Abstract
Perception by plants of so-called microbe-associated molecular patterns (MAMPs) such as bacterial flagellin, referred to as pattern-triggered immunity, triggers a rapid transient accumulation of reactive oxygen species (ROS). We previously identified two cell wall peroxidases, PRX33 and PRX34, involved in apoplastic hydrogen peroxide (H2O2) production in Arabidopsis (Arabidopsis thaliana). Here, we describe the generation of Arabidopsis tissue culture lines in which the expression of PRX33 and PRX34 is knocked down by antisense expression of a heterologous French bean (Phaseolus vulgaris) peroxidase cDNA construct. Using these tissue culture lines and two inhibitors of ROS generation, azide and diphenylene iodonium, we found that perxoxidases generate about half of the H2O2 that accumulated in response to MAMP treatment and that NADPH oxidases and other sources such as mitochondria account for the remainder of the ROS. Knockdown of PRX33/PRX34 resulted in decreased expression of several MAMP-elicited genes, including MYB51, CYP79B2, and CYP81F2. Similarly, proteomic analysis showed that knockdown of PRX33/PRX34 led to the depletion of various MAMP-elicited defense-related proteins, including the two cysteine-rich peptides PDF2.2 and PDF2.3. Knockdown of PRX33/PRX34 also led to changes in the cell wall proteome, including increases in enzymes involved in cell wall remodeling, which may reflect enhanced cell wall expansion as a consequence of reduced H2O2-mediated cell wall cross-linking. Comparative metabolite profiling of a CaCl2 extract of the PRX33/PRX34 knockdown lines showed significant changes in amino acids, aldehydes, and keto acids but not fatty acids and sugars. Overall, these data suggest that PRX33/PRX34-generated ROS production is involved in the orchestration of pattern-triggered immunity in tissue culture cells.
A current model for plant defense specifies various tiers of surveillance, starting with pattern-triggered immunity (PTI; or basal resistance). PTI involves the recognition of so-called microbe-associated molecular patterns (MAMPs) such as bacterial flagellin or peptidoglycan. However, some potential pathogens can overcome PTI by producing effectors that interfere with defense signaling. These effectors, in turn, can be recognized by resistance (R) proteins, leading to a second tier of resistance, which in some cases includes the hypersensitive response (HR) involving programmed cell death of infected host cells (Dangl and Jones, 2001; Chisholm et al., 2006; Jones and Dangl, 2006).
The use of plant cell tissue cultures has played an important role in the dissection of host defense mechanisms because they facilitate detailed analysis of the earliest events following MAMP recognition by receptors in the plasma membrane (Ramonell et al., 2002; Navarro et al., 2004). Indeed, the activation of receptors leading to rapid responses such as Ca2+ and H+ influx and K+ efflux were first characterized in tissue cultures (Bolwell and Daudi, 2009). These studies have contributed significantly to our understanding of the underlying biochemistry leading to the generation of reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide, and nitric oxide, as antimicrobial factors or as intracellular or intercellular signaling molecules. More recently, the hypersensitive response has been modeled in Arabidopsis (Arabidopsis thaliana) tissue culture cells treated with effector proteins (Kaffarnik et al., 2009).
The production of ROS in response to pathogen attack is one of the first measurable events in the plant defense response. Although there are many potential ROS generation systems, genetic analysis and biochemical studies using inhibitors of ROS-generating enzymes have established that two main categories of enzymes are involved in ROS production in response to pathogens: NADPH oxidases and class III cell wall peroxidases. The NADPH oxidase-dependent oxidative burst is diphenylene iodonium (DPI) sensitive and has been shown to be required for ROS production in Arabidopsis plants in response to infection by Peronospora parasitica or Pseudomonas syringae pv tomato DC3000 expressing the type III effector avrRpm1 (Torres et al., 2002). However, because there are several isoforms of the main subunit of the NADPH protein complex encoded by separate genes (rboh genes), it has been difficult to assess whether NADPH oxidases play an essential role in the plant defense response, although recently, rbohF loss-of-function mutants were shown to be more susceptible to P. syringae (Chaouch et al., 2012).
In addition to NADPH oxidases, class III cell wall peroxidases have been shown to play a key role in the generation of an oxidative burst (Bindschedler et al., 2006; Bolwell and Daudi, 2009; Daudi et al., 2012). The peroxidase-dependent oxidative burst, which, in contrast to the NADPH-generated burst, is sodium azide and cyanide sensitive but DPI insensitive, has been described as a three-component system (Bolwell et al., 2002) involving peroxidases, ion fluxes, and provision of a suitable substrate. The natural physiological substrates used by these peroxidases to generate ROS have not yet been identified. Peroxidase-dependent oxidative bursts have been described in Arabidopsis (Bindschedler et al., 2006; Davies et al., 2006), Daucus carota (Bach et al., 1993), French bean (Phaseolus vulgaris; Bolwell, 1995, 1999; Bolwell et al., 2001), Capsicum annum (Choi et al., 2007), and Lactuca sativa (Bestwick et al., 1997). Importantly, Arabidopsis plants compromised for the expression of at least two cell wall peroxidase-encoding genes, PRX33 and PRX34, exhibited dramatically enhanced susceptibility to a variety of fungal and bacterial pathogens (Bindschedler et al., 2006; Daudi et al., 2012). This latter phenotype of the peroxidase mutants potentially differentiates the role of peroxidases from that of the NADPH oxidases during plant defense. In contrast to the peroxidases, there are no published reports that NADPH oxidase mutants are highly susceptible to pathogen attack; however, a caveat is the potential redundancy of rboh genes (Torres et al., 2002).
Although the pathogen-elicited oxidative burst can be readily detected in planta, detailed physiological analysis of the oxidative burst is facilitated in elicitor-treated plant cell cultures. Transcriptional analysis of flagellin- and chitin-treated suspension-cultured cells (Ramonell et al., 2002; Navarro et al., 2004), as well as proteomic analysis of cell cultures treated with elicitor preparations derived from the maize (Zea mays) pathogen Fusarium moniliforme (Chivasa et al., 2006) or P. syringae (Kaffarnik et al., 2009), have shown that the transcript and protein profiles of elicitor-treated cell cultures are similar to those found in planta in response to pathogen attack. These cell culture studies have also linked various physiological responses with the subsequent generation of an oxidative burst. For example, work in French bean cells treated with an elicitor from Colletotrichum lindemuthianum or Arabidopsis cells treated with an elicitor from Fusarium oxysporum showed that cAMP, G proteins, and Ca2+ and K+ fluxes were required for peroxidase-mediated H2O2 generation (Bolwell, 1995; Bolwell et al., 1999, 2002; Bindschedler et al., 2001; Davies et al., 2006).
In previous work from our laboratories, transgenic lines of Arabidopsis were generated in which the expression of cell wall peroxidases were knocked down by antisense expression of an antisense heterologous French bean peroxidase cDNA (Bindschedler et al., 2006). In French bean, the oxidative burst is generated at least in part by a cell wall type III peroxidase referred to as French bean peroxidase 1 (FBP1; Bolwell, 1999; Blee et al., 2001; Bolwell et al., 2002). Transgenic Arabidopsis plants expressing antisense FBP1 cDNA showed reduced levels of mRNA corresponding to the class III peroxidase-encoding genes At3g49110 (PRX33) and At3g49120 (PRX34), reduced levels of H2O2 production in response to a F. oxysporum cell wall elicitor, and increased susceptibility to both bacterial and fungal pathogens (Bindschedler et al., 2006; Daudi et al., 2012). In this study, we utilized a similar RNA interference-mediated strategy in tissue culture cells. The uniformity of cultured cells and the ability to uniformly expose them to a variety of treatments facilitated exploration of the roles of peroxidases in response to MAMPs. This led to the findings that PRX33/PRX34 are required for a MAMP-elicited oxidative burst and for the expression of a variety of proteins involved in defense and cell wall expansion. Interestingly, H2O2 generated by PRX33/PRX34 also appears to activate the expression of the PRX33, PRX34, and RBOHD genes.
RESULTS
Generation of Arabidopsis Cell Cultures Expressing Antisense FBP1 cDNA
Arabidopsis (ecotype Landsberg erecta [Ler]) cell cultures expressing a full-length French bean peroxidase1 cDNA sequence in an antisense orientation (asFBP1; referred to below as LerasFBP1) were generated by Agrobacterium tumefaciens-mediated transformation as described in “Materials and Methods.” Several independent lines derived from single cells were selected from different transformation events, and the insertion sites of the transgenes were mapped in several lines determined to be carrying a single insertion as described in “Materials and Methods” (Supplemental Fig. S1). Two lines, IG-2, with a single insertion in chromosome 1, position 23.286.830, and IG-3, with a single insertion in chromosome 4, position 16.064.022, were selected for further analysis (Supplemental Fig. S2). The insertion sites in both lines were in presumptive noncoding regions of the genome (Supplemental Fig. S2). Similar to transgenic plants expressing the FBP1 cDNA (Bindschedler et al., 2006; Daudi et al., 2012), PRX33 and PRX34 mRNA levels in IG-2 and IG-3 were significantly lower than in the nontransformed Ler cell culture both before and after elicitation with a cell wall elicitor preparation from F. oxysporum (Table I).
