Abstract
While much research has been directed to harnessing the antimicrobial properties of exogenous NO, the possibility of bacteria developing resistance to such therapy has not been thoroughly studied. Herein, we evaluate potential NO resistance using spontaneous and serial passage mutagenesis assays. Specifically, Staphylococcus aureus, Methicillin-resistant S. aureus (MRSA), Staphylococcus epidermidis, Escherichia coli, and Pseudomonas aeruginosa were systematically exposed to NO-releasing 75mol% MPTMS-TEOS nitrosothiol particles at or below minimum inhibitory concentration (MIC) levels. In the spontaneous mutagenesis assay, bacteria that survived exposure to lethal concentrations of NO showed no increase in MIC. Similarly, no increase in MIC was observed in the serial passage mutagenesis assay after exposure of these species to sub-inhibitory concentrations of NO through 20 d.
Keywords: antimicrobial resistance, nitric oxide, resistance, spontaneous mutagenesis, serial passage mutagenesis
1.1 Introduction
Nitric oxide (NO) is an endogenous diatomic free radical implicated in several physiological processes including vasodilation, immune response, neurotransmission, and wound healing.[1] During infection, NO is released by macrophages and other immune cells at >1 μM concentrations where it serves as a broad spectrum biocidal agent.[1,2,3,4,5,6] Nitric oxide induces both nitrosative and oxidative stress that results in numerous toxic effects on bacteria, including direct modification of membrane proteins, lipid peroxidation, and DNA cleavage.[1,6,7,8] As such, the exogenous application of NO as a therapy has been the subject of intense interest during the past decade.[9,10,11,12,13,14,15]
Controlled NO storage and delivery using chemical NO donors has led to several pharmacological applications.[16] Example antimicrobial NO delivery vehicles include low molecular weight compounds (e.g., sodium nitroprusside, N-diazeniumdiolated proline, and S-nitroso-N-acetylpenicillamine),[17,18,19] macromolecular vehicles,[14,20,21,22,23,24] and polymeric coatings.[10,25,26,27,28,29,30,31,32,33] We have previously reported the bactericidal activity of NO-releasing silica nanoparticles and sol-gel-derived xerogel films against Pseudomonas aeruginosa at concentrations of minimal toxicity to mammalian cells.[11,13]
It is known that bacteria possess mechanisms for reducing the pharmacological effects of drugs such as antibiotics by directly removing the drug (i.e., efflux pumps), reduced drug diffusion via porin loss or modification, overproduction or alterations of drug target sites, or enzymatic drug degradation.[34,35,36,37,38,39,40] For example, Charrel et al. reported that some β-lactam antibiotic-resistant Enterobacter aerogenes were porin deficient, resulting in a high MIC for β-lactam even in the absence of increased β-lactamase production.[41] Recent research also indicates that select bacteria are capable of up-regulating NO scavengers[42,43,44,45,46] and/or altering respiration in response to endogenous NO.[47] An example is NO detoxification by flavohemoglobin, a protein that is up-regulated in E. coli in response to macrophage-produced NO.[45] Endogenous thiols such as mycothiol, a glutathione analog produce by mycobacteria, have also been shown to reduce the toxicity of NO and other oxygen species.[48,49] Enzymes including reductases and superoxide dismutase have been implicated to serve similar functions.[45,50] With respect to cellular respiration, Husain et al. reported arrested respiration in Salmonella with concomitant accumulation of nicotinamide adenine dinucleotide (NADH), thereby increasing the ability of the bacteria to resist oxidative stress.[47]
While the antimicrobial action of NO-releasing materials is established,[11,12,13,29,51,52,53] knowledge about the bacterial resistance to exogenous concentrations of NO remains scarce.[42,43,44,45,46,47,54] Miller et al. reported that S. aureus was not capable of developing resistance to exogenous gaseous NO; however, NO exposure was intermittent with discontinuous selective pressure against the NO-susceptible bacteria.[15,55,56,57] Herein, we report a thorough bacterial resistance study using both spontaneous mutation and serial passage mutagenesis assays with continuous exposure to physiologically relevant concentrations of NO from NO-releasing silica nanoparticles. Representative gram positive and gram negative bacteria were selected to provide preliminary resistance information as a function of bacteria classification and structure.
