast isolation and transient expression assays were carried out as described previously (Yoo et al., 2007). All effector constructs were generated by inserting the cDNA between the 35SC4PPDK promoter and the NOS terminator in a plant expression vector for protoplast transient assays (Baena-González et al., 2007; Cho and Yoo, 2011). Methods for NES1, NES2, and the ER retention signal were adapted from Xiao et al. (2001). All constructs were verified by DNA sequencing. All protoplast transient assays were performed with UBQ10-renillaLUC (UBQ10-rLUC) as an internal control. The reporter activities were calculated based on the fLUC-rLUC ratio and normalized to the values obtained without treatment or effector expression.
RNA Isolation and Transcript Measurement
For gene expression analysis, total RNA was isolated by the Trizol method (Invitrogen), and 1 μg of total RNA was used for cDNA synthesis. Gene expression was quantitatively measured using real-time PCR (CFX96 Real-Time System with C1000 Thermal Cycler; Bio-Rad) with the iQ SYBR Green dye-added PCR mix (Bio-Rad). Tubulin4 (At1g04820) and Elongation Initiation Factor4a (At3g13920) transcripts were used as controls with gene-specific primers as in Supplemental Table S1.
ChIP
About 100 seedlings of Col or wild-type OsSnRK1-expressing transgenic lines were prepared after submergence as described above. ChIP was performed as described previously (Cho et al., 2006). Seedlings were harvested and fixed in 1% formaldehyde in a vacuum, and the reaction was stopped by adding 0.3 m Gly in ice-cold Tris-buffered saline. After chromatin isolation, histone-bound DNA was fragmented to an average size of less than 500 bp by sonication (Bioruptor; Diagenode). After removing cellular debris, an aliquot of each sample was stored as an input DNA control and later subjected to reverse cross-linking. Immunoprecipitation was performed with anti-phospho-AMPKα antibody (Cell Signaling Tech) overnight at 4°C with rotation. The protein-DNA complexes were precipitated with Protein A beads (Roche) for 1 h at 4°C. After washing, elution of immunocomplexes, and reverse cross-linking, DNA was extracted using phenol-chloroform-alcohol. To detect enriched chromatin regions, real-time PCR (Bio-Rad) with the iQ SYBR Green dye-added PCR mix (Bio-Rad) was performed using gene-specific primer sets designed for ADH1 and PDC1 (Supplemental Table S1). For quantification of immunoprecipitated DNA, each ChIP sample was compared with its corresponding input control sample. At least three independent ChIP experiments were performed and analyzed using independent seedling sets.
Glc Response Assay
For sugar repression assays, seedlings were grown on full-strength MS agar medium containing 4% or 6% Glc (Sigma) or 6% mannitol (USB Corporation) for 5 d under a photoperiod of 16 h of light/8 h of dark (50 μmol m−2 s−1) at 23°C. For each experiment, seeds were stratified at 4°C for 4 d before plating. The results were confirmed through several replications.
Leaf Senescence Assay
Age-dependent leaf senescence was assayed by measuring cellular chlorophyll content. Chlorophyll was extracted from individual leaves using 100% ethanol (Lim et al., 2010), and the concentration per fresh weight of leaf tissue was calculated as described previously (Lichtenthaler, 1987). The photochemical efficiency of PSII was measured using a portable plant efficiency analyzer (Hansatech Instruments).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plant SnRK1 did not activate a general stress-responsive GST6 promoter activity.
Supplemental Figure S2. SnRK1 localized in the nucleus and affected gene expression.
Supplemental Figure S3. Transcript abundance of OsSnRK1, KIN10, and their mutated forms in stably transformed Arabidopsis transgenic plants.
Supplemental Figure S4. The GST6 gene expression was not induced by OsSnRK1 or KIN10 under flooding.
Supplemental Figure S5. Col and transgenic plants expressing WT and inactive forms of OsSnRK1 and KIN10 showed similar seedling growth.
Supplemental Figure S6. Detailed photo of Col and SnRK1 expressing transgenic seedling development under flooding.
Supplemental Figure S7. Comparison of SnRK1-DNA association with and without submergence.
Supplemental Table S1. Oligonucleotides for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Ms. Thi Bich Ngoc Pham for preparing plasmids and Dr. Jen Sheen for sharing DIN6-fLUC and KIN10 constructs.
References
- Ahuatzi D, Riera A, Peláez R, Herrero P, Moreno F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 282: 4485–4493 [DOI] [PubMed] [Google Scholar]
- Akimoto S, Mimuro M. (2007) Application of time-resolved polarization fluorescence spectroscopy in the femtosecond range to photosynthetic systems. Photochem Photobiol 83: 163–170 [DOI] [PubMed] [Google Scholar]
- Amodeo GA, Rudolph MJ, Tong L. (2007) Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 449: 492–495 [DOI] [PubMed] [Google Scholar]
- Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL. (2008) Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol 148: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Baek SH. (2011) When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell 42: 274–284 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bitrián M, Roodbarkelari F, Horváth M, Koncz C. (2011) BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J 65: 829–842 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. (2010) Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329: 1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW. (2009) Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Cho YH, Sheen J, Yoo SD. (2010) Low glucose uncouples hexokinase1-dependent sugar signaling from stress and defense hormone abscisic acid and C2H4 responses in Arabidopsis. Plant Physiol 152: 1180–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD. (2011) Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet 7: e1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CW, Davis RJ. (2006) Protein kinases: chromatin-associated enzymes? Cell 127: 887–890 [DOI] [PubMed] [Google Scholar]
- Coello P, Hey SJ, Halford NG. (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62: 883–893 [DOI] [PubMed] [Google Scholar]
- Crozet P, Jammes F, Valot B, Ambard-Bretteville F, Nessler S, Hodges M, Vidal J, Thomas M. (2010) Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J Biol Chem 285: 12071–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martínez-Barajas E, Coello P. (2009) SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol 149: 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 25: 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey SJ, Byrne E, Halford NG. (2010) The interface between metabolic and stress signaling. Ann Bot (Lond) 105: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. (2009) SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J 59: 316–328 [DOI] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho THD, Yu SM. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- León P, Sheen J. (2003) Sugar and hormone connections. Trends Plant Sci 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoid pigments of photosynthetic biomembranes. Methods Enzymol 18: 350–382 [Google Scholar]
- Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG. (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19: 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M. (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Poon IKH, Jans DA. (2005) Regulation of nuclear transport: central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L. (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou P, Zheng B, Hsu C-H, Sasaki AT, Cantley LC. (2011) A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab 13: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472: 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Watson N, Rodriguez C, Lodish HF. (2001) Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J Biol Chem 276: 39404–39410 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.