Table I. PRX33 and PRX34 mRNA levels in F. oxysporum-elicited IG-2 and IG-3 cell lines.
| PRX | Ler |
IG-2 |
IG-3 |
|||
| Basal | Elicited | Basal | Elicited | Basal | Elicited | |
| mRNA levels normalized to basal PRX33 or PRX34 mRNA levels in Ler | ||||||
| PRX33 | 1.0 | 24 ± 3.01 | 0.50 ± 0.09 | 0.35 ± 0.02 | 0.45 ± 0.04 | 0.46 ± 0.04 |
| PRX34 | 1.0 | 3.05 ± 1.0 | 0.26 ± 0.09 | 0.58 ± 0.30 | 0.27 ± 0.10 | 0.49 ± 0.07 |
| mRNA levels normalized to basal PRX33 mRNA levels in Ler | ||||||
| PRX34 | 1,728 ± 59.3 | 238 ± 4.7 | 861 ± 95.3 | 2,448 ± 358 | 1,095 ± 5.4 | 1,979 ± 96.9 |
Depletion of the PRX34 Protein in Cell Wall Preparations from the Transgenic Cell Lines
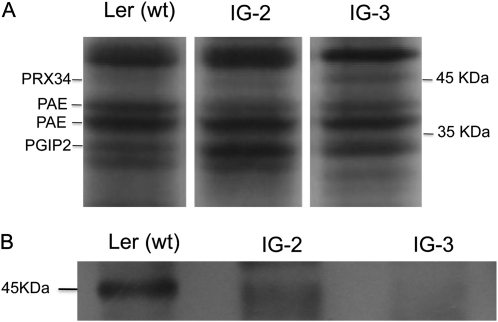
CaCl2 has been used previously to extract proteins including peroxidases from plant cell walls (Robertson et al., 1997; Blee et al., 2001; Chivasa et al., 2006). Nanoliquid chromatography-nanoelectrospray ionization-tandem mass spectrometry (nLC-nESI-MS/MS) analysis of CaCl2-extracted proteins from Ler cell cultures identified a weakly staining 45-kD band as PRX34 (Fig. 1A). Despite the incomplete silencing of the PRX34 transcript level in the IG-2 and IG-3 transgenic lines (Table I), this level of silencing was sufficient to reduce the cognate PRX34 protein to undetectable levels in IG-2 and IG-3 by nLC-nESI-MS/MS analysis. In contrast to PRX34, the levels of PRX33 in the nonelicited Ler tissue cultures was below the detection limits of MS/MS analysis, consistent with quantitative reverse transcription (qRT)-PCR analysis, which showed that PRX33 mRNA is present at a more than 1,000-fold lower level than PRX34 mRNA in the nonelicited Ler cultures (Table I). Diminished PRX34 protein levels in the IG-2 and IG-3 transgenic lines was confirmed by western-blot analysis. Fortuitously, a polyclonal antibody raised against horseradish peroxidase appears to cross-react with PRX34. A band is present in the Ler wild-type [Ler (wt)] cell culture extract with the same molecular mass as PRX34, but the abundance of this cross-reacting 45-kD protein is clearly reduced in the LerasFBP1 lines IG-2 and IG-3 (Fig. 1B).
Figure 1.
Identification of PRX34 by one-dimensional SDS-PAGE analysis and western-blot analysis of CaCl2-extracted cell wall proteins. After treatment, the cell cultures were filtered and then incubated for 30 min in 200 mm CaCl2, as described in “Materials and Methods.” Proteins were precipitated with 80% acetone, resuspended in sample buffer, loaded on an SDS-PAGE gel, and run overnight. The gels were stained with Coomassie blue for protein visualization. Proteins were identified by in-gel protein digestion and sequencing. A, The region between 40 and 55 kD, showing the presence of PRX34 in Ler (wt) and the absence of PRX34 in LerasFBP1 cells by lack of detection by nLC-nESI-MS/MS. B, Western-blot detection of PRX34 using a polyclonal antibody raised against horseradish peroxidase.
Consequences of the Knockdown of PRX33/PRX34 on ROS Production
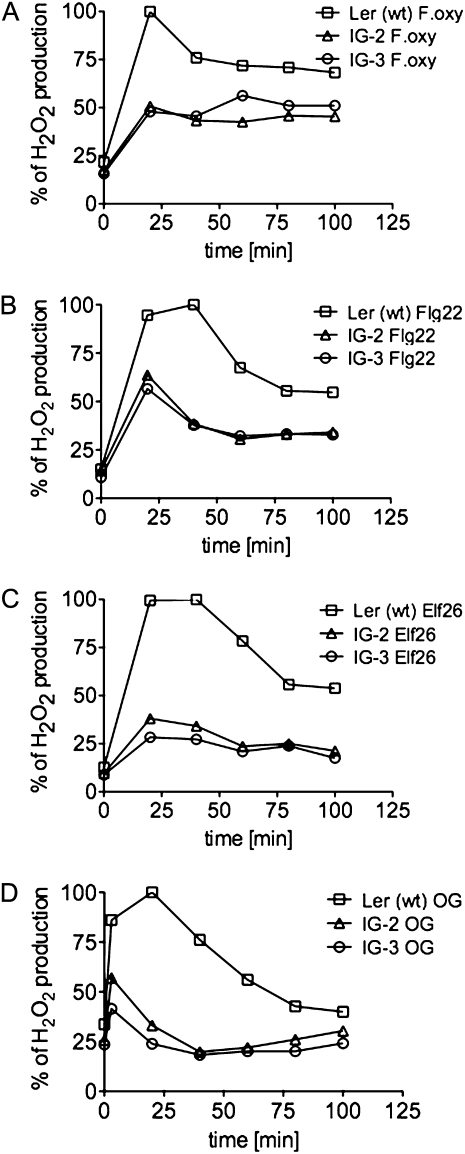
We tested the LerasFBP1 tissue culture lines IG-2 and IG-3 for ROS production following treatment with the F. oxysporum cell wall elicitor preparation, the synthetic peptide elicitors Flg22 and Elf26 (which correspond to conserved epitopes of bacterial flagellin and elongation factor EF-Tu, respectively; Felix et al., 1999; Kunze et al., 2004), or an oligogalacturonide preparation (Ferrari et al., 2007). All of these elicitors stimulated a robust oxidative burst in the nontransformed Ler cell culture, which was significantly reduced in the transgenic lines IG-2 and IG-3 (Fig. 2). The F. oxysporum cell wall preparation and Flg22 elicited 50% as much H2O2 in IG-2 and IG-3 as in Ler (wt), whereas Elf26 and oligogalacturonide elicited only 40% as much (for quantitation of the data in Fig. 2, see Supplemental Fig. S3A).
Figure 2.
H2O2 production in Ler (wt), IG-2, and IG-3 Arabidopsis cell cultures after treatment with bacteria and fungal elicitors. Arabidopsis Ler (wt), IG-2, and IG-3 were filtered and resuspended in fresh medium at 250 mg mL−1. After recovery, the cell cultures were elicited with 100 μg mL−1 F. oxysporum cell wall elicitor (A), 1 μm Flg22 (B), 1 μm Elf26 (C), or 100 μg mL−1 oligogalacturonide (OG; D). At 20-min intervals, H2O2 production was measured by the xylenol orange assay, as described in “Materials and Methods,” and normalized against Ler (wt). Representative graphs from three independent experiments are shown.
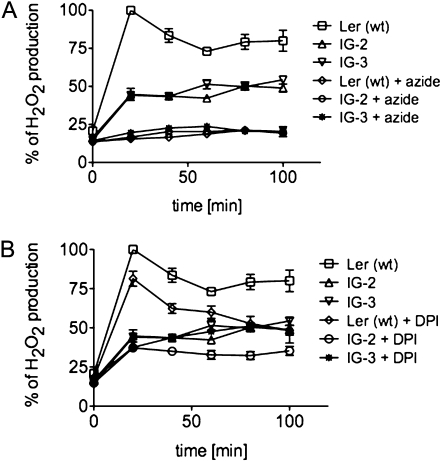
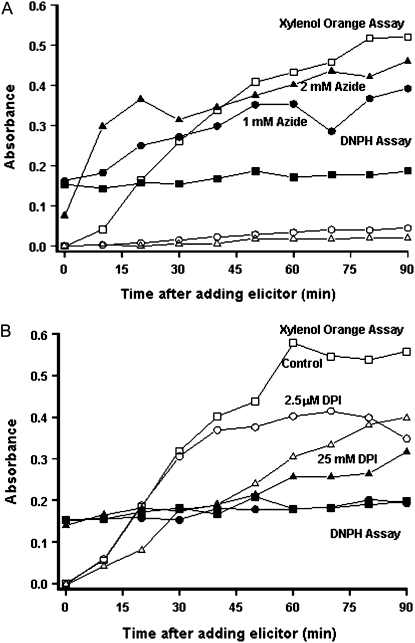
In both the Ler (wt) and the LerasFBP1 lines, treatment with 2 mm sodium azide, which blocks the peroxidase-mediated oxidative burst, completely abolished the H2O2 production elicited by the F. oxysporum cell wall preparation (Fig. 3A). In contrast, DPI, at a concentration of 0.2 μm, which is relatively specific for the inhibition of NADPH oxidases (Davies et al., 2006), only inhibited about 25% of the oxidative burst in both Ler (wt) and the LerasFBP1 lines (Fig. 3B; Supplemental Fig. S3B). These results are consistent with our previous data, which showed that Ler (wt) Arabidopsis cell cultures produce a sodium azide-sensitive but relatively DPI-insensitive oxidative burst in response to a F. oxysporum cell wall preparation (Bindschedler et al., 2006). Inhibition of an oxidative burst by 0.2 μm DPI, which causes 50% inhibition (I50) of the oxidative burst, is indicative of the involvement of an NADPH/NADH oxidase, whereas inhibition by azide with an I50 greater than 50 μm suggests a peroxidase-dependent mechanism (Bolwell et al., 1998; Frahry and Schopfer, 1998).
Figure 3.
Effect of sodium azide and DPI on H2O2 production after treatment with F. oxysporum cell wall elicitor in Ler (wt), IG-2, and IG-3 cell cultures. Arabidopsis Ler (wt), IG-2, and IG-3 were filtered and resuspended in fresh medium at 250 mg mL−1. After recovery, the cells were pretreated for 15 min with 2 mm sodium azide (A) or 0.2 μm DPI (B) followed by 100 μg mL−1 F. oxysporum extract. At 20-min intervals, H2O2 production was measured by the xylenol orange assay, as described in “Materials and Methods,” and normalized against Ler (wt). Errors bars represent sd of three independent experiments with two technical replicates each.
In addition to exhibiting lower elicited levels of H2O2 following MAMP treatment, we also observed approximately 20% lower basal levels of H2O2 in nonelicited IG-2 and IG-3 cultures (Supplemental Fig. S4). A similar level of reduction was also observed in the Ler (wt) culture after sodium azide treatment, but not after DPI treatment (Supplemental Fig. S4). Because the basal levels of H2O2 are not significantly affected by 0.2 μm DPI, which is relatively specific for NADPH oxidases, these data suggest that the residual basal levels of H2O2 that are not generated by peroxidases are most likely generated by cell metabolism rather than from the dismutation of NADPH oxidase-derived superoxide.
