Abstract
We report here that disruption of function of the ω-3 FATTY ACID DESATURASE7 (FAD7) enhances plant defenses against aphids. The suppressor of prosystemin-mediated responses2 (spr2) mutation in tomato (Solanum lycopersicum), which eliminates the function of FAD7, reduces the settling behavior, survival, and fecundity of the potato aphid (Macrosiphum euphorbiae). Likewise, the antisense suppression of LeFAD7 expression in wild-type tomato plants reduces aphid infestations. Aphid resistance in the spr2 mutant is associated with enhanced levels of salicylic acid (SA) and mRNA encoding the pathogenesis-related protein P4. Introduction of the Naphthalene/salicylate hydroxylase transgene, which suppresses SA accumulation, restores wild-type levels of aphid susceptibility to spr2. Resistance in spr2 is also lost when we utilize virus-induced gene silencing to suppress the expression of NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1), a positive regulator of many SA-dependent defenses. These results indicate that FAD7 suppresses defenses against aphids that are mediated through SA and NPR1. Although loss of function of FAD7 also inhibits the synthesis of jasmonate (JA), the effects of this desaturase on aphid resistance are not dependent on JA; other mutants impaired in JA synthesis (acx1) or perception (jai1-1) show wild-type levels of aphid susceptibility, and spr2 retains aphid resistance when treated with methyl jasmonate. Thus, FAD7 may influence JA-dependent defenses against chewing insects and SA-dependent defenses against aphids through independent effects on JA synthesis and SA signaling. The Arabidopsis (Arabidopsis thaliana) mutants Atfad7-2 and Atfad7-1fad8 also show enhanced resistance to the green peach aphid (Myzus persicae) compared with wild-type controls, indicating that FAD7 influences plant-aphid interactions in at least two plant families.
Fatty acid desaturases (FADs), which introduce double bonds into the aliphatic tails of fatty acids, influence plant susceptibility to a wide variety of stresses. They promote drought and salt tolerance and also mediate plant adaptation to temperature extremes (Upchurch, 2008). Several FADs are up-regulated in response to chilling and confer cold tolerance in a variety of plant species by increasing the production of trienoic fatty acids, which enhance membrane fluidity (Kodama et al., 1994; Berberich et al., 1998; Khodakovskaya et al., 2006; Wang et al., 2006; Zhou et al., 2010). Conversely, FAD activity and trienoic fatty acid levels decrease at high temperatures, and Arabidopsis (Arabidopsis thaliana) double mutants that are deficient in two chloroplast-localized ω-3 FADs (FAD7 and FAD8) display enhanced heat tolerance (Murakami et al., 2000). FADs also influence resistance to numerous biotic stresses. For example, the Arabidopsis Atfad7fad8 mutant shows increased vulnerability to the bacterial pathogen Pseudomonas syringae, whereas suppression of the homologous OsFAD7 and OsFAD8 genes in rice (Oryza sativa) results in enhanced resistance to the rice blast fungus Magnaporthe grisea (Yaeno et al., 2004; Yara et al., 2007). Thus, FADs appear to act as a sort of rolling fulcrum that can shift the balance between resistance to some stresses and susceptibility to others.
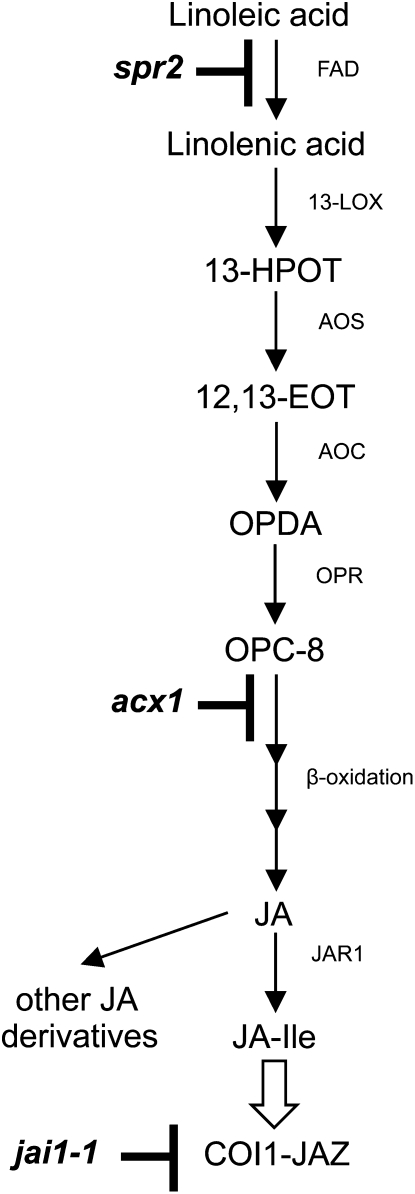
The influence of ω-3 FADs on biotic stress is due in part to their critical role in the biosynthesis of the defense hormone jasmonate (JA; Fig. 1). Jasmonoyl-l-Ile, which is a receptor-active form of JA (Howe and Jander, 2008; Fonseca et al., 2009; Sheard et al., 2010), activates many plant responses to wounding, insect attack, and certain pathogens. JA levels are enhanced in transgenic rice that overexpress FAD7 and are depleted by inhibition of ω-3 FAD activity in Arabidopsis and potato (Solanum tuberosum) plants (McConn et al., 1997; Martín et al., 1999; Song et al., 2004). Furthermore, wounding and other stresses up-regulate FAD7, suggesting that FADs may play a role in regulating JA accumulation (Nishiuchi et al., 1997). In addition to influencing the availability of precursors for JA synthesis, FADs also modulate salicylate signaling. Salicylic acid (SA) is a β-hydroxy-benzoic acid that is required for basal resistance, systemic acquired resistance, and effector-triggered immunity against many pathogens (Vlot et al., 2009) and can also contribute to plant defenses against aphids (Li et al., 2006). The SUPPRESSOR OF SA INSENSITIVITY2 (SSI2) gene, which encodes a stearoyl acyl carrier protein FAD that converts stearic acid (C18:0) to oleic acid (C18:1), inhibits SA signaling. Accumulation of SA is enhanced as a result of decreased levels of oleic acid in the Arabidopsis ssi2 mutant (Kachroo et al., 2001, 2004; Shah et al., 2001) and in rice and soybean (Glycine max) plants in which SSI2 homologs have been silenced (Kachroo et al., 2008; Jiang et al., 2009). In soybean, SA accumulation is also enhanced by transient suppression of FAD3, an ω-3 FAD localized in the endoplasmic reticulum (Singh et al., 2011), and in Arabidopsis, high constitutive levels of SA have been reported in the Atfad3fad7fad8 triple mutant (Mène-Saffrané et al., 2009). Thus, there is growing evidence that certain FADs inhibit SA accumulation.
Figure 1.
JA synthesis and perception in plants. Mutations in tomato that block JA synthesis or perception are represented in boldface. Abbreviations are as follows: AOC, allene oxide cyclase; AOS, allene oxide synthase; COI1, coronatine insensitive 1; EOT, epoxy-9,11,15-octadecatrienoic acid; JA-Ile, jasmonic acid-isoleucine conjugate; JAR1, jasmonic acid resistant 1; JAZ, jasmonate ZIM domain protein; HPOT, hydroperoxy-octadecatrienoic acid; LOX, lipoxygenase; OPC, 3-oxo-2(2′(Z)-pentenyl)-cyclopentane-1-octanoic acid; OPDA, 2-oxo-phytodienoic acid; OPR, 12-oxo-phytodienoic acid reductase. Black arrows represent biosynthetic steps, whereas the white arrow represents recognition of JA-Ile by the COI1/JAZ coreceptor (Sheard et al., 2010). This figure was modified from Schaller (2001).
Because loss of function of ω-3 FADs impairs JA accumulation, mutants deficient in these enzymes have been utilized to study JA-dependent defenses against chewing insects. Compared with wild-type Arabidopsis, a triple mutant with defects in FAD3, FAD7, and FAD8 was shown to be highly susceptible to the fungus gnat Bradysia impatiens (McConn et al., 1997). This susceptibility reflects the plant’s inability to synthesize JA, because treating the triple mutant with exogenous methyl jasmonate (MeJA) restored insect resistance. Loss of function of a FAD7 homolog in tomato (Solanum lycopersicum) also impairs plant defenses against chewing insects. The suppressor of prosystemin-mediated responses2 (spr2) mutant in tomato carries a point mutation in LeFAD7 that introduces a premature stop codon and is predicted to result in a total loss of function of the protein (Li et al., 2003). The foliage of this mutant has enhanced levels of linoleic acid (C18:2) and only approximately 10% of the linolenic acid (C18:3) content observed in wild-type tomato plants (Li et al., 2003). The spr2 mutation inhibits JA-dependent responses to the wound signal systemin and nearly eliminates the expression of the JA-responsive PROTEINASE INHIBITOR II (PI-II) gene, a well-characterized marker of induced resistance to insects (Howe and Ryan, 1999; Li et al., 2003). Furthermore, tobacco hornworm larvae (Manduca sexta) consume much more foliage and grow two to three times larger on spr2 plants than on wild-type controls, and M. sexta adults preferentially oviposit on spr2 (Li et al., 2003; Sánchez-Hernández et al., 2006). Thus, although there is considerable functional redundancy among FAD7, FAD8, and FAD3 in Arabidopsis and genes homologous to FAD8 and FAD3 are present in tomato (Yu et al., 2009; ITAG, 2011), FAD7 appears to play a dominant role in regulating induced resistance in tomato.
Whereas the contribution of the octadecanoid pathway to induced resistance against chewing insects and cell-content feeders is well established, its role in plant interactions with piercing-sucking herbivores requires further characterization (Thompson and Goggin, 2006). Aphids and whiteflies extract phloem sap through slender mouth parts that cause far less mechanical injury than the mandibles of chewing insects (Walling, 2008), and to date, they have not been reported to induce detectable levels of JA (Heidel and Baldwin, 2004; De Vos et al., 2005). The decoy hypothesis posits that these phloem-feeding insects limit the induction of JA-dependent defenses by inducing SA, which can interact antagonistically with JA signaling (Zhu-Salzman et al., 2004; de Vos et al., 2007). Although recent evidence suggests that SA accumulation may not be required for the repression of JA by whiteflies (Zhang et al., 2009), there is strong evidence that whitefly nymphs down-regulate genes associated with JA signaling (Kempema et al., 2007; Zhang et al., 2009). Furthermore, the development of whitefly nymphs on Arabidopsis is promoted by mutations that impair JA perception (coronatine insensitive1 [coi1]) or constitutively activate SA signaling (cim10; Zarate et al., 2007), and the spr2 mutation in tomato results in increased whitefly oviposition, although it does not affect nymphal development (Sánchez-Hernández et al., 2006). Thus, there is strong evidence that JA contributes to basal resistance against whiteflies but that whiteflies are also adapted to inhibit JA signaling in their hosts.