1.2 Material and Methods
1.2.1 Strains, media, and chemical reagents
3-Mercaptopropyltrimethoxysilane (MPTMS) and tetraethoxysilane (TEOS) were purchased from Gelest (Tullytown, PA). Bacteria were propagated at 37 °C in tryptic soy broth (TSB) and agar (TSA, Becton, Dickinson, Franklin Lakes, NJ). Sodium chloride, potassium chloride, sodium phosphate monobasic, methanol, ethanol, ammonium hydroxide, and hydrochloric acid were obtained from Fisher Scientific (Pittsburgh, PA). Sodium phosphate dibasic and sodium nitrite were obtained from Sigma Aldrich (St. Louis, MO). Escherichia coli O157:H7 (35150), Pseudomonas aeruginosa (19143), methicillin-susceptible Staphylococcus aureus (MSSA) (29213), methicillin-resistant Staphylococcus aureus (MRSA) (33591), and Staphylococcus epidermidis (35983) were obtained from American Type Culture Collection (ATCC, Manassas, VA). Distilled water was purified to 18.2 MΩ·cm with a Millipore Milli-Q Gradient A-10 water purification system (Bedford, MA).
1.2.2 Synthesis of mercaptosilane-based silica particles
Nitrosothiol particles (75 mol% MPTMS/TEOS) were synthesized following a procedure reported previously.[21] Briefly, 3-mercaptopropyltrimethoxysilane (MPTMS, 424 μL) and tetraethoxysilane (TEOS, 169 μL) were mixed and added dropwise via a Kent Scientific Genie Plus syringe pump at a flow rate of 0.5 mL min−1 through an 18.5 gauge needle to a solution of ethanol (16.3 mL), water (1.4 mL), and ammonium hydroxide (11 mL). The reaction was stirred for 2 h at room temperature and the particles collected by centrifugation at 3645g (10 min), washed twice with 40 mL EtOH, recollected, and dried overnight at ambient conditions.
1.2.3 Nitrosation of mercaptosilane-based silica particles
Thiols within the particles were nitrosated upon reaction with nitrous acid as follows. Particles (~200 mg) were first added to 4 mL methanol (MeOH). While stirring, 2 mL of hydrochloric acid (5 M) was added to the suspension. A 2 mL aqueous solution containing sodium nitrite (2× molar excess to thiol) and DTPA (500 μM) was then added to the particle suspension, and the mixture was stirred for 2 h in the dark on ice. Particles were collected by centrifugation at 3645g (5 min), washed with 40 mL chilled 500 μM DTPA(aq), recollected, washed with 40 mL chilled MeOH, recollected, and vacuum dried in the dark for 30 min. Particles were stored at −20 °C in vacuo until used.
1.2.4 Nitric oxide release characterization
Real-time NO release from 75 mol% MPTMS/TEOS particles was measured at 1 s intervals using a Sievers Chemiluminescence Nitric Oxide Analyzer (Boulder, CO). Particles were added to 25 mL deoxygenated TSB (37 °C) containing 50 μL antifoaming agent B (Sigma-Aldrich) to prevent frothing. Decomposition of the nitrosothiol to release NO was initiated by the heat of the solution and trace amounts of free copper likely present in the TSB solution. The solution was sparged with nitrogen (80 mL/min) with additional nitrogen was supplied to the reaction flask to match the collection rate of the NOA (200 mL min−1). The apparatus was covered with aluminum foil to prevent light-initiated nitrosothiol decomposition.
1.2.5 Minimum inhibitory concentration assay
Bacterial cultures were grown from an overnight stock in TSB to 108 colony forming units (cfu) mL−1 and diluted to 2 × 106 cfu ml−1. Bacteria were added to serial dilutions of nitrosothiol particles in a 96-well plate resulting in a final concentration of 106 cfu mL−1 bacteria. After incubating by shaking for 24 h at 37 °C, MIC values were determined as the lowest particle concentration not supporting bacterial growth (i.e., not turbid).
1.2.6 Spontaneous resistance assay
Bacterial cultures were grown from an overnight stock in TSB to ~109 cfu mL−1. A 1-mL aliquot of the 109 cfu mL−1 culture was added to NO-releasing particles at 2–8× the MIC measured for each bacterial species. Following a 24 h incubation at 37 °C in the dark with agitation, 1 mL of each concentration was plated on TSA (200 μL on 5 separate plates) and incubated overnight at 37 °C. Surviving colonies were propagated overnight at 37 °C in TSB, reinoculated and grown to 108 cfu mL−1. The MICs for propagated strains were determined using the above procedure and compared to the parent strain. Surviving colonies on TSA that could not be propagated in TSB were passaged on TSA for three days and then grown in TSB overnight at 37 °C. If overnight growth in TSA was successful, the MIC was then evaluated and compared to the parent strain. Otherwise, formation and settling of a bacterial precipitate did not allow an MIC assay to be performed.
1.2.7 Serial passage assay
Bacterial cultures were grown from an overnight stock in TSB to 108 cfu mL−1 and diluted to 2 × 106 cfu mL−1. The bacterial suspensions were then added to serial dilutions of nitrosothiol particles in a 96-well plate resulting in wells containing 106 cfu mL−1 bacteria and nitrosothiol particle concentrations of 2, 1, 0.5, 0.25, and 0.125× the MIC (n=3). After incubating by shaking for 24 h at 37 °C, MIC values were recorded, and an aliquot from the well containing the highest particle concentration that supported bacterial growth was diluted to 2 × 106 cfu mL−1. The MIC assay was performed using this bacterial suspension. The entire process was repeated for 20 exposure cycles.