RBOHD, PRX33, and PRX34 Expression Is Induced by H2O2
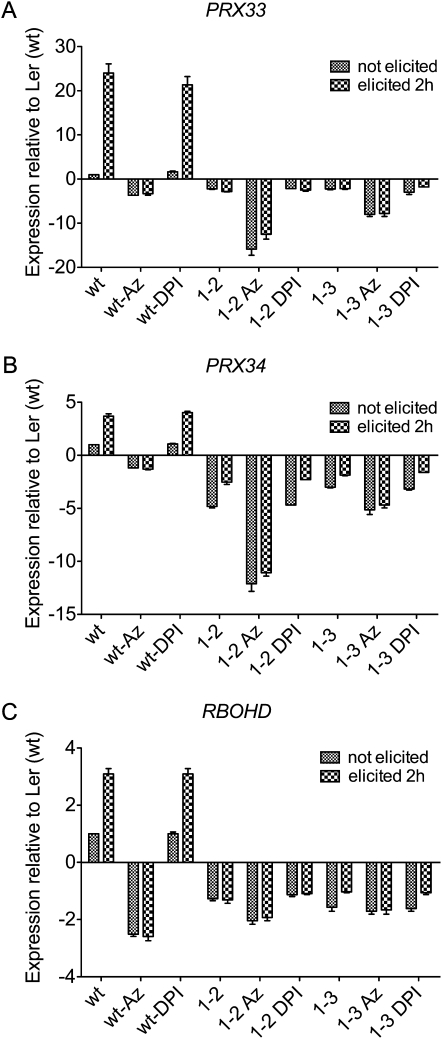
Figure 4 shows that the expression of PRX33 and PRX34 as well as the expression of RBOHD are significantly induced by F. oxysporum cell wall elicitor in the Ler (wt) tissue culture cells. Somewhat unexpectedly, however, both the basal and induced levels of RBOHD expression were reduced in IG-2 and IG-3, suggesting that RBOHD expression is dependent on the expression of PRX33 and PRX34 and that RBOHD expression may be regulated by the H2O2 generated by PRX33 and PRX34.
Figure 4.
qRT-PCR quantification of transcript levels of PRX33, PRX34, and RBOHD in Ler (wt), IG-2, and IG-3 lines. Expression levels of PRX33, PRX34, and RBOHD mRNAs in Ler (wt), IG-2, and IG-3 cell cultures were measured at 0 and 2 h following elicitation with purified cell wall extract from F. oxysporum. Cell cultures were pretreated with 2 mm sodium azide or 0.2 μm DPI and incubated for 15 min before elicitation. Data represent averages ± sd of three independent experiments.
We took advantage of the differential inhibitory activities of sodium azide and DPI and the LerasFBP1 tissue culture lines to further investigate the regulation of the PRX33, PRX34, and RBOHD genes. Consistent with the conclusion that RBOHD expression is regulated by H2O2, Figure 4C shows that F. oxysporum cell wall preparation-elicited activation of RBOHD is completely blocked by sodium azide but not by DPI pretreatment. Moreover, sodium azide pretreatment also appeared to significantly lower nonelicited (basal) levels of RBOHD. Also unexpectedly, sodium azide (but not DPI) markedly lowered the basal levels of PRX33 and PRX34 expression in IG-2 and IG-3 (Fig. 4, A and B).
The data in Figure 4 suggest that PRX33, PRX34, and RBOHD transcription is positively regulated by H2O2, allowing feed-forward increases in H2O2 production from the basal state during pathogen attack. The observation that RBOHD transcription as well as PRX33 and PRX34 transcription are regulated by H2O2 is particularly interesting, since it provides new evidence for a common regulatory mechanism for PRX- and RBOH-dependent H2O2 production. It was not likely that azide or DPI had a negative overall effect on transcription, since the levels of glyceraldehyde 3-phosphate dehydrogenase (GADPH), a commonly used “housekeeping” gene, were not affected after azide or DPI treatment (Supplemental Fig. S5).
Knockdown of PRX33 and PRX34 Affects the Expression of Key Genes Involved in PTI
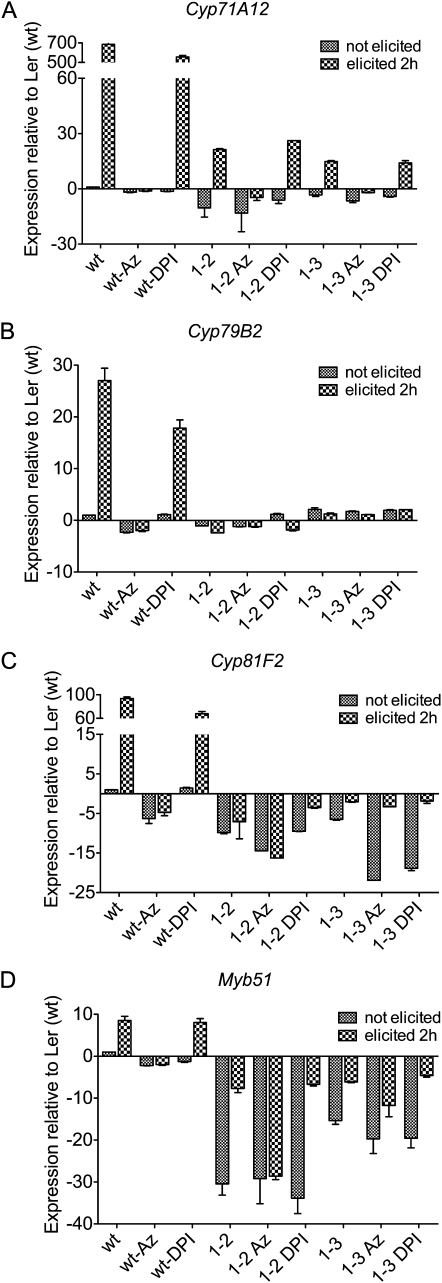
To further explore the roles of PRX33 and PRX34 in PTI, we analyzed the relative transcriptional activation of selected defense-related genes known to be up-regulated by MAMPs. MYB51 and the CYTOCHROME P450 genes 79B2 (CYP79B2) and 81F2 (CYP81F2), which are involved in indole glucosinolate (IGS) biosynthesis, are required for callose deposition in seedling cotyledons following MAMP treatment (Clay et al., 2009). In the Ler (wt) culture, MYB51, which encodes a transcription factor, was up-regulated up to 10-fold 2 h after elicitation with the F. oxysporum cell well preparation (Fig. 5D). Similarly, the IGS biosynthetic genes CYP79B2 and CYP81F2 were up-regulated 30- and 100-fold, respectively (Fig. 5, B and C). In contrast, there was no significant up-regulation of these three IGS-related genes in the LerasFBP1 lines. Similar results were obtained with CYP71A2, a gene involved in camalexin biosynthesis (Fig. 5A; Millet et al., 2010). When Ler (wt) cells were pretreated with DPI, specifically inhibiting NADPH oxidases, all four MAMP-activated genes were up-regulated by F. oxysporum elicitor, but at an approximately 25% lower level compared with Ler (wt). In contrast, in Ler (wt) cells pretreated with sodium azide, which inhibits both peroxidase-mediated and mitochondrial ROS production, none of the four genes were induced.
Figure 5.
qRT-PCR quantification of transcript levels of CYP79B2, CYP71A12, MYB51, and CYP81F2 in Ler (wt), IG-2, and IG-3 lines. Expression levels of CYP79B2, CYP71A12, MYB51, and CYP81F2 mRNAs in Ler (wt), IG-2, and IG-3 cell cultures were measured at 0 and 2 h following elicitation with purified cell wall extract from F. oxysporum. Cell cultures were pretreated with 2 mm sodium azide or 0.2 μm DPI and incubated for 15 min before elicitation. Data represent averages ± sd of three independent experiments.
Knockdown of PRX33/PRX34 Affects Proteins Involved in Pathogen Defense
We carried out proteomic analysis to determine the effects of the knockdown of PRX33 and PRX34 on CaCl2-extracted cell wall proteins involved in pathogen defense (Tables II and III; Supplemental Fig. S6). A number of proteins directly linked with plant defense were down-regulated in IG-2 and IG-3. One such group of proteins was the ROS-scavenging enzymes catalase (CAT3) and ascorbate peroxidase (APX1; Supplemental Fig. S6, bands 6 and 23), whose production is normally induced by ROS. These two enzymes were only detected in Ler (wt) cultures, consistent with the reduced levels of H2O2 observed in LerasFBP1 lines. Three other proteins involved in defense were also absent, plant defensins PDF2.2 and PDF2.3, and Gibberellic Acid-Stimulated Transcript1 (GAST1)-like protein (Supplemental Fig. S6, bands 15 and 13). GAST1-like protein is a Cys-rich peptide involved in plant defense as part of a two-component protein complex that is cross-linked with a 42-kD chitin-binding Pro-rich protein and immobilizes invading pathogens in the cell wall (Bindschedler et al., 2006). PDF2.2 and PDF2.3 are low-molecular-weight Cys-rich protease inhibitors that prevent the degradation of cell wall proteins by microbial proteases. Down-regulation of PDF2.2 and PDF2.3 is consistent with previous results showing that genes such as At5g44420, which encode other members of this protease inhibitor family, are also down-regulated in PRX33/PRX34 knockdown plants (Bindschedler et al., 2006). In contrast to the defense-related proteins described above, at least one defense-related protein, polygalacturonase-inhibiting protein (PGIP2; Devoto et al., 1998; Ferrari et al., 2006), was clearly more abundant in nonelicited IG-2 and IG-3 cells than in the Ler (wt) culture (Supplemental Fig. S6, band 30).