Further work is needed to test the decoy hypothesis in plant-aphid interactions. Aphids can in some cases up-regulate genes associated with JA signaling (Thompson and Goggin, 2006; Gao et al., 2007; Kusnierczyk et al., 2007; Kuśnierczyk et al., 2008), and several studies suggest that JA-dependent defenses hinder aphid infestation. Artificial JA treatment enhances aphid resistance in several plant species (Omer et al., 2001; Bruce et al., 2003, 2008; Zhu-Salzman et al., 2004; Cooper and Goggin, 2005); furthermore, aphid population growth on Arabidopsis is enhanced by the coi1 mutation, which inhibits JA perception, and is suppressed by the cev1 mutation, which promotes constitutive JA and ethylene signaling (Ellis et al., 2002; Mewis et al., 2005). On the other hand, there is evidence that SA also contributes to plant defenses against aphids, in contrast to its putative role as a decoy response in interactions between whiteflies and Arabidopsis. Although analyses of aphid population growth on Arabidopsis mutants with altered SA signaling have given equivocal results (Thompson and Goggin, 2006; de Vos et al., 2007), in tomato, SA induction by aphids has been shown to be an important component of effector-mediated immunity and may also contribute to basal defense against aphids (Li et al., 2006). Therefore, further work is needed to elucidate the relative contributions of JA and SA to plant defenses against aphids. In addition to the impact of FADs on JA synthesis and SA signaling, there are also other routes through which this group of enzymes may influence plant-aphid interactions. At least two FADs are known to influence plant defenses against aphids either directly or indirectly. In zonal geranium (Pelargonium × hortorum), a Δ914:0 FAD mediates resistance to aphids and mites through its role in the synthesis of toxic anacardic acids (Schultz et al., 1996). In Arabidopsis, loss of function of another FAD, SSI2, enhances aphid resistance, and petiole exudates from ssi2 mutants have antibiotic effects on aphids (Pegadaraju et al., 2005; Louis et al., 2010). Although the ssi2 mutant has high constitutive levels of SA, aphid resistance in ssi2 does not appear to require this hormone because resistance is retained in the ssi2 Naphthalene/salicylate hydroxylase (NahG) double mutant, and NahG inhibits SA accumulation (Pegadaraju et al., 2005). Instead, Pegadaraju and coworkers (2005) propose that aphid resistance in this mutant is due to hypersenescence. These studies indicate that FADs may influence a plant’s susceptibility to aphids through a diversity of mechanisms. Therefore, the goal of this study was to further investigate the role of FADs in plant interactions with aphids.
In contrast to previous observations that ω-3 FADs are required for resistance to chewing insects, we report here that loss of function of FAD7 in the spr2 mutant in tomato confers resistance to the sap-feeding potato aphid, Macrosiphum euphorbiae. To our knowledge, this is the first report of an ω-3 FAD inhibiting insect resistance, and this finding suggests that FAD7 can mediate tradeoffs between plant defenses against different herbivores. Aphid resistance in spr2 does not appear to depend upon impaired JA signaling, because exogenous MeJA fails to restore aphid susceptibility in spr2. Furthermore, aphid resistance is not observed in other mutants blocked in JA synthesis or perception. Instead, aphid resistance in the spr2 mutant requires SA accumulation and is dependent upon NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1). Aphid resistance is also observed in Arabidopsis mutants deficient in FAD7 and a double mutant deficient in FAD7 and FAD8, indicating that the impact of FAD7 on aphid resistance may be conserved in other species.
RESULTS
Aphid Infestations Are Reduced on spr2 but Not on Other JA Mutants
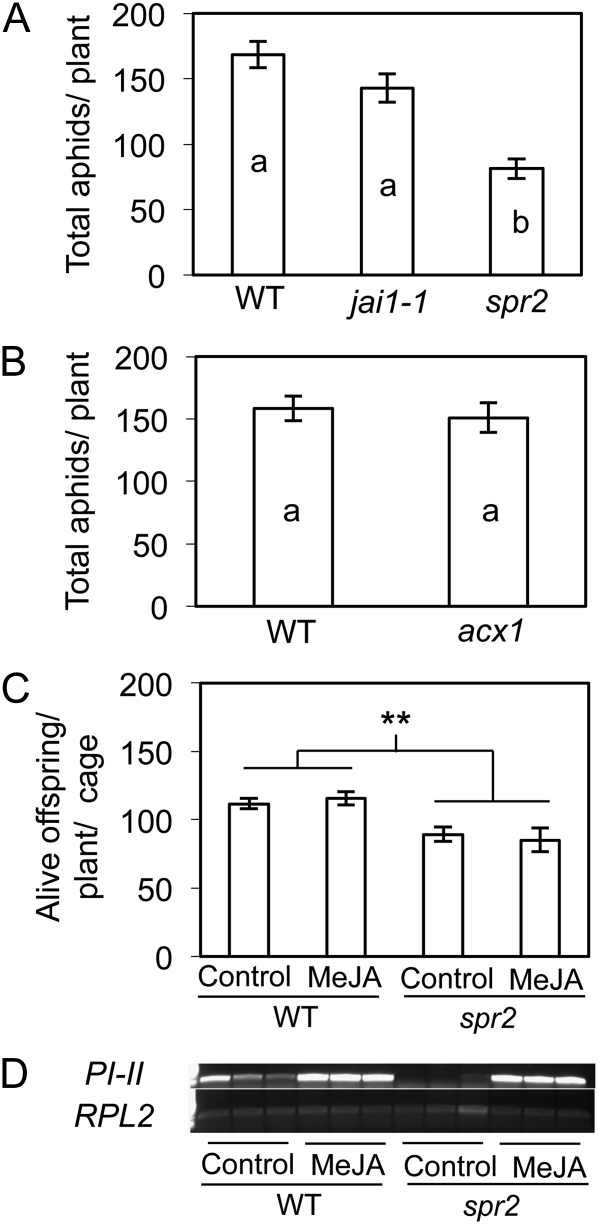
Population growth of M. euphorbiae was significantly lower on the spr2 mutant (P < 0.0001; Fig. 2A), which carries a loss-of-function mutation in LeFAD7, than on wild-type tomato plants (cv Castlemart) or on jasmonic acid insensitive1 (jai1-1), a mutant line impaired in JA-Ile perception due to a deletion mutation in LeCOI1 (Fig. 1). Aphids in this assay were not confined to cages, and so final aphid numbers were influenced by aphid host acceptance as well as by aphid survival and fecundity on the different plant genotypes. Suppression of the JA pathway in spr2 and jai1-1 was confirmed by the absence of wound-induced proteinase inhibitor accumulation in these plants (data not shown). To determine whether the effect of spr2 on aphid performance is due to the inhibition of JA synthesis, we also examined aphid performance on acx1, which carries a mutation that results in a loss of function of acyl-CoA oxidase 1A, thereby blocking the β-oxidation step of JA synthesis (Li et al., 2005; Fig. 1). Like spr2, both jai1-1 and acx1 are deficient in JA-dependent defenses and are highly susceptible to chewing insects (Li et al., 2001, 2003, 2004, 2005; Chen et al., 2005a; Sánchez-Hernández et al., 2006). Neither jai1-1 nor acx1, however, had a significant effect on aphid population growth compared with wild-type plants (Fig. 2, A and B; P < 0.05). Furthermore, application of MeJA did not impact aphid population growth on spr2, even though, as expected, it up-regulated the defensive gene PI-II (Fig. 2, C and D). These findings support prior assertions that JAs are not required for plant defenses against aphids (Bhattarai et al., 2007) and indicate that the effects of spr2 on aphids are likely independent of its effects on JA synthesis.
Figure 2.
Aphid infestations are reduced on spr2 but are unaffected by MeJA treatment or by other mutations that impair JA signaling. A and B, Wild-type (WT; cv Castlemart) and mutant (spr2, jai1-1, and acx1) tomato plants were inoculated with 15 aphids per plant, which were not confined to cages and were free to leave the plants. The total number of remaining aphids and their progeny per plant were counted 5 d after inoculation and analyzed by one-way ANOVA. Mean separations were performed using Student’s t test. Values ± se labeled with different letters differ significantly at α = 0.05. C, Wild-type and spr2 plants were treated with MeJA (75 μm) and inoculated with aphids 24 h after treatment (five aphids per cage; three clip cages per plant; 10 plants per treatment group). Live offspring were counted 6 d after inoculation, and the average numbers of offspring per cage per plant were analyzed by two-way ANOVA. ** Significant main effect of genotype at P < 0.0001. D, Expression of JA-responsive PI-II was monitored by RT-PCR 24 h after MeJA treatment. Expression of constitutive RPL2 is presented as a loading control. n = 10 for A, n = 8 for B, and n = 10 for C.
Loss of Function of FAD7 Confers Aphid Resistance in Both Tomato and Arabidopsis
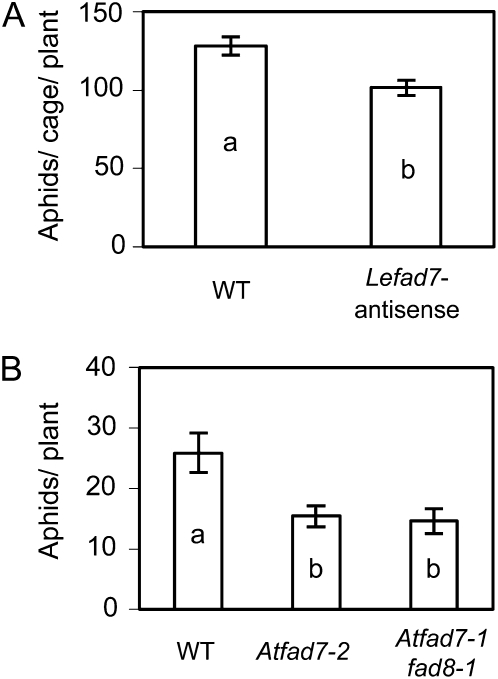
The fact that spr2 is resistant to aphids suggests that FAD7 suppresses aphid resistance. To further demonstrate the role of FAD7 in aphid resistance, aphid population growth was measured on a previously described tomato line that has low linolenic acid (C18:3) and high linoleic acid (C18:2) content as a result of antisense silencing of LeFAD7 (Liu et al., 2006, 2010). Offspring production of potato aphids caged on the antisense line was significantly lower (P = 0.0013) than on the wild-type control (cv L402; Fig. 3A). Moreover, loss of function of FAD7 has similar effects on aphid infestations on Arabidopsis. Unlike its homolog in tomato, AtFAD7 in Arabidopsis shows considerable functional redundancy with AtFAD8 (Gibson et al., 1994; McConn et al., 1994); therefore, the Atfad7-1fad8-1 double mutant was tested in addition to the Atfad7-2 single mutant. Plants were challenged with the green peach aphid (Myzus persicae) because Arabidopsis is not a host for the potato aphid. Seven days after inoculation, both Atfad7-2 and Atfad7-1fad8-1 plants had approximately 42% fewer aphids than the wild-type control (ecotype Columbia; P = 0.0083; Fig. 3B). The alternative mutant allele Atfad7-1, which is in the Columbia-glabra1 background, also conferred aphid resistance (data not shown).
Figure 3.