Table II. Identifier numbers of proteins identified by in-gel trypsin digestion of CaCl2-extracted cell wall proteins of Arabidopsis Ler (wt) and LerasFBP1 cells.
nd, Not determined; ni, not identified.
| Band No. | Arabidopsis Genome Initiative No. | Description | Score | Peptide No.a | Coverage | Relative Abundanceb |
|||
| IG-2 | IG-3 | ||||||||
| % | fold change | sd | fold change | sd | |||||
| Ler (wt) unelicited | |||||||||
| 1 | ni | ni | – | – | – | 0.73 | 0.025 | 0.76 | 0.069 |
| 2 | At1g32960 | ATSBT3.3 (subtilase) | 442 | 11 | 15 | 0.96 | 0.059 | 1.01 | 0.103 |
| 3 | At5g20950 | Glycosyl hydrolase family 3 | 931 | 19 | 31 | 1.47 | 0.039 | 1.56 | 0.088 |
| 4 | At4g12420 | SKS5 (copper ion binding/oxidoreductase) | 673 | 15 | 25 | nd | nd | nd | nd |
| At5g49360 | BXL1 (β-xylosidase 1) | 393 | 9 | 11 | |||||
| 5 | At3g18080 | AtBGLU44 (glycosyl hydrolase family 1) | 771 | 23 | 36 | 1.23 | 0.073 | 1.38 | 0.205 |
| 6 | At1g78850 | Curculin-like (Man-binding) lectin | 810 | 17 | 37 | nd | nd | nd | nd |
| At1g20620.1 | CAT3 (catalase) | 217 | 6 | 10 | |||||
| 7 | At1g70710 | Endo-1,4-β-glucanase (ATCEL1) | 635 | 13 | 28 | 1.27 | 0.094 | 1.32 | 0.377 |
| 8 | At1g71380.1 | ATCEL3/ATGH9B3 (glycosyl hydrolase 9B3) | 916 | 17 | 34 | 1.09 | 0.103 | 1.00 | 0.092 |
| 9 | At5g08380 | ATAGAL1 (α-galactosidase 1) | 536 | 11 | 27 | nd | nd | nd | nd |
| At3g49120.1 | ATPERX34 (peroxidase 34) | 408 | 9 | 25 | |||||
| 10 | At4g19410.1 | Pectinacetylesterase | 295 | 8 | 20 | 0.74 | 0.048 | 0.77 | 0.087 |
| 11 | At5g06870.1 | PGIP2 (polygalacturonase-inhibiting protein 2) | 739 | 16 | 51 | 1.81 | 0.272 | 1.69 | 0.435 |
| At4g37520 | PRXR2 (peroxidase ATP9a) | 134 | 3 | 11 | nd | nd | nd | nd | |
| 21 | At5g64120 | PRX71 (peroxidase 71) | 524 | 8 | 29 | ||||
| 22 | At3g14310 | ATPME3 (pectin methylesterase 3) | 865 | 16 | 26 | 0.62 | 0.241 | 0.63 | 0.267 |
| 12 | At3g45960.1 | ATEXLA3 (expansin-like A3) | 543 | 11 | 43 | 1.12 | 0.021 | 0.93 | 0.005 |
| 19 | At4g38400.1 | ATEXLA2 (expansin-like A2) | 505 | 12 | 36 | nd | nd | nd | nd |
| At3g45970.1 | ATEXLA1 (expansin-like A1) | 381 | 9 | 31 | |||||
| 23 | At1g07890.1 | APX1 (ascorbate peroxidase 1) | 364 | 8 | 42 | nd | nd | nd | nd |
| 24 | At1g02930.1 | ATGSTF6 (glutathione-S transferase 6) | 288 | 7 | 24 | nd | nd | nd | nd |
| 25 | At4g11650 | Osmotin | 135 | 3 | 9 | nd | nd | nd | nd |
| 26 | At4g38740 | ROC1 (rotamase CyP 1; peptidyl-prolyl cis-trans isomerase) | 340 | 8 | 43 | nd | nd | nd | nd |
| 27 | At3g52960 | Peroxiredoxin type 2 | 120 | 4 | 16 | nd | nd | nd | nd |
| 13 | At1g75750 | GAST1-like protein | 438 | 7 | 46 | nd | nd | nd | nd |
| 14 | At2g02130.1 | Plant defensin PDF2.3 (low-molecular-weight Cys-rich 68) | 117 | 2 | 31 | 0.61 | 0.006 | 0.45 | 0.091 |
| At2g02100.1 | Plant defensin PDF2.2 (low-molecular-weight Cys-rich 69) | 108 | 2 | 41 | |||||
| At3g18280 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | 103 | 4 | 31 | |||||
| LerasFBP1 unelicited | |||||||||
| 29 | At2g16430.2 | ATPAP10 (acid phosphatase) | 877 | 20 | 53 | nd | nd | nd | nd |
| 30 | At5g06870.1 | PGIP2 (polygalacturonase-inhibiting protein 2) | 831 | 16 | 55 | nd | nd | nd | nd |
| 32 | At3g18280 | Protease inhibitor/seed storage/lipid transfer protein (LTP) | 128 | 5 | 31 | nd | nd | nd | nd |
Peptides identified are described in Supplemental Table S2.
The relative abundance of the most intense bands was calculated from SDS-PAGE gels using ImageJ.
Table III. Down-regulated and up-regulated cell wall proteins in LerasFBP1 lines in comparison with Ler (wt).
| Band No. | Description |
| Down-regulated proteins in IG-2 and IG-3 | |
| 6 | CAT3 (catalase) |
| 10 | PAE (pectinacetylesterase) |
| 22 | PME3 (pectin methylesterase 3) |
| 23 | APX1 (ascorbate peroxidase 1) |
| 13 | GAST1-like protein |
| 14 | Plant defensin PDF2.3 (low-molecular-weight Cys-rich 68) |
| 14 | Plant defensin PDF2.2 (low-molecular-weight Cys-rich 69) |
| Up-regulated proteins in IG-2 and IG-3 | |
| 29 | ATPAP10 (acid phosphatase) |
| 11 | PGIP2 (polygalacturonase-inhibiting protein 2) |
Knockdown of PRX33/PRX34 Affects Proteins Involved in Cell Wall Remodeling
The knockdown of PRX33/PRX34 in IG-2 and IG-3 not only affected the expression of defense-related proteins but also affected several cell growth-related proteins (Table III) such as purple acid phosphatase (PAP10), which was clearly present in IG-2 and IG-3 but was not detected in Ler (wt) plants (Supplemental Fig. S6, band 29). PAPs are involved in the induction of β-glucan synthases in the cell wall (Kaida et al., 2009, 2010). PGIPs, which, as discussed above, are also up-regulated in IG-2 and IG-3, play an important role in cell growth in addition to pathogen defense. By inhibiting the cell wall-loosening activity of endogenous polygalacturonases, PGIPs inhibit cell expansion in a similar way to peroxidase-mediated cross-linking of cell wall components. Up-regulation of PGIPs, therefore, could be a consequence of enhanced cell growth due to lower H2O2 levels in IG-2 and IG-3. Interestingly, the leaves of PRX33/PRX34 knockdown plants are approximately 30% larger than those of wild-type plants (Daudi et al., 2012), consistent with the hypothesis that the up-regulation of PGIP2 and PAP10 contributes to the stiffening and remodeling of the cell wall.
In contrast to the up-regulation of PGIP2 and PAP10, pectinacetylesterase and pectinmethylesterase (Supplemental Fig. S6, bands 10 and 22, respectively) were down-regulated in IG-2 and IG-3. The down-regulation of these proteins, which are involved in the breakdown of pectin, may inhibit the role that the pectin matrix plays in maintaining cell wall integrity by cross-linking with the cellulose and xyloglucan structural framework (Carpita and Gibeaut, 1993). These lines of evidence suggest that peroxidases are directly (by cross-linking) or indirectly (by regulating H2O2 levels) regulating cell growth (Passardi et al., 2005; Cosio and Dunand, 2009).
Knockdown of PRX33/PRX34 Leads to a Differential Accumulation of Organic Acids during ROS Production
To help identify potential substrates that generate H2O2 via peroxidases in response to pathogen attack, we profiled the CaCl2-extracted supernatant solutions (see “Materials and Methods”) from wild-type and IG-2 and IG-3 cultures for metabolites that accumulated during the apoplastic oxidative burst in response to elicitation by the F. oxysporum cell wall preparation. Previous metabolomic studies of cell wall fractions of French bean and Arabidopsis cell wall cultures revealed that glycerol, malate, citrate, succinate, malonate, fumarate, palmitate, and stearate accumulated after elicitation with fungal extracts during the apoplastic oxidative burst, where peroxidases are actively producing H2O2 (Bolwell, 1999; Bolwell et al., 2002). Organic acids have also been found in P. syringae-infected plants (Rico and Preston, 2008).
In the experiments reported here, CaCl2-extracted metabolites from tissue culture cells were profiled as bis(trimethylsilyl)trifluoroacetamide (BSTFA), ethyl chloroformate (ECF), or 2,4-dinitrophenylhydrazine (DNPH) derivatives using gas chromatography (GC)-MS and/or HPLC. Typical profiles are shown in Supplemental Figures S7 and S8. HPLC allowed the identification of aldehydes and ketones as DNPH derivatives, ECF identified organic acids and amino acids, and BSTFA identified fatty acids and sugars. CaCl2-extracted metabolites of Ler (wt) and IG-2 and IG-3 were compared before and 30 min after elicitation with the F. oxysporum cell wall preparation.
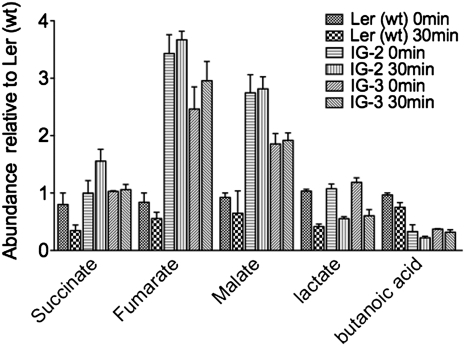
In the case of the Ler (wt) cell cultures, there was no accumulation of any of the organic acids identified previously. However, in IG-2 and IG-3, malate and fumarate accumulated to 4-fold higher levels than in the wild-type culture (Fig. 6). In contrast, the levels of lactate decreased by 50% upon elicitation in the Ler (wt) and LerasFBP1 lines. Even though succinate, malate, and fumarate are part of the citric acid cycle in mitochondria and the glyoxylic acid cycle in peroxisomes, changes in the accumulation of malate, fumarate, and lactate in the apoplast could indicate a perturbation of malate dehydrogenase and lactate dehydrogenase (LDH) enzyme activities, which has long been known to be involved in providing NADH for H2O2 production by peroxidases (Halliwell, 1977, 1978). Interestingly, both malate dehydrogenase and LDH have been described as present in cell walls (Chivasa et al., 2002). Moreover, changes in the accumulation of small organic acids could also be due to a perturbation of cell metabolism. Thus, changes in the activity of enzymes such as glycolate oxidase could directly affect constitutive H2O2 levels (Noctor et al., 2002; Fahnenstich et al., 2008).
Figure 6.
Quantification of small organic acids present in the CaCl2 extracts of Ler (wt), IG-2, and IG-3 cell cultures before and after elicitation. Ler (wt), IG-2, and IG-3 cell cultures were elicited with purified cell wall extract from F. oxysporum. After elicitation, the cell cultures were filtered and then incubated for 30 min in 200 mm CaCl2. The supernatant was then recovered, derivatized with ECF or BSTFA, and analyzed by GC-MS, as described in “Materials and Methods.” Data represent averages ± sd of the normalization against Ler (wt).
Alternative Sources of H2O2
The use of DNPH as a derivatization reagent showed that when peroxidases and possibly other heme proteins are inhibited with sodium azide, keto acids and aldehydes accumulated in Ler (wt) cultures after treatment with a cell wall extract from F. oxysporum (Fig. 7; Supplemental Fig. S9A). HPLC and LC-MS/MS analysis of these samples showed that the main compound accumulating after sodium azide treatment was acetaldehyde (Supplemental Fig. S9B), which suggests that some of the ROS production under these conditions could be derived from intracellular sources. As previously mentioned, sodium azide treatment will not only inhibit peroxidases but heme proteins in general. Thus, it is possible that sodium azide treatment causes a nonspecific perturbation of plant cell metabolism. On the other hand, the rapid accumulation of acetaldehyde and the known inhibitory activity of sodium azide in aerobic respiration suggest that mitochondria could be one of the intracellular sources of ROS (Gleason et al., 2011). Also, as these cell cultures were maintained under continuous light and the cultures were green, photosynthesis-derived H2O2 could also be a source of ROS during the oxidative burst.
Figure 7.
Accumulation of H2O2, keto acids, and aldehydes following elicitation in Ler (wt) cell cultures. Arabidopsis Ler (wt) cell cultures were resuspended in new medium and pretreated with sodium azide (1 or 2 mm; A) or DPI (2.5 or 25 μm; B) and then elicited with 100 μg mL−1 F. oxysporum extract. Samples were collected every 10 min, and after centrifugation, the supernatants were collected. H2O2 was measured using the xylenol orange assay, and keto acids and aldehydes were quantified by DNPH derivatization as described in “Materials and Methods.”
DISCUSSION
By generating Arabidopsis cell culture lines in which the expression of two genes encoding cell wall peroxidases, PRX33 and PRX34, is knocked down by heterologous expression of an antisense heterologous French bean peroxidase cDNA, we have shown that these peroxidases are required for a variety of MAMP-activated responses, including the production of an oxidative burst and the expression of a variety of MAMP-activated genes. We also took advantage of the ability to easily treat tissue culture cells with chemical inhibitors to investigate the relative roles of peroxidases and NADPH oxidases in MAMP-elicited responses. These studies showed that sodium azide, which primarily inhibits peroxidases but not NADPH oxidases, blocks MAMP-mediated responses in Arabidopsis tissue culture cells, whereas DPI, which preferentially inhibits NADPH oxidases, does not. Unexpectedly, we also found that transcription of PRX33 and PRX34, as well as RBOHD, which encodes an NADPH oxidase, is positively regulated by H2O2, indicating a feed-forward mechanism for enhancing the level of the defense response during the course of pathogen attack.
We also took advantage of the tissue culture system to carry out proteomic and metabolomic analyses. The proteomic studies showed that knocking down the expression of PRX33 and PRX34 had a significant impact on the levels of a variety of cell wall-associated proteins involved in defense or cell expansion. The goal of the metabolomic studies was to identify the substrate for peroxidase-mediated production of H2O2. Although the substrate was not identified, the metabolomic data suggest that there is a direct link between malate/lactate cell wall metabolism and the peroxidase-dependent oxidative burst. Thus, malate dehydrogenases and/or LDHs may be involved in the oxidative burst by generating NADH, which could be used as a substrate by cell wall-associated peroxidases. The presence of these enzymes in the cell wall has been described previously (Gross et al., 1977; Halliwell, 1977, 1978).
Relative Roles of PRX33 and PRX34 during the Oxidative Burst
Given that antisense expression of the FBP1 cDNA knocks down the expression of both PRX33 and PRX34 and that it is likely that some expression remains in the knockdown lines, it is not possible to reach any definitive conclusions about the respective roles of PRX33 and PRX34 in this experimental system. On the other hand, our previous observations in Arabidopsis leaves showed that PRX34 is expressed at higher levels than PRX33 (Bindschedler et al., 2006; Daudi et al., 2012). Similarly, PRX34 is expressed at a substantially higher level than PRX33 in Ler (wt) cell cultures, and only the PRX34 protein was detectable in CaCl2 extracts by nLC-MS/MS analysis. Considering that the oxidative burst starts within 5 to 10 min after elicitation and that only PRX34 was detected in the apoplast, it seems likely that PRX34 plays a major role in the generation of H2O2 during the oxidative burst. While PRX33 was not detected in nonelicited cultures, the relatively strong up-regulation of PRX33 (24-fold) following elicitation compared with PRX34 (3-fold) suggests that it may play an important role after the oxidative burst, although the absolute level of PRX34 mRNA was still approximately 200-fold higher than PRX33 mRNA after elicitation (Table I). In addition, because DPI treatment leads to a modest but significant reduction in the oxidative burst, it appears likely that NADPH oxidases also participate in H2O2 generation during the oxidative burst. Interestingly, RBOHD transcript levels are also up-regulated upon elicitation, apparently in response to H2O2 production.
It is not likely that PRX33 and PRX34 have different enzymatic functions, due to their high sequence similarity. PRX33 and PRX34 probably originated from a recent tandem duplication, sharing 91% similarity at the DNA level and 94% similarity at the amino acid level. However, the promoter regions of the two genes only show 35% similarity (Cosio and Dunand, 2009). Thus, the roles of these two peroxidases appear most likely to be determined by their expression and localization patterns rather than their enzymatic specificity.
Other Sources of H2O2
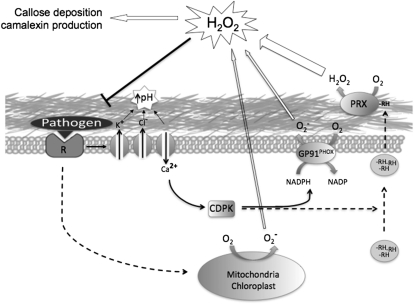
Taking advantage of the differential effects of sodium azide and DPI on peroxidase and NADPH activity, we estimated that peroxidases account for approximately 50% of the H2O2 produced during the oxidative burst, whereas NADPH oxidases and possibly mitochondria and chloroplasts contribute the remaining approximately 50% (Fig. 8). The postulated contribution of mitochondria to H2O2 production is based on our finding that acetaldehyde accumulates after sodium azide treatment. While we cannot rule out other possible intracellular ROS sources, the role of mitochondria in ROS production has recently been described (Gleason et al., 2011). In mitochondria, sodium azide inhibits complex IV (Moller, 2001), which leads to the indirect inhibition of complexes II and III (Moller, 2001; Hargreaves et al., 2007). Mutations in SUCCINATE DEHYDROGENASE1-1, which disable complex II, not only result in diminished ROS production in response to pathogens but enhanced susceptibility to Rhizoctonia solari and P. syringae pv tomato DC3000 (Gleason et al., 2011).
Figure 8.
Proposed model of the different components of the oxidative burst response in higher plants. Upon recognition of a pathogen, a cascade of signaling events is initiated, leading to an increase of extracellular pH, release of a reductant (RH), and activation of the cell wall peroxidases, mitochondrial ROS production, and NADPH oxidases through calcium-dependent protein kinase (CDPK)-dependent phosphorylation. Activation of these mechanisms ultimately results in an H2O2 signal that activates defense response pathways.
It is also important to keep in mind when interpreting the azide and DPI experiments described in this paper that azide inhibits other enzymes besides peroxidases and that DPI is not a specific inhibitor of NADPH oxidases. For example, because the cell cultures in our experiments were maintained in continuous light and were green, and because azide is a potent inhibitor of photosynthesis (Yuan and Daniels, 1956), it is also possible that some intracellular ROS could be derived from photosynthesis. Indeed, photorespiration is an important source of ROS (Noctor et al., 2002). Similarly, DPI is a nonspecific flavin-binding compound that can inhibit a variety of flavoenzymes in addition to NADPH oxidases (Riganti et al., 2004).
Roles of Peroxidases and NADPH Oxidases in PTI
Interestingly, MAMP-elicited gene expression was more severely affected in the PRX33- and PRX34-depleted tissue culture lines than the oxidative burst. There was very little or no MAMP-elicited up-regulation of a variety of genes tested, including several genes involved in the biosynthesis of indoleglucosinolates (CYP79B2, CYP81F2, and MYB51) or camalexin (CYP71A12). Similar results were obtained with the activation of PRX33, PRX34, and RBOHD. These latter data suggest that not all ROS are equally capable of activating gene expression, depending perhaps on the nature of the ROS, its location, or the timing of its production. Consistent with these data, DPI treatment, which is relatively specific for NADPH oxidases, had a minimal effect on MAMP-elicited gene expression, whereas treatment with sodium azide completely inhibited any induction of expression, suggesting that peroxidase and mitochondrial sources of H2O2 are essential for triggering these signaling pathways whereas NADPH oxidases are not. Similar conclusions concerning an active role for PRX33 and PRX34 (Daudi et al., 2012) and the lack of involvement of NADPH oxidases (Galletti et al., 2008; Daudi et al., 2012) in activating MAMP-elicited responses have been published previously.