Loss of function of FAD7 confers aphid resistance in both tomato and Arabidopsis. A, Adult potato aphids were confined to individual leaflets of intact tomato plants using clip cages (five aphids per cage; three cages per plant; 12 plants per genotype), and the total number of aphids was recorded after 6 d. Cultivar L402 was used as the untransformed wild-type (WT) control. B, Adult green peach aphids were confined on individual Arabidopsis plants using sleeve cages (two aphids per plant; 18 plants per genotype), and the total number of aphids per plant was recorded after 7 d. Aphid numbers were analyzed by one-way ANOVA, and mean separations were performed using Student’s t test. Values ± se labeled with different letters differ significantly at α = 0.05.
Loss of Function of FAD7 Reduces Aphid Infestations by Decreasing Aphid Settling, Survival, and Fecundity
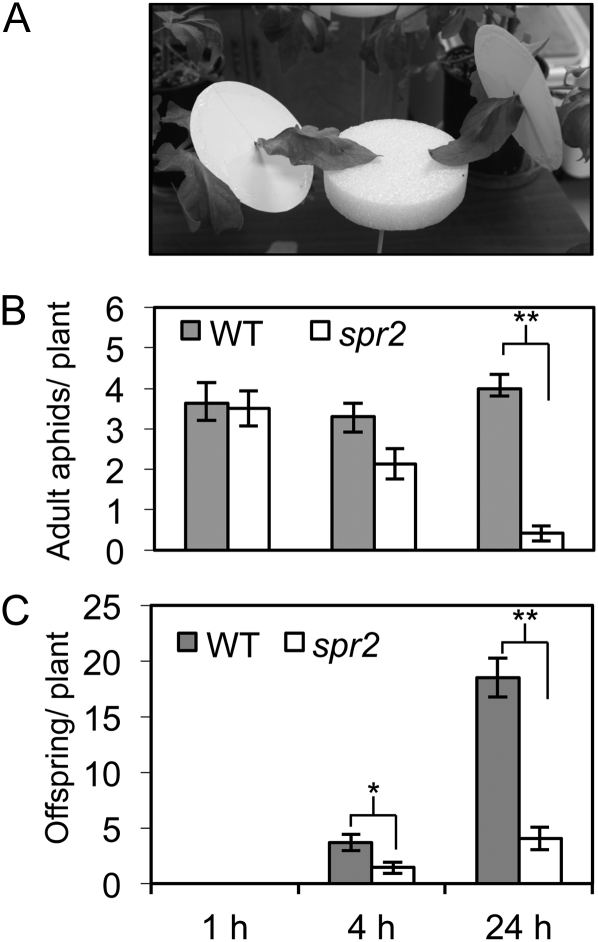
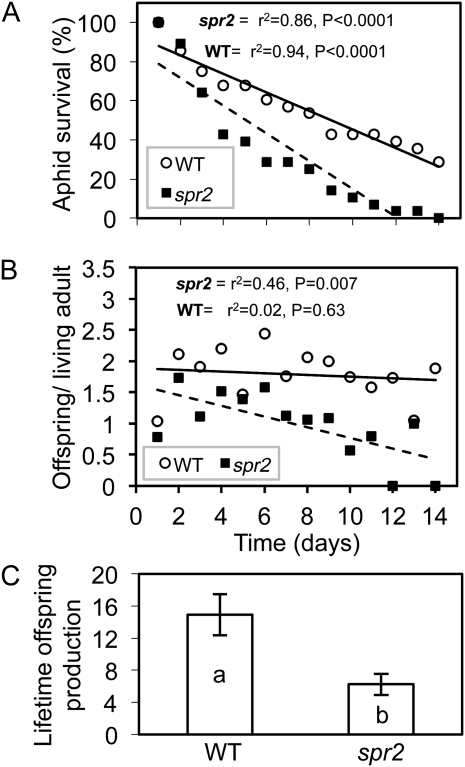
Host plant defenses against aphids may act by deterring aphids from settling on the plants (i.e. antixenosis) or by reducing their survival and/or offspring production (i.e. antibiosis). To determine if loss of function of FAD7 results in antixenotic effects, adult potato aphids were placed between intact spr2 and wild-type tomato plants on choice arenas that allowed them to go back and forth between plants (Fig. 4A). Because aphid reproduction is an indicator of host acceptance and is typically initiated shortly after identifying a suitable host plant, offspring production as well as adult position were monitored at 1, 4, and 24 h after release. At 1 h, adults were distributed roughly equally between the two genotypes (Fig. 4B). However, at 4 and 24 h, after the aphids had a more lengthy opportunity to feed on the plants, adults preferentially congregated (Fig. 4B) and reproduced (Fig. 4C) on wild-type plants, and 24 h after inoculation, total aphid numbers were almost five times higher on wild-type plants than on spr2 plants (pairwise t test, P < 0.001). This suggests that, compared with wild-type plants, spr2 mutants either lack important cues that promote host acceptance or produce deterrents that actively repel aphids. To measure the potential antibiotic effects of spr2 independent of any effect on host preference, adult females were individually caged on spr2 and wild-type plants, and adult survival was monitored daily until all aphids on the mutant genotype were dead (14 d). Offspring were also counted and removed daily to assess reproduction. The mortality rate was more than 50% higher on spr2 plants than on wild-type plants (Fig. 5A), and the average number of days that adult aphids survived on this mutant (5 ± 0.5 d) was significantly lower (P < 0.05) than on wild-type controls (8 ± 1 d). Aphid fecundity, as measured by the number of offspring divided by the number of surviving adult females, also significantly declined over time on spr2, whereas fecundity remained stable on wild-type plants (Fig. 5B). As a result of these decreases in fecundity and longevity, lifetime offspring production (Fig. 5C) was more than 50% lower on spr2 than on wild-type plants (P < 0.01). Therefore, choice and no-choice tests demonstrate that loss of function of FAD7 has both antixenotic effects that inhibit aphid settling and antibiotic effects that reduce survival and fecundity.
Figure 4.
Loss of function of FAD7 reduces aphid host acceptance. A, Wingless adult aphids (10 adults per arena) were placed on choice arenas between paired 6-week-old plants of spr2 and the wild-type (WT) control (cv Castlemart). The majority of aphids moved off the choice arena onto the plants within minutes of release. Aphids were free to move back and forth between the two plants. B and C, The number of adults on each plant (B) and the offspring they produced (C) were counted at 1, 4, and 24 h after aphids were placed on the arenas. Marked pairwise comparisons denote significant differences according to paired t tests at α = 0.05 (*) or α = 0.001 (**). Error bars indicate ± se (n = 10 pairs).
Figure 5.
Loss of function of FAD7 decreases aphid survival and fecundity. Newly emerged adult female aphids were caged on spr2 or wild-type (WT; cv Castlemart) plants (one aphid per cage; two cages per plant; 14 plants per genotype), and the cages were monitored daily to track the survival (A) and daily offspring production (B) of each aphid as well as their lifetime totals for offspring production (C). Regression analyses were performed to estimate aphid mortality rates and changes over time in daily fecundity. Lifetime offspring production was analyzed by one-way ANOVA, and values ± se having different letters are significantly different at α = 0.05.
Loss of Function of FAD7 Enhances P4 Expression and Local SA Induction in Response to Aphid Feeding
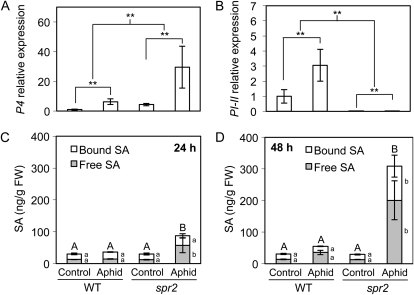
The pathogenesis-related gene P4 in tomato (GenBank gene identifier 544185) is homologous to PR1a in tobacco (Nicotiana tabacum) and Arabidopsis and is up-regulated in response to exogenous SA and its analog benzothiadiazole (Van Kan et al., 1995; Fidantsef et al., 1999; Schuhegger et al., 2006) as well as in response to the ethylene mimic ethephon (Van Kan et al., 1995; Chao et al., 1999). The proteinase inhibitor gene PI-II (GenBank gene identifier 543955) is transcriptionally activated in response to JAs and also ethylene (Farmer et al., 1992; Ohtsubo et al., 1999). Reverse transcription-quantitative PCR (RT-qPCR) was used to assess the effects of aphid feeding on the transcript abundance of P4 and PI-II in locally infested foliage of spr2 and wild-type tomato plants (cv Castlemart) 48 h after aphid inoculation. Aphid feeding on wild-type plants up-regulated the expression of P4 from 6- to 39-fold (Fig. 6A; Supplemental Fig. S1) and had a relatively modest effect on PI-II, causing either a slight up-regulation (Fig. 6B) or no significant change in expression (Supplemental Fig. S1). This is consistent with prior reports that the induction of PI-II by aphids is relatively weak and transient compared with the induction of P4 (Fidantsef et al., 1999; Martinez de Ilarduya et al., 2003). As predicted by previous observations (Li et al., 2006), PI-II expression was negligible in spr2 (Fig. 6B). In contrast, P4 expression was approximately four to five times higher in spr2 plants when compared with the respective wild-type treatments (Fig. 6A), suggesting that SA signaling might be enhanced in this mutant. SA and JA are known to interact antagonistically under certain circumstances (for review, see Pieterse et al., 2009), so it is possible that by suppressing JA synthesis, the spr2 mutation relieves the SA pathway from repression by JA. However, unlike spr2, the jai1-1 mutation did not dramatically enhance P4 accumulation in response to aphids (Supplemental Fig. S1).
Figure 6.
Loss of function of FAD7 enhances the expression of the SA-responsive gene P4 and local SA accumulation in response to aphid feeding but suppresses the expression of the JA-responsive gene PI-II. A and B, Wild-type (WT; cv Castlemart) and spr2 tomato plants were challenged with the potato aphid (100 aphids confined to a single leaf with a sleeve cage) or mock inoculated with empty cages, and P4 and PI-II transcript abundance was analyzed 48 h after inoculation. Expression values were calculated by RT-qPCR relative to the wild-type mock-inoculated control, normalized using the RPL2 gene, and analyzed by two-way ANOVA. Error bars represent ± se (n = 4). ** P < 0.001. C and D, Wild-type (cv Castlemart) and spr2 tomato plants were challenged with potato aphids or mock inoculated with empty cages (60 aphids confined to the three terminal leaflets with a sleeve cage; five plants per genotype per time point). At 24 and 48 h after inoculation, total, free, and bound SA were quantified by HPLC in infested or mock-inoculated leaflets. Values were analyzed by ANOVA, and mean separations were performed using Student’s t test. Bars of the same pattern ± se with different lowercase letters are significantly different at α = 0.05; bars with different uppercase letters show significant differences in total SA content (free + bound). FW, Fresh weight.
To explore the possibility of enhanced SA signaling in spr2, local accumulation of SA in response to aphid feeding was measured in spr2 and wild-type plants 24 and 48 h after aphid infestation. In locally infested tissue, the total, free, and bound SA levels significantly increased in response to aphid feeding in the spr2 mutant but not in wild-type tomato (Fig. 6, C and D). These results indicate that loss of function of FAD7 enhances local SA induction and PR gene expression in response to aphids.
Aphid Resistance Conferred by Loss of Function of FAD7 Requires SA Accumulation and Is Dependent upon NPR1
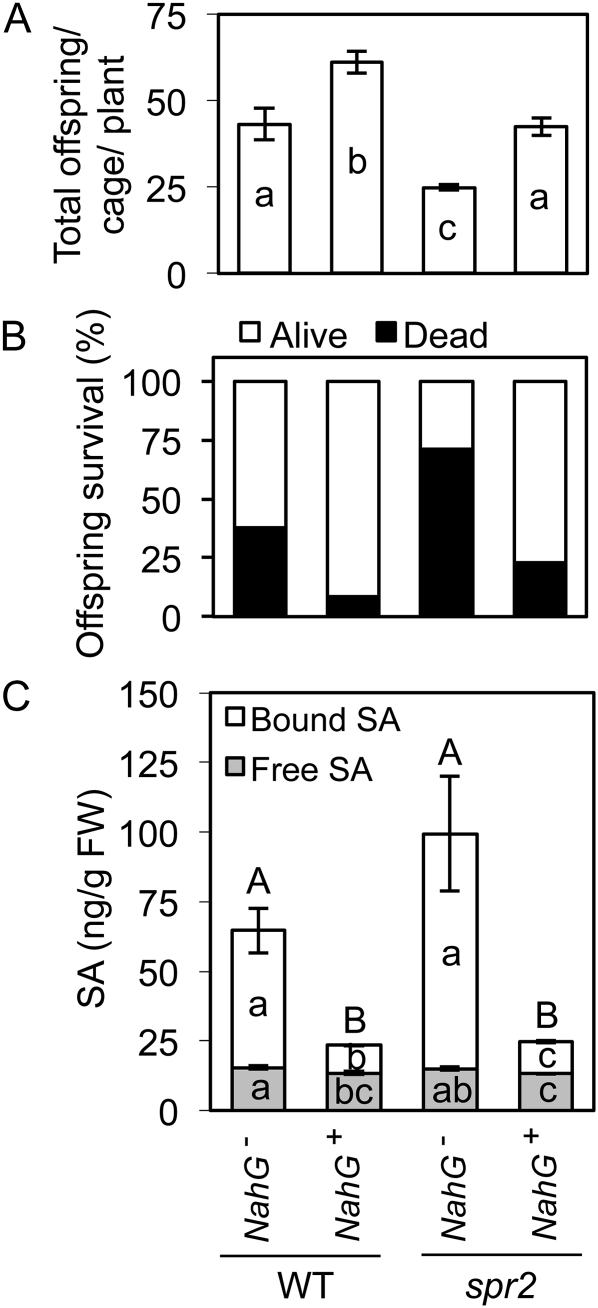
To determine if SA has a causal role in aphid resistance in spr2, this mutant tomato line was crossed to a transgenic line that carries NahG, a bacterial gene encoding salicylate hydroxylase, which degrades SA to catechol (Gaffney et al., 1993). The NahG transgene has been shown to reduce SA accumulation in plants and abrogate SA-dependent defenses (Gaffney et al., 1993). Segregating plants from the (spr2 × NahG)F2 generation were screened by PCR to select four phenotypic bulks that varied in the presence or absence of NahG (NahG+ or NahG−) and of a functional copy of LeFAD7 (wild type or spr2). When all four bulks were challenged with the potato aphid, offspring production (Fig. 7A) and survival (Fig. 7B) were reduced on the spr2 single mutant bulk (spr2/NahG−) compared with the wild-type bulk (WT/NahG−), providing further evidence that aphid resistance in the spr2 mutant line is due to the presence of the mutation at the Lefad7 locus. Furthermore, in the double mutant bulk (spr2/NahG+), the presence of NahG compromised aphid resistance, restoring offspring production to wild-type levels (Fig. 7A) and significantly reducing offspring mortality (Fig. 7B). Li et al. (2006) have also shown that the NahG transgene causes a modest increase in aphid longevity in wild-type tomato, although overexpression of NahG in Arabidopsis did not alter short-term aphid population growth (Pegadaraju et al., 2005), possibly because of the shorter duration of the bioassay on Arabidopsis (2 d) compared with tomato (17 d). Infiltration of 1 mm catechol to spr2 did not influence aphid population growth or mortality (Supplemental Fig. S2), suggesting that the effects of NahG on aphids were due to SA depletion rather than catechol accumulation.
Figure 7.
Aphid resistance conferred by loss of function of FAD7 is compromised by the NahG transgene. The spr2 and NahG tomato lines were crossed, the F1 progeny were self-pollinated, and the F2 generation was screened by PCR for the presence or absence of the NahG transgene, the wild-type (WT) LeFAD7 allele, and the spr2 mutation in LeFAD7. Four phenotypic bulks were selected: (1) WT/NahG− = plants carrying at least one copy of the wild-type LeFAD7 allele and lacking the NahG transgene; (2) WT/NahG+ = plants carrying the wild-type LeFAD7 allele and NahG; (3) spr2/NahG− = plants homozygous for the spr2 mutation in LeFAD7 but lacking NahG; and (4) spr2/NahG+ = double mutant plants homozygous for the spr2 mutation and carrying NahG. All four bulks were inoculated with potato aphids (five aphids per cage; three cages per plant; 14–17 plants per bulk), and 6 d after inoculation, total offspring (dead and alive) were counted to measure adult fecundity (A) and offspring survival (B). One day after the aphids were counted, total, free, and bound SA content was measured in six randomly selected samples per bulk (C). The average number of total, dead, and living aphids per cage per plant was Box Cox transformed (Box and Cox, 1964) to stabilize variances, all values were analyzed by one-way ANOVA, and means were separated using Student’s t tests. Bars of the same pattern ± se with different letters differ significantly at α = 0.05. Uppercase letters above the bars in C denote significant differences in total (free + bound) SA content. FW, Fresh weight.
The foliar fatty acid content of the four bulks was also compared by gas chromatography to confirm that this was not impacted by NahG. Consistent with previous reports (Li et al., 2003), all plants that were homozygous for the spr2 mutation had higher C16:2 and C18:2 fatty acid content and lower C16:3 and C18:3 content compared with plants that carried the wild-type FAD7 allele (Supplemental Fig. S3). The fatty acid profile of the double mutants [(spr2/NahG+)F4] was also equivalent to that of the spr2 single mutant plants [spr2 or (spr2/NahG−)F4], indicating that the observed differences in aphid resistance between these two bulks were not due to changes in fatty acid content. Therefore, the effects of NahG on aphid resistance in the spr2 background can be attributed to suppressed SA accumulation, which was confirmed by HPLC in all bulks that carried the NahG transgene (Fig. 7C).
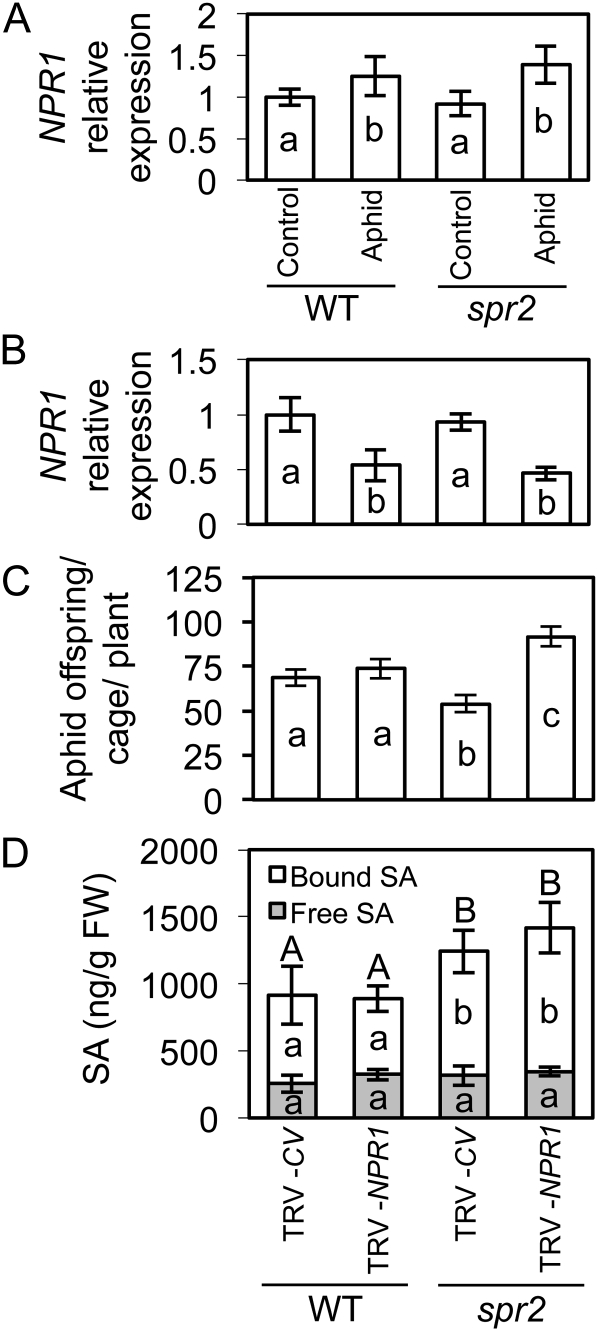
We also examined the contribution of a tomato ortholog of NPR1, a positive regulator of many SA-dependent defenses (Zhang et al., 1999; Dong, 2004). The NPR1 transcript was up-regulated by aphid feeding in both wild-type and spr2 plants (Fig. 8A). Virus-induced gene silencing (VIGS) was performed in both genotypes, and the ability of our silencing construct to suppress NPR1 transcript accumulation was confirmed by RT-qPCR (Fig. 8B). Whereas silencing of NPR1 did not significantly influence aphid population growth on wild-type plants, aphid numbers were nearly 70% higher on spr2 plants that received the NPR1 silencing construct than on mutant plants infiltrated with the control vector (Fig. 8C). Total SA levels were higher in spr2 than in wild-type plants but were not significantly altered by silencing of NPR1 (Fig. 8D); therefore, we hypothesize that silencing NPR1 compromised aphid resistance by suppressing defenses downstream of this regulator.
Figure 8.
NPR1 is up-regulated by aphid feeding and contributes to aphid resistance in the spr2 mutant. A, Expression of the NPR1 gene was measured 48 h after inoculation in spr2 and wild-type (WT; cv Castlemart) plants infested with aphids by RT-qPCR using RPL2 as the reference gene. B to D, VIGS using TRV was performed to suppress the expression of NPR1 in tomato, and a construct of similar size that does not silence any endogenous genes in tomato was used as a control vector (TRV-CV). Silencing of NPR1 was corroborated by RT-qPCR using RPL2 as the reference gene (B). Plants were challenged with the potato aphid (four aphids per cage; four cages per plant; eight plants per treatment group), and the total number of live adults and offspring was recorded 6 d after inoculation (C). Local total, free, and bound SA was measured 1 d after aphid count (D). Values were analyzed by ANOVA, and mean separations were performed using Student’s t test. Values ± se with different letters are statistically different at α = 0.05. Uppercase letters above bars in C denote significant differences in total (free + bound) SA content. (n = 7, 8, 6, and 8 respectively). FW, Fresh weight.