Proteomic Changes in the Transgenic Cell Lines
One somewhat unexpected observation was the decreased levels of pectinacetylesterases and pectinmethylesterases and the increased levels of PGIPs in the PRX33/PRX34 knockdown tissue culture lines. Pectinacetylesterases and pectinmethylesterases control the degree of acetyl and methyl esterification of cell wall polygalacturonans, and PGIPs inhibit cell expansion by inhibiting the cell wall-loosening activity of polygalacturonases that hydrolyze pectin. Although significant changes in the cross-linking of Hyp-rich glycoproteins following elicitor treatment have not been observed (Brisson et al., 1994; Wojtaszek et al., 1995; Brown et al., 1998), it is possible that the knockdown of basal levels of PRX33 and PRX34 leads to cell expansion by diminishing peroxidase-catalyzed cross-linking of cell wall polymers, which in turn affects the regulation of genes involved in cell expansion. A similar compensatory mechanism has also been observed in mutants lacking the cellulose synthase gene CESA3, where a reduction of cellulose content affected the pectin and xyloglucan composition of the cell wall (Caño-Delgado et al., 2003; Bosca et al., 2006). These data together with our proteomic data suggest a sensing mechanism that monitors the integrity of the cell wall. Consistent with this model, previous studies in French bean cell cultures and legumes have shown that Fusarium cell wall elicitor causes peroxidative cross-linking and immobilization of Pro- and Cys-rich polypeptides within the wall (Bradley et al., 1992; Wojtaszek et al., 1995; Wojtaszek, 1997; Bindschedler et al., 2006). Also consistent with this model is the finding that leaves of ecotype Columbia plants expressing asFBP1 are about 30% larger than wild-type leaves (Bindschedler et al., 2006; Daudi et al., 2012).
Interestingly, although it appears that PRX33 and PRX34 are essential for the production of H2O2 in response to various MAMPs, their postulated role in peroxidative cell wall cross-linking utilizes H2O2 as a substrate. The ability of peroxidases to both generate H2O2 and utilize H2O2 as a substrate is well documented, although little is known about the conditions that determine which reaction is catalyzed in vivo (Dunford, 1993).
Metabolic Changes in the Transgenic Cell Lines
The peroxidase-dependent oxidative burst has been described as a three-component system, which consists of the peroxidases, a pH change, and the release of the substrate (Bolwell, 1999; Bolwell et al., 2002). Although the substrates utilized by peroxidases to generate an oxidative burst in vivo have not been identified, clues to the nature of such a substrate or substrates may be found in the accumulation of stearate, palmitate, glycerol, malate, citrate, and succinate in CaCl2 extracts of French bean cell cultures in response to fungal elicitors, suggesting that there is a direct role of cell wall organic and fatty acids in the oxidative burst (Bolwell, 1999; Bolwell et al., 2002). However, it is important to note that strong reductants such as these metabolites are volatile and highly unstable, consequently making them very difficult to characterize. Therefore, despite considerable efforts, it was not possible to specifically identify an individual substrate of the cell wall peroxidases in this study. On the other hand, the metabolomic studies have provided evidence of the pathways that underlie the provision of the substrate, thereby providing clues about the nature and identity of the in vivo substrate.
In our study, the major differences between Ler (wt) and LerasFBP1 CaCl2 extracts following elicitation were observed in organic acids, mainly malate and fumarate, which were up to 4-fold higher in the LerasFBP1 lines than in the wild type. In contrast, lactate levels decreased in response to elicitation in both Ler (wt) and LerasFBP1 cells. The accumulation of organic acids has been linked with the activity of NADH peroxidases. Malate, fumarate, and lactate are intermediates in the production of NADH, which is a substrate for the generation of H2O2 by peroxidases (Gross et al., 1977; Halliwell, 1977, 1978). Specifically, malate is oxidized to oxaloacetate by malate dehydrogenase and lactate is oxidized to pyruvate by LDH to produce NADH from NAD+ (Gross et al., 1977; Halliwell, 1977, 1978). Thus, the decrease in the accumulation of lactate could be due to the activity of a LDH, which may provide NADH as a reducing agent for the peroxidases. Even though a LDH was not identified in proteomic aspects of this study, previous work with this cell culture demonstrated that LDH is present in the cell wall of Arabidopsis Ler cell cultures (Slabas et al., 2004).
The role of organic acids in the cell wall is still unknown. However, changes in the accumulation of organic acids upon elicitation suggest that they play an active role in plant defense. In addition, succinate, malate, and fumarate are part of the citric acid cycle in mitochondria and the glyoxylic acid cycle in peroxisomes. Thus, another possibility for the changes in the accumulation of these metabolites following elicitation is that they are a consequence of exocytosis events that are known to occur in plant cell-pathogen interactions (Brown et al., 1998) and may involve peroxisomes that move toward and accumulate at the site of infection (Scheel, 1998; Lipka et al., 2005; An et al., 2006; Hardham et al., 2008).
CONCLUSION
Our data suggest that the class III cell wall peroxidases PRX33 and PRX34 play key roles in both pathogen defense and cell growth. The production of H2O2 by PRX33 and PRX34 appears to be required for both basal and MAMP-elicited transcription of PRX33, PRX34, RBOHD, and other defense-related genes, suggesting a positive feed-forward self-amplifying activation of defense responses. The requirement of PRX33 and PRX34 for the transcription of defense-related genes is reflected in proteomic data showing that several defense proteins, including PDF2.2, PDF2.3, and GAST-1, are absent or down-regulated in the nonelicited LerasFBP1 lines. With respect to cell growth, in the absence of PRX33 and PRX34, the lack of H2O2 leads to increased cell size as a consequence of decreased levels of polymer cross-linking in the cell wall. The higher levels of PGIP observed in the LerasFBP1 lines may be due to a compensatory feedback system involved in the inhibition of cell wall loosening.
MATERIALS AND METHODS
Materials
Naphthalene acetic acid, kinetin, 3,3-diaminobenzidine, and xylenol orange were obtained from Sigma-Aldrich.
Plant Material and Elicitors
Arabidopsis (Arabidopsis thaliana) ecotype Ler suspension-culture cell lines were a gift from Prof. A.R. Slabas (Durham University) and were maintained in Murashige and Skoog basal salts with minimal organics medium (Sigma-Aldrich) containing Suc (30 mg mL−1), naphthalene acetic acid (0.5 μg mL−1), and kinetin (0.05 μg mL−1) and buffered to pH 5.6 to 5.7 with sodium hydroxide. The cultures were kept under low light intensity in a 16-h-light/8-h-dark regime. Fusarium oxysporum cell wall elicitor was generated according to Davies et al. (2006). The synthetic peptides Flg22 and Elf26 were synthesized by the Massachusetts General Hospital Peptide/Protein Core Facility and used at 0.5 μm. Purified oligogalacturonides were prepared as described (Ferrari et al., 2007) and were used at 50 μg mL−1. Sodium azide and DPI pretreatments were carried out at 2 mm and 0.2 μm, respectively, unless otherwise stated.
Cell Culture Transformation
Ler cell cultures were transformed by Agrobacterium tumefaciens-mediated transformation according to Ferrando et al. (2000) with modifications. Briefly, A. tumefaciens strain LB4404 containing the vector pBin19 with the antisense sequence of the French bean (Phaseolus vulgaris) peroxidase (asFBP1; Bindschedler et al., 2006) was grown on Luria-Bertani medium containing 10 μg mL−1 rifampicin and 50 μg mL−1 kanamycin to an optical density at 600 nm of 0.8. Cells were pelleted by centrifugation and resuspended in sterile Murashige and Skoog medium, and approximately 1 × 1010 A. tumefaciens cells were added to 100 mL of a freshly subcultured Arabidopsis Ler (wt) cell culture and kept in the dark under constant agitation. Three days after cocultivation, the suspension culture was removed from the shaker for 5 min to allow the cells to sediment, and the medium was removed and replaced with fresh medium containing 60 μg mL−1 kanamycin and 200 μg mL−1 timentin. The medium was replaced once every week for the following 5 weeks to eliminate nontransformed cells from the population of cells in the culture. Once the cells began to grow and generated a healthy, green coloration, 10 mL of cells was subcultured in 100 mL of fresh medium containing 60 μg mL−1 kanamycin.
PCR-Based Genome Walking
PCR-based genome walking was carried out following the protocol described by Siebert et al. (1995). Briefly, tissue culture cells were filtered under gentle vacuum through a single layer of filter paper (Whatman no. 1; Sigma-Aldrich) to remove the supernatant and then frozen in liquid nitrogen. Before genomic DNA (gDNA) extraction, frozen samples were ground using a mortar and pestle and gDNA was extracted from plant material using the cetyl-trimethyl-ammonium bromide method (Moore and Dowhan, 2002). After quantification, 2.5 μg of gDNA was digested overnight with ScaI. Digested gDNA was purified by phenol-chloroform extraction and resuspended in 20 μL of deionized water. An adaptor was prepared by making a 25 μm solution containing both the long and short adaptor primers (Supplemental Table S1) and incubating the mixture for 2 min at 100°C. The adaptor was ligated to 5 μL of digested gDNA with concentrated T4 DNA ligase (Biolabs), and the reaction was incubated overnight at 16°C. Further incubation at 70°C for 5 min inactivated the ligase. The sample was then diluted 10-fold in TE buffer (10 mm Tris [pH 7.5] and 1 mm EDTA) to a final volume of 100 μL. For the PCR amplification of specific sequences in the genome, four sets of primers were used: adaptor primer 1 (AP1), nested adaptor primer 2 (AP2), gene-specific primer 1 (GSP1), and gene-specific primer 2 (GSP2), as described in Supplemental Table S1. The amplification of the sequence of interest was carried out by two consecutive nested PCRs. For the first reaction, 1 μL of the diluted gDNA-adaptor sample was used with the AP1/GSP1 primers and a thermocycler program as follows: seven cycles of 25 s at 95°C followed by 3 min at 72°C, then 32 cycles of 25 s at 95°C and 3 min at 67°C, and finally one cycle of 7 min at 72°C. The first PCR was then diluted 1:50, and the dilution was used to do a nested reaction with the AP2/GSP2 primers and a thermocycler program as follows: five cycles of 25 s at 95°C followed by 3 min at 72°C, then 20 cycles of 25 s at 95°C and 3 min at 67°C, and finally one cycle of 7 min at 72°C. The PCR products of the nested PCR were analyzed on a 1% agarose gel, and the products were purified from the gel using the QIAquick gel extraction kit (Qiagen) and sequenced (Eurofins).