DISCUSSION
Although JA mediates induced resistance against many chewing insects and cell-content feeders, our results suggest that, in tomato, JA does not contribute to antibiotic defenses against a phloem feeder, the potato aphid. Mutations that block JA synthesis (spr2 and acx1) or perception (jai1-1) in tomato fail to enhance aphid population growth (Fig. 2), despite the fact that these mutations improve host suitability for other herbivores (Li et al., 2003, 2004, 2005). Instead, the mutant line spr2 reduces the settling behavior, survival, and fecundity of the potato aphid. This contrasts sharply with a prior report that oviposition by another phloem-feeding insect, Bemisia tabaci, is enhanced on spr2 (Sánchez-Hernández et al., 2006). Our results support prior assertions that plant responses to whiteflies and aphids differ (Kempema et al., 2007) and indicate that mechanisms of effective basal host plant resistance may vary even within a single feeding guild of insects such as phloem feeders.
Several lines of evidence establish that aphid resistance is due to the loss of function of FAD7, independent of the host genetic background. We observed the same aphid-resistant phenotype in the FAD7 antisense suppression line generated in tomato cv L402 (Fig. 3A) and in the Arabidopsis FAD mutants (Fig. 3B) that we observed in the spr2 line, which was developed by chemical mutagenesis in tomato cv Castlemart (spr2). Moreover, the spr2 aphid-resistant phenotype was recovered in the segregating tomato (spr2 × NahG)F2 population when plants were selected based on the presence or absence of the fad7 mutation (Fig. 7, A and B), directly linking the loss of function of FAD7 to aphid resistance. Aphid resistance in spr2 does not appear to be the result of impaired JA signaling, since aphid performance did not change in the jai1-1 or acx1 mutant and application of MeJA did not compromise resistance in spr2 (Fig. 2). The presence of aphid resistance in the Arabidopsis Atfad7-1 and Atfad7-2 mutants also lends strong support for the hypothesis that the impact of FAD7 on aphids is not mediated by JA. At 22°C to 23°C, these mutants retain approximately 50% to 75% of wild-type levels of C18:3 (Browse et al., 1986; McConn et al., 1994; McConn and Browse, 1996), which is likely in excess of the amount needed to produce JA in response to stress; moreover, the absence of male sterility in these lines also suggests the ability to synthesize JA (McConn and Browse, 1996). Together, these findings demonstrate that FAD7 inhibits aphid resistance in both tomato and Arabidopsis and that its impact on aphids is likely independent of JA.
Aphid resistance in plants with impaired FAD7 function could potentially be due to altered fatty acid metabolism. The most abundant fatty acids in tomato foliage are C18:3, C18:2, C16:3, and C16:0 fatty acids, in descending order of abundance. Compared with wild-type plants, the spr2 mutant is characterized by high C18:2 (approximately four times the wild-type level), very low C18:3 (less than 10% of the wild-type level), and undetectable C16:3 levels as well by slight increases in C18:1 and C16:2 fatty acids (Supplemental Fig. S3; Li et al., 2003). Potentially, aphid resistance in spr2 may be due to (1) an increase in oleic acid (C18:1) or its derivatives; (2) an increase in dienoic fatty acids (primarily C18:2) and their derivatives; or (3) a decrease in trienoic fatty acids (C18:3 and C16:3) and their products. It is unlikely that these changes would have direct nutritional consequences for aphids, because phloem sap appears to contain primarily C16:0 rather than dienoic or trienoic fatty acids (Madey et al., 2002) and aphids can survive and reproduce on artificial diets entirely lacking in fatty acids (Douglas and Simpson, 2003). However, fatty acids have been proposed to participate in defense signaling either directly or indirectly (Kachroo and Kachroo, 2009) and are also precursors for the synthesis of azelaic acid and numerous oxylipins that contribute to plant immunity (Blée, 2002; Jung et al., 2009). For example, a study in potato demonstrated that aphids strongly induce the production of 9-hydroperoxy-octadecadienoic acid, a derivative of linoleic acid synthesized by 9-lipoxygenases (Gosset et al., 2009). Potentially, increases in 9-hydroperoxy-octadecadienoic acid or other oxylipin derivatives of linoleic acid may contribute to aphid resistance in spr2. Alternatively, fatty acid desaturation could influence plant defenses against aphids by altering the composition of the plant’s cuticle, which has recently been shown to play a role in defense signaling (Kachroo and Kachroo, 2009; Xia et al., 2009, 2010).
Another possibility to consider is that FAD7 enzymatic activity might influence aphid host selection behavior or performance through its influence on plant volatile profiles. Emission of volatile terpenes is more than 2-fold lower in spr2 than in wild-type plants, probably as a result of reduced JA levels (Sánchez-Hernández et al., 2006). Typically, terpenoids have repellent effects on aphids and other insects (Aharoni et al., 2003; Bleeker et al., 2009), but it is conceivable that one or more terpenes that are reduced in spr2 could contribute to aphid attraction. The spr2 mutation in tomato also alters the profile of C6 volatile organic compounds (VOCs) generated from linoleic acid (C18:2) and linolenic acid (C18:3) via the hydroperoxide lyase pathway (HPL), resulting in a dramatic increase in hexanal and hexanol production and a decrease in (Z)-3-hexenal and (Z)-3-hexanol (Canoles et al., 2006; Sánchez-Hernández et al., 2006). Similar shifts in volatile profiles were also observed in Atfad7 Arabidopsis plants (Zhuang et al., 1996), although overall production of C6 volatiles is reported to be extremely low in the Columbia ecotype (Duan et al., 2005; Chehab et al., 2008). HPL appears to contribute to aphid resistance in potato (Vancanneyt et al., 2001), and several VOCs have direct antibiotic effects on aphids in vitro (Hildebrand et al., 1993). However, several lines of evidence suggest that HPL-derived VOCs are not essential to aphid resistance in plants with impaired FAD7 function. In Arabidopsis, we observed aphid resistance in Atfad7 mutants developed in a Columbia background (Fig. 3B), even though this ecotype carries a mutation that inhibits HPL activity and C6 volatile production (Duan et al., 2005). Overexpression of HPL in Arabidopsis also does not alter the host preference, population increase, or weight gain of the green peach aphid, even though it results in a more than 40-fold increase in C6 volatile production (Chehab et al., 2008). Furthermore, in tomato, antisense suppression of lipoxygenase C had no detectable effect on aphid host acceptance or population growth, despite dramatic reductions in VOC emissions (H. Klee and F. Goggin, unpublished data). Therefore, it is unlikely that plant volatile profiles alone are responsible for the effects of FAD7 on aphids.
While inhibition of FAD7 enzymatic activity clearly alters the production of C6 volatiles and many other fatty acid-derived compounds with roles in signaling and defense, it is also possible that the effects of FAD7 on aphid resistance may be independent of fatty acid metabolism. This would be consistent with the fact that Atfad7-2 and Atfad7-1fad8-1 confer aphid resistance even though these mutations cause relatively modest changes in fatty acid content at moderate temperatures (Browse et al., 1986; McConn and Browse, 1996). Instead, the FAD7 protein itself may influence aphid resistance, possibly through interactions with other chloroplast-localized proteins.
Whether through its desaturase activity or through other protein functions, our data indicate that wild-type FAD7 suppresses local SA-dependent defenses against aphids in tomato. Aphid resistance in the tomato spr2 mutant is associated with higher than normal levels of local SA (Fig. 6, C and D) and mRNA encoding the pathogenesis-related protein P4 (Fig. 6A) in aphid-infested foliage. The NahG transgene, which suppresses SA accumulation, restores wild-type levels of aphid susceptibility to spr2 (Fig. 7, A and B). Resistance in spr2 is also compromised when we utilize VIGS to suppress the expression of NPR1 (Fig. 8C). NPR1 is a key regulator of SA-dependent defenses (Zhang et al., 1999; Dong, 2004). Interestingly, silencing NPR1 in Nicotiana attenuata also reduces free fatty acid levels and thereby inhibits the induction of 13-hydroperoxy-octadecatrienoic acid and JA in response to wounded or simulated insect herbivory (Kallenbach et al., 2010). Further work is needed to explore the potential interaction between NPR1 on fatty acid metabolism. Additional studies are also necessary to determine if aphid resistance in Arabidopsis FAD mutants is also SA dependent, particularly in light of recent conflicting reports about the potential impacts of FAD7 on SA signaling in this species (Chaturvedi et al., 2008; Xia et al., 2010). However, our findings are consistent with other studies indicating that SA can contribute to plant defenses against aphids in other tomato genotypes. Suppression of SA accumulation increases aphid longevity in the wild-type cv Moneymaker and compromises aphid resistance in another cultivar (Motelle) that carries the Mi-1.2 aphid resistance gene (Li et al., 2006). Application of the SA analog benzothiadiazole also reduces aphid population growth on the Moneymaker cultivar (Cooper et al., 2004; Boughton et al., 2006; Li et al., 2006). Furthermore, tobacco mosaic virus infection reduces plant susceptibility to aphids in wild-type tomato but not in transgenic plants impaired in SA accumulation, which suggests that the SA-mediated defense responses against pathogens in tomato are also effective against aphids (Rodriguez-Saona et al., 2010).
There are several potential routes through which loss of function of FAD7 may enhance SA accumulation in tomato in response to aphid infestation. SA synthesis from chorismate occurs in the plastid (Wildermuth et al., 2001), where FAD7 is also localized; thus, it is possible that the FAD7 protein or a metabolite whose abundance is affected by FAD7 activity modulates a plastid component involved in SA biosynthesis. Although aphid resistance appears to be independent of JA itself, it is also conceivable that SA signaling might be enhanced by a decrease in the abundance of intermediates in JA synthesis [e.g. 13-hydroperoxy-octadecatrienoic acid, 12,13-epoxy-9,11,15-octadecatrienoic acid, 2-oxo-phytodienoic acid, or 3-oxo-2(2′(z)-pentenyl)-cyclopentane-1-octanoic acid 8; Fig. 1], because the SA and JA pathways can interact antagonistically under certain conditions (Bostock, 2005). Alternatively, FAD7 could potentially influence SA signaling indirectly by influencing the accumulation of reactive oxygen species (Yaeno et al., 2004; Mène-Saffrané et al., 2009). Mène-Saffrané et al. (2009) propose that trienoic fatty acids serve as important antioxidants and that constitutive SA levels are enhanced in the Arabidopsis Atfad3fad7fad8 mutant as a result of increased accumulation of reactive oxygen species. On the other hand, Yaeno and coworkers (2004) propose that linolenic acid promotes the reactive oxygen burst in response to pathogens by activating NADPH oxidase and that decreased linolenic acid levels in the Atfad7fad8 mutant result in decreased accumulation of hydrogen peroxide and superoxide in response to P. syringae. Clearly, further work is needed to investigate how ω-3 FADs impact reactive oxygen species accumulation and how this, in turn, may influence SA signaling.
CONCLUSION
Plants must defend themselves against a broad array of pests, including insect herbivores that utilize a diversity of feeding strategies to exploit their hosts. Plant defenses vary with the nature of the attacker and are coordinated by a highly conserved group of plant hormones, including JA and SA. Our study demonstrates that loss of function of FAD7 in the spr2 mutant in tomato enhances basal resistance to the potato aphid, a phloem-feeding herbivore with piercing-sucking mouth parts. Although JA synthesis and JA-dependent defenses against caterpillars and whiteflies are compromised in this mutant, decreased JA levels in these plants are not directly responsible for aphid resistance. Instead, resistance is linked to enhanced SA accumulation and requires SA signaling mediated by the NPR1 gene. Thus, loss of function of FAD7 enhances defenses against aphids and represses defenses against other insects through independent effects on SA and JA signaling. Our results provide novel insights into the contribution of FADs to plant defenses against aphids and to tradeoffs in resistance to different insects between and within feeding guilds.
MATERIALS AND METHODS
Plant and Insect Materials
Eight tomato (Solanum lycopersicum) genotypes were used in this study: the mutant lines spr2, jai1-1, and acx1 and the corresponding wild-type background cv Castlemart; the transgenic line Lefad7-antisense and the untransformed control cv L402; and NahG and its untransformed wild-type control, cv Moneymaker. In addition, crosses were performed between spr2 and NahG (described further below). All genotypes were grown in LC1 Sunshine potting mix (Sungro Horticulture) supplemented with 15-9-12 Osmocote Plus slow-release fertilizer (Scotts-MiracleGro). The NahG transgenic line was kindly provided by Dr. Jonathan Jones (Sainsbury Laboratory). Plants were maintained under stable greenhouse conditions (approximately 21°C–27°C, 16-h/8-h light/dark photoperiod) and watered with a dilute nutrient solution containing 1,000 μL L−1 CaNO3 (Hydro Agri North America), 500 μL L−1 MgSO4 (Giles Chemical), and 500 μL L−1 4-18-38 Gromore fertilizer (Gromore). Arabidopsis (Arabidopsis thaliana) ecotype Columbia (CS60000) and mutants Atfad7-2 (CS8042) and Atfad7-1fad8-1 (CS8036) were obtained from the Arabidopsis Biological Resource Center. The Atfad7gl1-1 and Atgl1-1 mutants were kindly donated by Dr. Jyoti Shah (University of North Texas). The plants were maintained in a Conviron growth chamber (Controlled Environments; 23°C, 65% relative humidity, 16-h/8-h light/dark photoperiod) and grown in LC1 Sunshine potting mix supplemented with 15-9-12 Osmocote Plus fertilizer, fertilized weekly with 24-8-16 MiracleGro all-purpose plant food (Scotts-MiracleGro). The potato aphid (Macrosiphum euphorbiae) was maintained in Conviron growth chambers (20°C, 16-h/8-h light/dark photoperiod) on a combination of tomato seedlings (cv UC82), potato (Solanum tuberosum), and jimson weed (Datura stramonium). The green peach aphid (Myzus persicae) was maintained at room temperature (approximately 23°C, 16-h/8-h light/dark photoperiod) on cabbage (Brassica oleracea var Capitata) seedlings.
JA Treatment
Wild-type tomato (cv Castlemart) and spr2 plants each were sprayed with 75 μm MeJA or water and covered with a plastic bag for 2 h to allow MeJA penetration. Twenty-four hours after treatment, 10 plants per treatment group were used to measure population growth of the potato aphid (described below), and tissue from three additional plants was collected to confirm the induction of JA-dependent defenses through semiquantitative RT-PCR analysis of PI-II expression (see below).
Gene Expression Analysis
Semiquantitative RT-PCR
To analyze PI-II expression in MeJA-treated plants, leaf samples were flash frozen in liquid nitrogen 24 h after treatment and stored at −80°C until RNA extraction. Total RNA was extracted from each leaf sample using TRIzol reagent (Invitrogen), and RNA concentration and quality were assessed using a Nanodrop spectrophotometer (Thermo Scientific). Total RNA was DNase treated with TURBO DNA-free (Ambion) followed by RT of 0.5 μg of RNA using oligo(dT)18 primers and SuperScript II reverse transcriptase (Invitrogen) in a 20-μL reaction volume. Semiquantitative PCR was performed using 50 ng of cDNA as template and 0.2 μm final concentration of each primer under the following conditions: a 5-min initial denaturation at 95°C; 22 amplification cycles (denaturation at 95°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 45 s); and a final extension at 72°C for 5 min using GoTaq green master mix (Promega). The endogenous, constitutively expressed gene Ribosomal Protein L2 (RPL2) was used as a loading control. Primers were PI-II (GenBank accession no. AY129402 for mRNA sequence) forward (5′-CCCACGTTCAGAAGGAAGTC-3′) and reverse (5′-TGAACGGGGACATCTTGAAT-3′) and RPL2 (GenBank accession no. X64562) forward (5′-GAGGGCGTACTGAGAAACCA-3′) and reverse (5′-CTTTTGTCCAGGAGGTGCAT-3′). PCR products were visualized on a 1% agarose gel stained with GelRed dye (Biotium).
RT-qPCR
To measure the impact of potato aphid infestation on P4, PI-II, and NPR1 expression, wild-type (cv Castlemart) and spr2 plants were inoculated with aphids, which were confined to a single leaf on each plant using large organza sleeve cages (100 aphids per cage). Control plants were mock inoculated with empty cages, and leaf tissue was collected 48 h after inoculation (four plants per treatment group). NPR1 expression was also analyzed in a second set of plants with a lower inoculum level (25 aphids per cage; three plants per treatment group), and because NPR1 expression did not differ between the two inoculum levels, data from these two sets of plants were pooled for analysis (seven plants per treatment group). RNA extraction and RT were performed as described above. The RT reaction products were diluted to 40 μL, and a 2-μL aliquot (a 25-ng RNA equivalent) was used as the template for real-time PCR. For each of the four biological replicates, two technical replicates were included in the quantitative PCR experiments. Mock reactions lacking reverse transcriptase were also included for each RNA sample to confirm the absence of DNA contamination. Real-time qPCR was carried out using the QuantiTect SYBR Green PCR kit (Qiagen) scaled for a 20-μL reaction volume, with final primer concentrations of 0.5 μm. The Mx3000P real-time PCR system (Stratagene) was used for PCR and fluorescence detection. The PCR conditions were as follows: 15 min of activation at 95°C; 40 amplification cycles (denaturation at 94°C for 15s, annealing at 59°C for 30s, and extension at 72°C for 30s); and a final data acquisition step to generate disassociation curves (95°C for 1 min and 55°C for 30s). Dissociation curves were examined for all samples to confirm that each primer set generated a single amplification product. Primer pairs used for RT-qPCR were as follows: P4 (GenBank accession no. M69247 for mRNA sequence) forward (5′-CAACTCAAGAGCGGGTAGTTG-3′) and reverse (5′-CCACACATTTTTCCACCAACAC-3′) and LeNPR1 (GenBank accession no. AY640378.1) forward (5′-CTCCAGGCGGTAAGGAAA-3′) and reverse (5′-CAAATAGGCGAGCACACTGA-3′); see above for RPL2 and PI-II primer sets. To estimate the efficiency of amplification for each of these primer sets, RT-qPCR was performed on serial dilutions of a set of cDNA standards, and the PCR efficiencies were calculated using the E = 10[−1/Ct slope] methodology (Rasmussen, 2000). Relative gene expression was calculated using the methodology of Pfaffl (2001). Data for our genes of interest were normalized to the expression levels of RPL2, and relative gene expression for each treatment group was calculated relative to the untreated wild-type control group in each experiment. For statistical analysis, the relative expression values for each treatment group were log2 transformed to stabilize variances. Data were analyzed by two-way ANOVA, and means for significant effects at α = 0.05 were separated using Student’s t test with JMP version 8.0 (SAS Institute).
SA Quantification
To measure the influence of potato aphid infestation on SA accumulation, spr2 and wild-type (cv Castlemart) tomato plants were challenged with potato aphids by introducing 60 aphids into an organza sleeve cage enclosing the terminal three leaflets of a single leaf, selected from the sixth or seventh node position up from the oldest true leaf (one cage per plant; five plants per treatment group per time point). Control plants were mock inoculated with empty cages. The inoculated leaflets (approximately 250 g of tissue per plant) were collected 24 and 48 h after inoculation (local tissues), and a similar amount of tissue was also sampled from an upper unwounded leaf (eighth or ninth node) of each plant to measure the systemic SA accumulation. SA was extracted and quantified by HPLC as described previously (Branch et al., 2004). In brief, SA was measured using an Agilent 1100 HPLC device with fluorometric detection. The column was a 4.6- × 75-mm Agilent RR XDB C18 used with an isocratic mobile phase composed of 75% 20 mm formate, pH 3.8, 20% methanol, and 5% acetonitrile at a flow rate of 0.75 mL min−1 at 35°C. Bound SA was measured after converting to free SA by acid hydrolysis. Recovery rates were determined using o-anisic acid as an internal standard and were typically greater than 60%. Free and bound SA levels were analyzed using three-way ANOVA, and means separations were performed using Student’s t test with JMP version 8.0.
Development and Characterization of Tomato spr2/NahG Double Mutants
Crosses were performed using spr2 as the maternal parent and NahG as the pollen donor. The spr2 flowers were emasculated 1 d prior to anthesis and crossed manually with NahG pollen the next day. Then, the (spr2 × NahG)F1 hybrid plants were self-pollinated to obtain the (spr2 × NahG)F2 population. Since the spr2 mutation in LeFAD7 and the NahG transgene were found in different genetic backgrounds (cv Castlemart and cv Moneymaker, respectively), a modified bulked segregant analysis approach was performed on the F2 generation to determine the effects of NahG and the spr2 mutation on SA levels and aphid resistance. This method compensates for segregation at other loci and is a rapid alternative to the development of near-isogenic lines (Michelmore et al., 1991). Segregating plants from the (spr2 × NahG)F2 generation were screened by PCR for the presence or absence of the wild-type LeFAD7 allele, the spr2 mutation, and the NahG transgene. Four phenotypic bulks of at least 14 plants each were selected: (1) plants carrying at least one copy of the wild-type LeFAD7 allele and lacking the NahG transgene; (2) plants carrying the wild-type LeFAD7 allele and NahG; (3) plants homozygous for the spr2 mutation in LeFAD7 but lacking NahG; and (4) double mutant plants homozygous for the spr2 mutation and carrying NahG. Since plants within each phenotypic bulk had the same genotype at the LeFAD7 and NahG loci but had random variations at other unlinked loci, comparisons among bulks eliminate the potential effects of genetic background, and differences in aphid resistance and SA levels among bulks could be attributed to LeFAD7, NahG, and the interaction between these genes. Approximately 400 (spr2 × NahG)F2 plants were PCR screened for the presence of the LeFAD7 wild-type allele or the spr2 mutation using single-nucleotide polymorphism primers (wild-type LeFAD7 allele, forward [5′-ATATTGGGCGGAGATGTGAA-3′] and reverse [5′-AACCACATTCTGATAGAACC-3′]; spr2 mutation, forward [5′-CTAACTAAAATGGCAAGTTGA-3′] and reverse [5′-TACCCTCAATGCCCAACAAT-3′]). Then, selected plants were PCR screened for the presence of the NahG transgene (forward [5′-GTAGCCATGTGCTGGAAGGT-3′] and reverse [5′-CCTCACTGGAAAGGTGAGGA-3′]). DNA was isolated using the REDExtract-N-Amp plant PCR kit (Sigma), and touchdown PCR (Korbie and Mattick, 2008) was performed to increase amplification sensitivity and specificity using the following conditions: initial denaturation = 95°C for 5 min; phase I = 95°C for 45 s, 65°C to 56°C for 45 s (reducing 1°C per cycle), and 72°C for 45 s; phase II = 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s (20 cycles); and final extension at 72°C for 5 min. PCR products were visualized on 1% agarose gels. For each of the four bulks selected through PCR screening, six plants were used for SA measurement as described above and 14 to 17 plants were used for an aphid bioassay (described below).