H2O2 Detection
H2O2 production in elicited Arabidopsis cells was measured using a xylenol orange assay as described previously, in which hydroperoxides are reduced by ferrous ions in acid solution, forming a ferric product-xylenol orange complex that is detected spectrophotometrically at 560 nm (Gay et al., 1999; Bindschedler et al., 2001).
CaCl2 Treatment of Arabidopsis Cell Culture
Tissue culture cells were filtered under gentle vacuum through a single layer of filter paper to remove the supernatant. Then, the cells were resuspended in ice-cold 200 mm calcium chloride buffer, pH 5.0, at a concentration of 2 mL g−1 dry tissue and incubated under gentle agitation for 30 min at 4°C. After the incubation, cells were filtered and the supernatant was collected for further analysis.
DNPH Derivatization and Analysis by HPLC
Low-Mr carboxylic acids and aldehydes present in the CaCl2 extracts of cell cultures were derivatized with DNPH. The derivatization was performed as described by Qureshi et al. (1982) with some modifications. Briefly, 20 mL of cells was elicited with 100 μg mL−1 F. oxysporum elicitor. At specific time points, cells were pelleted by centrifugation and 2 mL of supernatant medium was collected. To 300 μL of the supernatant medium, 75 μL of 0.2 n HCl was added. After a second centrifugation, 300 μL was incubated for 15 min at room temperature after adding 50 μL of 5 mm DNPH solution in 2 n HCl. For total quantification, 150 μL of deionized water and 0.5 mL of potassium phosphate buffer, pH 12.5, were added to the derivatized samples. Then, the samples were cleared by centrifugation for 3 min at maximum speed and transferred to 1-mL cuvettes. Finally, the A450 was read 15 to 60 min after addition of the phosphate buffer. For HPLC analysis, DNPH derivatives were extracted twice with 300 μL of ethyl acetate. The combined organic phases were extracted with 300 μL of 10% Na2CO3 in order to separate the nonreactive DNHP and aldehydes. The aqueous phase was cleaned once with 300 μL of ethyl acetate. The aqueous layer containing derivatized keto acids was recovered, and 200 μL was taken for HPLC analysis. The remaining organic layer containing derivatized aldehydes was diluted (1:1) with water, and 200 μL was used in HPLC analysis. Twenty microliters of sample was then injected into a Hewlett-Packard 1050 series high-performance liquid chromatograph equipped with a C18 reverse-phase column (250 × 4.6 mm, 5-μm particle size; Phenomenex Sphereclone) and a Milton Roy detector. The samples were analyzed using an isocratic solvent system according to Qureshi et al. (1982).
ECF Derivatization
Cultured cells were elicited with 100 μg mL−1 F. oxysporum elicitor. At 0 and 25 min, the supernatant was derivatized with a modification of the protocol described by Huang et al. (1993). Briefly, 1 mL of each sample was placed in a 15-mL glass tube. Then, 300 μL of ethanol, 100 μL of pyridine, and 100 μL of ECF were added. After vortexing, the samples were incubated in an ultrasonic bath for 5 min. Following that, 50 mg of NaCl was added and the samples were vortexed. Once the NaCl was dissolved, 75 μL of chloroform and 25 μL of toluene were added and the samples were vortexed for 1 min. Subsequently, all samples were centrifuged for 1 min at 3,000 rpm. One hundred microliters of the chloroform-toluene fraction was transferred to a GC vial for GC-MS analysis.
BSTFA Derivatization
Arabidopsis cells were elicited with 100 μg mL−1 F. oxysporum elicitor. At different time points, 10 mL of medium was vacuum filtered and 25 mg of cells was collected in an Eppendorf tube and frozen in liquid nitrogen. Samples were thawed on ice and extracted for 10 min in an ultrasonic bath at room temperature in 200 μL of chloroform containing tritert-butylbenzene and 4-phenylbutyric acid as internal standards. A water:methanol solution (1:3, v/v) was prepared, and 800 μL of the solution was added. The samples were extracted in an ultrasonic bath for 20 min. Following that, all samples were centrifuged for 10 min at 5,000 rpm. Three hundred microliters of the supernatant was taken in a sterile Eppendorf tube, and the samples were concentrated to dryness in a SpeedVac at 45°C. To the dry samples, 20 μL of methoxyamine hydrochloride (20 mg mL−1) was added, and the samples were incubated overnight at room temperature to allow methoxymation of aldehydes and keto groups. Subsequently, 20 μL of BSTFA (containing 1% trimethylchlorosilane) was added, and the samples were incubated at room temperature for 1 h. Finally, 20 μL of hexane was added, and the samples were transferred to a GC vial for GC-MS analysis.
GC-MS Conditions
Two microliters of the derivatized sample was injected in splitless mode by a Hewlett-Packard 7673 autosampler into a Hewlett-Packard 5890 series II gas chromatograph equipped with a 25-m × 0.22-mm (i.d.) HT5 column. The gas chromatograph was coupled to a Hewlett-Packard 5970 series mass selective detector. The data obtained were analyzed with Chemstation software (Agilent).
Preparation of Proteins for MS Analysis
Proteins were extracted from the cell walls of intact cells by the CaCl2 method and processed for SDS-PAGE as described previously (Wojtaszek et al., 1995). Selected bands from SDS-PAGE gels were excised, cut into 1-mm2 pieces, and placed in nonstick Eppendorf tubes. To remove the Coomassie blue, gel pieces were sequentially washed twice with ammonium bicarbonate (ABC) followed by three to five consecutives washes in ABC:acetonitrile (ACN; 1:1, v/v). The supernatant was removed after each washing step, and gel pieces were dried in a SpeedVac for 10 min without heating. The dry gel pieces were resuspended in 10 mm dithiothreitol in ABC and incubated for 1 h at 56°C in order to reduce the proteins. The resulting free thiol (without sulfhydryl) protein groups were alkylated by adding an equal volume of 55 mm iodoacetamide in ABC and incubating the reaction for 1 h in the dark. The gel pieces were then washed three times with ACN:ABC (1:1, v/v) followed by one wash with 100% ethanol. The dehydrated gel pieces were dried in a SpeedVac for 10 min without heating. Dry gel pieces were rehydrated in 15 μL of 12.5 μm trypsin (Promega) and 25 μL of ABC. The samples were incubated for 16 h at 37°C. After incubation, the supernatant containing the digested peptides was removed and transferred to a new tube. The remaining gel pieces were then washed with 50% ACN in 3% trifluoroacetic acid for 10 min, and the resulting supernatant was combined with the previous one. The gel pieces were washed again in the same solution followed by an incubation of 5 min in an ultrasonic bath, and the resulting supernatant was combined with the previous one. Finally, the gel pieces were washed in 100% ACN for 2 min. The combination of all the supernatants was concentrated to 20 μL in a SpeedVac and stored at −80°C until protein analysis.
Protein Sequencing
For multidimensional protein identification, nLC-nESI-MS/MS analysis was performed using an Eksigent MDLC HPLC device attached to a Thermo Scientific 7 Tesla LTQ-FT Ultra mass spectrometer operating in positive ion mode operating in the positive ion mode. Tryptic peptides were filtered through a 0.45-μm nylon filter, and 25-μL samples were injected in the HPLC-MS system. A CVC Microtech analytical C18 column (Biobasic C18, 5-μm particle size, 300-Å pore size, 10 cm x 75 μm) was used to separate peptides at a flow rate of 300 nL/min using water with 0.1% formic acid as phase A and ACN with 0.1% formic acid as phase B with the following gradient: 0 min, 1% B; at 60 min, 75% B; washing at 100% B for 10 min; and reequilibrating the column at initial conditions for 10 min. The mass spectrometer was operated with the Thermo nanospray source without sheath or auxiliary gas, an ion-spray voltage of 1800 V, and a capillary temperature of 200°C. The mass spectrometer was operated in data-dependent acquisition mode with a high resolution (100,000 at 400 mass-to-charge ratio) survey scan (mass-to-charge ratio, 350–1600), and data-dependent low-resolution MS/MS on the six most abundant ions with dynamic exclusion after two MS/MS experiments. Database searches of nLC-nESI-MS/MS mass spectra with Mascot (version 2.3, Matrix Sciences) included the following parameters: precursor mass tolerance, 1.2 D; product ion mass tolerance, 0.6 D; peptide charge of +1, +2, and +3; and trypsin for digesting enzyme with up to three missed cleavages allowed. Viridiplantae proteins were searched using the nonredundant National Center for Biotechnology Information database (National Center for Biotechnology Information no. 20090430). Mascot reported that a score of 43 was required for 95% confidence in the peptide identification, whereas manual examination of the measured delta mass between measured mass and that calculated for the assigned peptide (delta; Supplemental Table S2) revealed that nearly all assignments were achieved at less than 5 ppm, substantially increasing confidence in the peptide identifications.
PCR and qRT-PCR
Total RNA was extracted from plant material using the Qiagen RNeasy Mini Kit (Qiagen). RT from total RNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen). PCR was carried out using GoTaq DNA Polymerase (Promega) with gDNA or cDNA as the template. The product of each reaction was then analyzed by running the samples under constant voltage on an agarose gel containing 1× SYBR Safe DNA gel stain (Invitrogen). The amplification products were visualized under UV light. qRT-PCR was carried out using a Rotor-Gene Q real-time machine (Qiagen) and SYBR Green Jump Start (Sigma). The program used was as follows: 2 min at 94°C, followed by 45 cycles of 15 s at 94°C, 60 s at 58°C, and 20 s at 72°C. The run finished with a melting curve from 72°C to 95°C with 1°C increments every 5 s The expression values were normalized against tubulin β-9 chain. The sequences of the primers used for each gene are listed in Supplemental Table S1.