Silencing of NPR1
VIGS
The tobacco rattle virus (TRV) vector pYL156 was kindly provided by Dr. Dinesh-Kumar (Yale University). An insert of 414 bp corresponding to nucleotides 1,124 to 1,537 of the LeNPR1 gene (GenBank accession no. AY640378.1) was cloned (forward primer, 5′-ATATAGAATTCCTGCTCCAAAGGATCGGTTA-3′; reverse primer, 5′-ATATACTCGAGCAGACAAGTCATCAGCATCCA-3′) and inserted into pYL156 using EcoRI and XhoI sites. A construct (TRV-CV) carrying a 396-bp insert corresponding to nucleotides 544 to 939 of the GUS reporter gene (GenBank accession no. S69414.1) was used as a control vector (Hartl et al., 2008; Wu et al., 2011), and another construct that carries a 408-bp DNA fragment corresponding to nucleotides 1,175 to 1,583 of the Phytoene Desaturase (PDS) gene (GenBank accession no. M88683.1) was used as a visual reporter to monitor the onset of VIGS. The NPR1 silencing construct TRV-NPR1, TRV-CV, and TRV-PDS were introduced into 2-week-old spr2 and wild-type tomato plants by agroinfiltration as described by Wu et al. (2011) and maintained at 20°C/16 h of light. Plants were used for an insect bioassay (described below) 21 d after agroinfiltration, when widespread bleaching symptoms were observed in plants infiltrated with TRV-PDS. One day after the bioassay was scored, aphids were gently removed from the plants using paint brushes and leaf tissue was collected for RNA extraction and SA quantification by HPLC (performed as described above).
Confirmation of Gene Silencing
Leaf tissue for every cage was flash frozen in liquid nitrogen and stored at −80°C. RNA was isolated from eight randomly selected samples for each treatment group, and RT-qPCR was performed following the methods described above. Primers for gene expression analysis were designed so that they would not overlap with the inserts in the VIGS constructs (LeNPR1 forward [5′-CTCCAGGCGGTAAGGAAA-3′] and reverse [5′-CAAATAGGCGAGCACACTGA-3′]).
Aphid Performance Bioassays
Potato Aphid Performance on Tomato
All insect bioassays were performed when tomato plants were between 4 and 5 weeks old, and, unless otherwise specified, they were conducted in growth chambers (23°C, 16-h/8-h light/dark photoperiod). Four types of bioassays were used to measure different aspects of aphid performance. Uncaged population growth assays were conducted to compare aphid infestations on wild-type plants, spr2, jai1-1, and acx1 (Fig. 2, A and B). Each plant was inoculated with 15 aphids that were not confined to cages and were able to move from leaf to leaf on the same plant or to reject the host by dropping off the plant. Total aphid numbers per plant were counted 5 d after inoculation and reflected the net effects of the host plants on aphid host acceptance, survival, and fecundity. Clip-cage assays were also used to measure differences in short-term aphid population growth in response to MeJA treatment (Fig. 2C), antisense suppression of FAD7 (Fig. 3A), the separate and combined effects of spr2 and NahG (Fig. 7, A and B), and VIGS silencing of NPR1 (Fig. 8). A fixed number of young adult aphids were confined to a single leaflet using a clip cage (four to five per cage and three to four cages per plant; exact numbers for each assay are reported in the figure legends), and aphid reproduction and survival were assessed 6 d after inoculation. Values for the individual cages (subreplicates) were averaged to obtain single data points for each replicate plant. This assay design allowed comparison of aphid population growth in the absence of host choice. Aphid numbers for both of these assay types were analyzed by ANOVA, and where appropriate, mean separations were performed with Student’s t tests using JMP version 8.0. A third assay type allowed precise measurement of the longevity and daily offspring production of individual adult aphids on wild-type and spr2 plants (Fig. 5). Newly emerged wingless adults (less than 24 h within emergence to adulthood) were confined to clip cages (one aphid per cage; two subreplicate cages per plant), and every 24 h adult survival was monitored and offspring were counted and removed from the cages. Plants were maintained in a greenhouse (21°C–27°C, 16-h/8-h light/dark photoperiod) due to the large amount of space required for this assay and were monitored for 14 d, until all aphids on the spr2 genotype were dead. The average aphid life spans (days lived), total offspring production, and daily fecundity (total offspring production per days lived) were analyzed by one-way ANOVA. Additionally, regression analyses were performed to estimate the aphid daily survival rate and fecundity with JMP version 8.0. A fourth assay type was utilized to compare aphid host preference between wild-type and spr2 plants (Fig. 4). Choice tests were performed by placing 10 newly emerged wingless adult aphids on a Styrofoam choice arena (15 cm diameter) between paired leaflets (sixth or seventh node position) on intact wild-type and spr2 plants (10 replicate pairs). The leaflets were isolated from the rest of the plant by placing 15-cm round barriers made of glossy photo paper and coated with Tanglefoot tangle-trap insect trap coating (Contech Enterprises) around the base of the leaflet petiole (Fig. 4A). Aphids typically moved from the arena onto a leaflet within minutes of release and would then either remain on the leaflet and begin reproduction or move to the other leaflet via the arena. The number of aphids on each leaflet or remaining on the arena was counted at 1, 4, and 24 h after release and analyzed by paired t tests with JMP version 8.0. Aphids that were on the plastic arena between the plants or that crawled down the plastic platform without settling on a leaflet were not included in the analysis.
Green Peach Aphid Population Growth on Arabidopsis
Newly emerged wingless adults were confined to plastic sleeve cages that cover the entire plant (two aphids per plant; 18 plants per genotype) at growth stage 5.10 (Boyes et al., 2001). Seven days after infestation, the total number of aphids was counted and analyzed by one-way ANOVA. Means were separated using Student’s t test with JMP version 8.0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Loss of JA sensitivity in the jai1-1 mutant does not enhance the expression of SA-responsive gene P4 but suppresses the expression of the JA-responsive gene PI-II.
Supplemental Figure S2. Effect of catechol on aphid performance.
Supplemental Figure S3. Impact of spr2 and NahG on foliar fatty acid profiles.
Supplementary Material
Acknowledgments
We thank Dr. Dinesh Kumar and Dr. Kyle Willis for providing materials for VIGS, Dr. Jyoti Shah and Dr. Jonathan Jones for providing seeds, Dr. Burt Bluhm for providing facilities for gas chromatography, and John Guerber and Clinton Trammel for their help with greenhouse and growth chamber maintenance. We also thank our anonymous reviewers for helpful feedback.
References
- Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FWA, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ. (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich T, Harada M, Sugawara K, Kodama H, Iba K, Kusano T. (1998) Two maize genes encoding ω-3 fatty acid desaturase and their differential expression to temperature. Plant Mol Biol 36: 297–306 [DOI] [PubMed] [Google Scholar]
- Bhattarai KK, Xie Q-G, Pourshalimi D, Younglove T, Kaloshian I. (2007) Coil-dependent signaling pathway is not required for Mi-1-mediated potato aphid resistance. Mol Plant Microbe Interact 20: 276–282 [DOI] [PubMed] [Google Scholar]
- Blée E. (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7: 315–322 [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Diergaarde PJ, Ament K, Guerra J, Weidner M, Schütz S, de Both MTJ, Haring MA, Schuurink RC. (2009) The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol 151: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43: 545–580 [DOI] [PubMed] [Google Scholar]
- Boughton AJ, Hoover K, Felton GW. (2006) Impact of chemical elicitor applications on greenhouse tomato plants and population growth of the green peach aphid, Myzus persicae. Entomol Exp Appl 120: 175–188 [Google Scholar]
- Box GEP, Cox DR. (1964) An analysis of transformations. J Royal Stat Soc Series B Methodol 26: 211–252 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch C, Hwang CF, Navarre DA, Williamson VM. (2004) Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol Plant Microbe Interact 17: 351–356 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt P, Somerville C. (1986) A mutant of Arabidopsis deficient in c(18:3) and c(16:3) leaf lipids. Plant Physiol 81: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA, Martin JL, Pickett JA, Pye BJ, Smart LE, Wadhams LJ. (2003) cis-Jasmone treatment induces resistance in wheat plants against the grain aphid, Sitobion avenae (Fabricius) (Homoptera: Aphididae). Pest Manag Sci 59: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Chamberlain K, Woodcock CM, Mohib A, Webster B, Smart LE, Birkett MA, Pickett JA, Napier JA. (2008) cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic interactions with aphids and their parasitoids. Proc Natl Acad Sci USA 105: 4553–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoles MA, Beaudry RM, Li C, Howe G. (2006) Deficiency of linolenic acid in Lefad7 mutant tomato changes the volatile profile and sensory perception of disrupted leaf and fruit tissue. J Am Soc Hortic Sci 131: 284–289 [Google Scholar]
- Chao WS, Gu YQ, Pautot V, Bray EA, Walling LL. (1999) Leucine aminopeptidase RNAs, proteins, and activities increase in response to water deficit, salinity, and the wound signals systemin, methyl jasmonate, and abscisic acid. Plant Physiol 120: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J. (2008) Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, Dehesh K. (2008) Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS ONE 3: e1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. (2005a) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102: 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WC, Jia L, Goggin FL. (2004) Acquired and R-gene-mediated resistance against the potato aphid in tomato. J Chem Ecol 30: 2527–2542 [DOI] [PubMed] [Google Scholar]
- Cooper WR, Goggin FL. (2005) Effects of jasmonate-induced defenses in tomato on the potato aphid, Macrosiphum euphorbiae. Entomol Exp Appl 115: 107–115 [Google Scholar]
- de Vos M, Kim JH, Jander G. (2007) Biochemistry and molecular biology of Arabidopsis-aphid interactions. Bioessays 29: 871–883 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux J-P, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Douglas AE, Simpson SJ. (2003) The nutritional physiology of aphids. Adv Insect Physiol 31: 73–140
- Duan H, Huang MY, Palacio K, Schuler MA. (2005) Variations in CYP74B2 (hydroperoxide lyase) gene expression differentially affect hexenal signaling in the Columbia and Landsberg erecta ecotypes of Arabidopsis. Plant Physiol 139: 1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Turner JG. (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Johnson RR, Ryan CA. (1992) Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol 98: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidantsef AL, Stout MJ, Thaler JS, Duffey SS, Bostock RM. (1999) Signal interactions in pathogen and insect attack: expression of lipoxygenase, proteinase inhibitor II, and pathogenesis-related protein P4 in the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol 54: 97–114 [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Gao LL, Anderson JP, Klingler JP, Nair RM, Edwards OR, Singh KB. (2007) Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago truncatula. Mol Plant Microbe Interact 20: 82–93 [DOI] [PubMed] [Google Scholar]
- Gibson S, Arondel V, Iba K, Somerville C. (1994) Cloning of a temperature-regulated gene encoding a chloroplast ω-3 desaturase from Arabidopsis thaliana. Plant Physiol 106: 1615–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosset V, Harmel N, Göbel C, Francis F, Haubruge E, Wathelet J-P, du Jardin P, Feussner I, Fauconnier M-L. (2009) Attacks by a piercing-sucking insect (Myzus persicae Sultzer) or a chewing insect (Leptinotarsa decemlineata Say) on potato plants (Solanum tuberosum L.) induce differential changes in volatile compound release and oxylipin synthesis. J Exp Bot 60: 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Merker H, Schmidt DD, Baldwin IT. (2008) Optimized virus-induced gene silencing in Solanum nigrum reveals the defensive function of leucine aminopeptidase against herbivores and the shortcomings of empty vector controls. New Phytol 179: 356–365 [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Baldwin IT. (2004) Microarray analysis of salicylic acid- and jasmonic acid-signalling in responses of Nicotiana attenuata to attack by insects from multiple feeding guilds. Plant Cell Environ 27: 1362–1373 [Google Scholar]
- Hildebrand DF, Brown GC, Jackson DM, Hamilton-Kemp TR. (1993) Effects of some leaf-emitted volatile compounds on aphid population increase. J Chem Ecol 19: 1875–1887 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Ryan CA. (1999) Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITAG (2011) The official annotation for the tomato genome. International Tomato Annotation Group. http://solgenomics.net/organism/solanum_lycopersicum/genome (September 2, 2011)
- Jiang C-J, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H. (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Interact 22: 820–829 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. (2009) Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Fu D-Q, Havens W, Navarre D, Kachroo P, Ghabrial SA. (2008) An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol Plant Microbe Interact 21: 564–575 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P. (2009) Fatty acid-derived signals in plant defense. Annu Rev Phytopathol 47: 153–176 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Venugopal SC, Lapchyk L, Falcone D, Hildebrand D, Kachroo P. (2004) Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc Natl Acad Sci USA 101: 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. (2010) Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol 152: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempema LA, Cui X, Holzer FM, Walling LL. (2007) Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs: similarities and distinctions in responses to aphids. Plant Physiol 143: 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodakovskaya M, McAvoy R, Peters J, Wu H, Li Y. (2006) Enhanced cold tolerance in transgenic tobacco expressing a chloroplast ω-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta 223: 1090–1100 [DOI] [PubMed] [Google Scholar]
- Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K. (1994) Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3-fatty acid desaturase in transgenic tobacco. Plant Physiol 105: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie DJ, Mattick JS. (2008) Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc 3: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Kuśnierczyk A, Winge P, Jørstad TS, Troczyńska J, Rossiter JT, Bones AM. (2008) Towards global understanding of plant defence against aphids: timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ 31: 1097–1115 [DOI] [PubMed] [Google Scholar]
- Kusnierczyk A, Winge P, Midelfart H, Armbruster WS, Rossiter JT, Bones AM. (2007) Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae. J Exp Bot 58: 2537–2552 [DOI] [PubMed] [Google Scholar]
- Li C, Liu G, Xu C, Lee GI, Bauer P, Ling HQ, Ganal MW, Howe GA. (2003) The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15: 1646–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, et al. (2005) Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17: 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li C, Howe GA. (2001) Genetic analysis of wound signaling in tomato: evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol 127: 1414–1417 [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I. (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 19: 655–664 [DOI] [PubMed] [Google Scholar]
- Liu X, Yang J, Li B, Yang X, Meng Q. (2010) Antisense expression of tomato chloroplast ω-3-fatty acid desaturase gene (LeFAD7) enhances the tomato high-temperature tolerance through reductions of trienoic fatty acids and alterations of physiological parameters. Photosynthetica 48: 59–66 [Google Scholar]
- Liu X-Y, Yang J-H, Li B, Yang X-M, Meng Q-W. (2006) Antisense-mediated depletion of tomato chloroplast ω-3-fatty acid desaturase enhances thermal tolerance. J Integr Plant Biol 48: 1096–1107 [DOI] [PubMed] [Google Scholar]
- Louis J, Leung Q, Pegadaraju V, Reese J, Shah J. (2010) PAD4-dependent antibiosis contributes to the ssi2-conferred hyper-resistance to the green peach aphid. Mol Plant Microbe Interact 23: 618–627 [DOI] [PubMed] [Google Scholar]
- Madey E, Nowack LM, Thompson JE. (2002) Isolation and characterization of lipid in phloem sap of canola. Planta 214: 625–634 [DOI] [PubMed] [Google Scholar]
- Martín M, León J, Dammann C, Albar JP, Griffiths G, Sánchez-Serrano JJ. (1999) Antisense-mediated depletion of potato leaf ω3 fatty acid desaturase lowers linolenic acid content and reduces gene activation in response to wounding. Eur J Biochem 262: 283–290 [DOI] [PubMed] [Google Scholar]
- Martinez de Ilarduya O, Xie Q, Kaloshian I. (2003) Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol Plant Microbe Interact 16: 699–708 [DOI] [PubMed] [Google Scholar]
- McConn M, Browse J. (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Hugly S, Browse J, Somerville C. (1994) A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast ω-3-desaturase. Plant Physiol 106: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L, Dubugnon L, Chételat A, Stolz S, Gouhier-Darimont C, Farmer EE. (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J Biol Chem 284: 1702–1708 [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC. (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol 138: 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K. (2000) Trienoic fatty acids and plant tolerance of high temperature. Science 287: 476–479 [DOI] [PubMed] [Google Scholar]
- Nishiuchi T, Hamada T, Kodama H, Iba K. (1997) Wounding changes the spatial expression pattern of the Arabidopsis plastid ω-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell 9: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo N, Mitsuhara I, Koga M, Seo S, Ohashi Y. (1999) Ethylene promotes the necrotic lesion formation and basic PR gene expression in TMV-infected tobacco. Plant Cell Physiol 40: 808–817 [Google Scholar]
- Omer AD, Granett J, Karban R, Villa EM. (2001) Chemically-induced resistance against multiple pests in cotton. Int J Pest Manage 47: 49–54 [Google Scholar]
- Pegadaraju V, Knepper C, Reese J, Shah J. (2005) Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol 139: 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Rasmussen R. (2000) Quantification on the LightCycler. In S Meuer, C Wittwer, K Nakagawara, eds, Rapid Cycle Real-Time PCR: Methods and Applications. Springer, Heidelberg, pp 21–34
- Rodriguez-Saona CR, Musser RO, Vogel H, Hum-Musser SM, Thaler JS. (2010) Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds. J Chem Ecol 36: 1043–1057 [DOI] [PubMed] [Google Scholar]
- Sánchez-Hernández C, López MG, Délano-Frier JP. (2006) Reduced levels of volatile emissions in jasmonate-deficient spr2 tomato mutants favour oviposition by insect herbivores. Plant Cell Environ 29: 546–557 [DOI] [PubMed] [Google Scholar]
- Schaller F. (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11–23 [PubMed] [Google Scholar]
- Schuhegger R, Ihring A, Gantner S, Bahnweg G, Knappe C, Vogg G, Hutzler P, Schmid M, Van Breusegem F, Eberl L, et al. (2006) Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ 29: 909–918 [DOI] [PubMed] [Google Scholar]
- Schultz DJ, Cahoon EB, Shanklin J, Craig R, Cox-Foster DL, Mumma RO, Medford JI. (1996) Expression of a delta 9 14:0-acyl carrier protein fatty acid desaturase gene is necessary for the production of ω 5 anacardic acids found in pest-resistant geranium (Pelargonium × hortorum). Proc Natl Acad Sci USA 93: 8771–8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Nandi A, Klessig DF. (2001) A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J 25: 563–574 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Fu DQ, El-Habbak M, Navarre D, Ghabrial S, Kachroo A. (2011) Silencing genes encoding omega-3 fatty acid desaturase alters seed size and accumulation of Bean pod mottle virus in soybean. Mol Plant Microbe Interact 24: 506–515 [DOI] [PubMed] [Google Scholar]
- Song J, Lee DE, Jung S, Kim H, Han O, Cho BH, Lee IJ, Back K. (2004) Characterization of transgenic rice plants expressing an Arabidopsis FAD7. Biol Plant 48: 361–366 [Google Scholar]
- Thompson GA, Goggin FL. (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57: 755–766 [DOI] [PubMed] [Google Scholar]
- Upchurch RG. (2008) Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett 30: 967–977 [DOI] [PubMed] [Google Scholar]
- Vancanneyt G, Sanz C, Farmaki T, Paneque M, Ortego F, Castañera P, Sánchez-Serrano JJ. (2001) Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc Natl Acad Sci USA 98: 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kan JAL, Cozijnsen T, Danhash N, De Wit PJ. (1995) Induction of tomato stress protein mRNAs by ethephon, 2,6-dichloroisonicotinic acid and salicylate. Plant Mol Biol 27: 1205–1213 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Walling LL. (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ming F, Pittman J, Han Y, Hu J, Guo B, Shen D. (2006) Characterization of a rice (Oryza sativa L.) gene encoding a temperature-dependent chloroplast omega-3 fatty acid desaturase. Biochem Biophys Res Commun 340: 1209–1216 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wu C, Jia L, Goggin FL. (2011) The reliability of virus-induced gene silencing experiments using tobacco rattle virus in tomato is influenced by the size of the vector control. Mol Plant Pathol 12: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Gao QM, Yu K, Lapchyk L, Navarre D, Hildebrand D, Kachroo A, Kachroo P. (2009) An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5: 151–165 [DOI] [PubMed] [Google Scholar]
- Xia Y, Yu K, Navarre DA, Seebold K, Kachroo A, Kachroo P. (2010) The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol 154: 833–846 [DOI] [PMC free article] [PubMed]
- Yaeno T, Matsuda O, Iba K. (2004) Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J 40: 931–941 [DOI] [PubMed] [Google Scholar]
- Yara A, Yaeno T, Hasegawa M, Seto H, Montillet J-L, Kusumi K, Seo S, Iba K. (2007) Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of omega-3 fatty acid desaturases. Plant Cell Physiol 48: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Yu C, Wang H-S, Yang S, Tang X-F, Duan M, Meng Q-W. (2009) Overexpression of endoplasmic reticulum omega-3 fatty acid desaturase gene improves chilling tolerance in tomato. Plant Physiol Biochem 47: 1102–1112 [DOI] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PJ, Zheng SJ, van Loon JJA, Boland W, David A, Mumm R, Dicke M. (2009) Whiteflies interfere with indirect plant defense against spider mites in Lima bean. Proc Natl Acad Sci USA 106: 21202–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Fan WH, Kinkema M, Li X, Dong XN. (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang M-J, Zhao S-T, Hu J-J, Lu M-Z. (2010) Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgenic Res 19: 647–654 [DOI] [PubMed] [Google Scholar]
- Zhuang H, Hamilton-Kemp TR, Andersen RA, Hildebrand DF. (1996) The impact of alteration of polyunsaturated fatty acid levels on C6-aldehyde formation of Arabidopsis thaliana leaves. Plant Physiol 111: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn J-E, Koiwa H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.