Statistical Analyses
For all experiments, at least three independent replicates were performed (unless otherwise stated). se values are shown. All graphs and statistical calculations were done in PRISM version 4.0 (GraphPad Software).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mapping of the insertion sites in IG-2 and IG-3.
Supplemental Figure S2. Insertion sites of the transformation cassette of LerasFBP1 lines IG-2 and IG-3.
Supplemental Figure S3. H2O2 production in Ler (wt), IG-2, and IG-3 cell cultures after MAMP elicitor treatment.
Supplemental Figure S4. Basal H2O2 levels in Ler (wt), IG-2, and IG-3.
Supplemental Figure S5. qRT-PCR quantification of transcript levels of GADPH in Ler (wt) after elicitation with F. oxysporum extract pretreated with sodium azide or DPI.
Supplemental Figure S6. Coomassie blue-stained SDS-PAGE gel of CaCl2-extracted proteins from Ler (wt) and LerasFBP1 cell cultures in response to F. oxysporum elicitor.
Supplemental Figure S7. GC-MS chromatogram of ECF derivatives of elicited Ler (wt) cell culture.
Supplemental Figure S8. GC-MS chromatogram of ECF derivatives of elicited LerasFBP1 cell culture.
Supplemental Figure S9. HPLC analysis and quantification of DNPH derivatives.
Supplemental Table S1. List of primers used in this study for qRT-PCR, RT-PCR, and genome-walking experiments.
Supplemental Table S2. List of peptides identified by in-gel trypsin digestion of CaCl2-extracted cell wall proteins of Arabidopsis Ler (wt) and LerasFBP1 cells.
Supplementary Material
Acknowledgments
We thank Mrs. Safina Khan for technical support.
References
- An Q, Ehlers K, Kogel K-H, van Bel AJE, Hückelhoven R. (2006) Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 172: 563–576 [DOI] [PubMed] [Google Scholar]
- Bach M, Schnitzler JP, Seitz HU. (1993) Elicitor-induced changes in Ca2+ influx, K+ efflux, and 4-hydroxybenzoic acid synthesis in protoplasts of Daucus carota L. Plant Physiol 103: 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick CS, Brown IR, Bennett MH, Mansfield JW. (1997) Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Minibayeva F, Gardner SL, Gerrish C, Davies DR, Bolwell GP. (2001) Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytol 151: 185–194 [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Whitelegge JP, Millar DJ, Bolwell GP. (2006) A two component chitin-binding protein from French bean: association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett 580: 1541–1546 [DOI] [PubMed] [Google Scholar]
- Blee KA, Jupe SC, Richard G, Zimmerlin A, Davies DR, Bolwell GP. (2001) Molecular identification and expression of the peroxidase responsible for the oxidative burst in French bean (Phaseolus vulgaris L.) and related members of the gene family. Plant Mol Biol 47: 607–620 [DOI] [PubMed] [Google Scholar]
- Bolwell GP. (1995) Cyclic AMP, the reluctant messenger in plants. Trends Biochem Sci 20: 492–495 [DOI] [PubMed] [Google Scholar]
- Bolwell GP. (1999) Role of active oxygen species and NO in plant defence responses. Curr Opin Plant Biol 2: 287–294 [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F. (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot 53: 1367–1376 [PubMed] [Google Scholar]
- Bolwell GP, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F, Rowntree EG, Wojtaszek P. (1999) Recent advances in understanding the origin of the apoplastic oxidative burst in plant cells. Free Radic Res (Suppl) 31: S137–S145 [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Daudi A. (2009) Reactive oxygen species in plant-pathogen interactions. In LA del Rio, A Puppo, eds, Reactive Oxygen Species in Plant Signaling. Springer-Verlag, Berlin, pp 113–133
- Bolwell GP, Davies DR, Gerrish C, Auh CK, Murphy TM. (1998) Comparative biochemistry of the oxidative burst produced by rose and French bean cells reveals two distinct mechanisms. Plant Physiol 116: 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell PP, Page A, Piślewska M, Wojtaszek P. (2001) Pathogenic infection and the oxidative defences in plant apoplast. Protoplasma 217: 20–32 [DOI] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I, Trethowan J, Kerry M, Mansfield J, Bolwell GP. (1998) Localization of components of the oxidative cross-linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non-pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J 15: 333–343 [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G. (2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Hamilton JM, Pringle RS, Ndimba BK, Simon WJ, Lindsey K, Slabas AR. (2006) Proteomic analysis of differentially expressed proteins in fungal elicitor-treated Arabidopsis cell cultures. J Exp Bot 57: 1553–1562 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR. (2002) Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 23: 1754–1765 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK. (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio C, Dunand C. (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60: 391–408 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, Bolwell GP. (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern triggered immunity. Plant Cell 24: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Bindschedler LV, Strickland TS, Bolwell GP. (2006) Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: implications for basal resistance. J Exp Bot 57: 1817–1827 [DOI] [PubMed] [Google Scholar]
- Devoto A, Leckie F, Lupotto E, Cervone F, De Lorenzo G. (1998) The promoter of a gene encoding a polygalacturonase-inhibiting protein of Phaseolus vulgaris L. is activated by wounding but not by elicitors or pathogen infection. Planta 205: 165–174 [DOI] [PubMed] [Google Scholar]
- Dunford B. (1993) Kinetics of peroxides reactions: horseradish, barley, Corpinus cinereus, lignin and manganese. In KG Welinder, S Rasmussen, C Penel, H Greppin, eds, Plant Peroxidases: Biochemistry and Physiology. University of Geneva, Geneva, pp 113–124
- Fahnenstich H, Scarpeci TE, Valle EM, Flügge UI, Maurino VG. (2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol 148: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Ferrando A, Farràs R, Jásik J, Schell J, Koncz C. (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Vairo D, Cervone F, De Lorenzo G. (2006) Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol Plant Microbe Interact 19: 931–936 [DOI] [PubMed] [Google Scholar]
- Frahry G, Schopfer P. (1998) Hydrogen peroxide production by roots and its stimulation by exogenous NADH. Physiol Plant 103: 395–404 [Google Scholar]
- Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S. (2008) The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay C, Collins J, Gebicki JM. (1999) Hydroperoxide assay with the ferric-xylenol orange complex. Anal Biochem 273: 149–155 [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB. (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GG, Janse C, Elstner EF. (1977) Involvement of malate, monophenols, and the superoxide radical in hydrogen peroxide formation by isolated cell walls from horseradish (Armoracia lapathifolia Gilib.). Planta 136: 271–276 [DOI] [PubMed] [Google Scholar]
- Halliwell B. (1977) Generation of hydrogen peroxide, superoxide and hydroxyl radicals during the oxidation of dihydroxyfumaric acid by peroxidase. Biochem J 163: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. (1978) Lignin synthesis: the generation of hydrogen peroxide and superoxide by horseradish peroxidase and its stimulation by manganese (II) and phenols. Planta 140: 81–88 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Takemoto D, White RG. (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol 8: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves IP, Duncan AJ, Wu L, Agrawal A, Land JM, Heales SJR. (2007) Inhibition of mitochondrial complex IV leads to secondary loss complex II-III activity: implications for the pathogenesis and treatment of mitochondrial encephalomyopathies. Mitochondrion 7: 284–287 [DOI] [PubMed] [Google Scholar]
- Huang ZH, Wang J, Gage DA, Watson JT, Sweeley CC, Husek P. (1993) Characterization of N-ethoxycarbonyl ethyl esters of amino acids by mass spectrometry. J Chromatogr A 635: 271–281 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaffarnik FA, Jones AM, Rathjen JP, Peck SC. (2009) Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics 8: 145–156 [DOI] [PubMed] [Google Scholar]
- Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS. (2009) Activation of β-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol 150: 1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Serada S, Norioka N, Norioka S, Neumetzler L, Pauly M, Sampedro J, Zarra I, Hayashi T, Kaneko TS. (2010) Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol 153: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Moore D, Dowhan D. (2002) Purification and concentration of DNA from aqueous solutions. Curr Protoc Mol Biol Chapter 2, Unit 2.1A
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JDG. (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH. (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot (Lond) 89: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24: 255–265 [DOI] [PubMed] [Google Scholar]
- Qureshi AA, Elson CE, Lebeck LA. (1982) Application of high-performance liquid chromatography to the determination of glyoxylate synthesis in chick embryo liver. J Chromatogr A 249: 333–345 [DOI] [PubMed] [Google Scholar]
- Ramonell KM, Zhang B, Ewing RM, Chen Y, Xu D, Stacey G, Somerville S. (2002) Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol Plant Pathol 3: 301–311 [DOI] [PubMed] [Google Scholar]
- Rico A, Preston GM. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact 21: 269–282 [DOI] [PubMed] [Google Scholar]
- Riganti C, Gazzano E, Polimeni M, Costamagna C, Bosia A, Ghigo D. (2004) Diphenyleneiodonium inhibits the cell redox metabolism and induces oxidative stress. J Biol Chem 279: 47726–47731 [DOI] [PubMed] [Google Scholar]
- Robertson D, Mitchell GP, Gilroy JS, Gerrish C, Bolwell GP, Slabas AR. (1997) Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from plants. J Biol Chem 272: 15841–15848 [DOI] [PubMed] [Google Scholar]
- Scheel D. (1998) Resistance response physiology and signal transduction. Curr Opin Plant Biol 1: 305–310 [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabas AR, Ndimba B, Simon WJ, Chivasa S. (2004) Proteomic analysis of the Arabidopsis cell wall reveals unexpected proteins with new cellular locations. Biochem Soc Trans 32: 524–528 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. (1997) Oxidative burst: an early plant response to pathogen infection. Biochem J 322: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P, Trethowan J, Bolwell GP. (1995) Specificity in the immobilisation of cell wall proteins in response to different elicitor molecules in suspension-cultured cells of French bean (Phaseolus vulgaris L.). Plant Mol Biol 28: 1075–1087 [DOI] [PubMed] [Google Scholar]
- Yuan EL, Daniels F. (1956) The effects of some inhibitors on the rates of photosynthesis and respiration by Chlorella. J Gen Physiol 39: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.