1.3 Results and Discussion
The bacterial species used in these studies were selected because they are frequently found in a clinical environment. While gaseous NO has proven useful for pulmonary treatment, it is generally not a good candidate for an antimicrobial therapeutic. The short half-life (<10 s) of NO in physiological milieu prevents its delivery to common infection sites such as an indwelling medical device (i.e., catheter) or deep wound. As such, nanoparticles chemically modified to store and release NO have been studied as candidate antimicrobials.[12,13,14] We have previously described particles that release NO over extended periods (from minutes to days), allowing more targeted NO delivery, thus ensuring more lethal concentrations of NO. Indeed, Hetrick and coworkers reported excellent efficacy of NO-releasing particles against both planktonic and biofilm-based bacteria.[12,13] To date, the only studies that have examined bacterial resistance to exogenous NO have used NO gas from cylinders.[15,55,56,57] Martinez and Baquero demonstrated that the development of resistant bacteria depends on antibiotic exposure parameters (i.e., concentration and kinetics).[58] Thus, we utilized chemically-stored NO release for these studies to more fully evaluate resistance potential. In particular, nitrosothiol-based NO-releasing particles were selected because their extended NO release capabilities (>24 h) facilitate continuous selective pressure for resistant mutants whereas low molecular weight N-diazeniumdiolate NO donors tend to release their NO payload more quickly, especially in aqueous media.[13,21]
The NO release profile of 75 mol% MPTMS-TEOS particles (635 ± 63 nm diameter) in TSB at 37 °C is shown in Figure 1. To mimic the conditions used during the bacteria assays, NO release measurements were conducted in the absence of light such that NO production was limited to thermal decomposition and not photolytic cleavage. Upon addition to the assay media (2 mg mL−1 final particle concentration) (TSB,37 °C), a bolus of NO was released at ~740 ppb mg−1 s−1. This level of NO decreased with time, ultimately dropping to ~11 ppb mg−1 s−1 after 24 h. Over the course of the assay, a total of 0.90 μmol mg−1 was released per mg of particles. Both the maximum instantaneous and the total NO released from the particles in TSB were slightly lower than reported previously in PBS (1205 ppb mg−1 s−1 and 1.17 μmol mg−1, respectively), which is likely due to reactions between NO and proteins present in TSB.[21]
Figure 1.
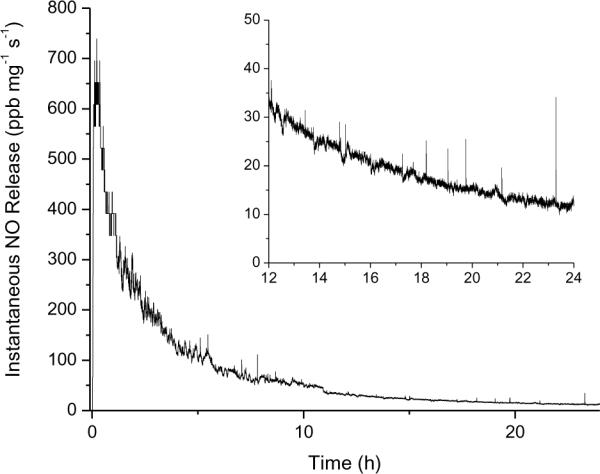
Representative NO release from 75 mol% MPTMS/TEOS particles in TSB at 37 °C. [Inset: Enlarged view of NO release during 12–24 h.]
1.3.1 Minimum inhibitory concentration determinations
Minimum inhibitory concentrations were used to rapidly determine the efficacy of the NO release and monitor for the emergence of resistance.[58,59,60,61] As shown in Table 1, the MICs were used for both the spontaneous and serial passage mutagenesis assays. The MIC of 75 mol% MPTMS/TEOS particles for each bacterial species was determined under growth conditions (TSB, 37 °C) over 24 h. The measured MICs ranged from 3.13 to 6.25 mg mL−1 across all bacterial species (Table 1).Of note, the MICs for both methicillin-susceptible and -resistant S. aureus were half that of S. epidermidis and the two Gram negative species, E. coli and P. aeruginosa. Representative Gram positive and Gram negative bacteria, S. aureus and P. aeruginosa, were also exposed to control (non-nitrosated) particles at concentrations equivalent to the MIC for nitrosated particles. No inhibition was observed, indicating that NO, rather than the particles, are responsible for the antimicrobial activity.
Table 1.
Minimum inhibitory concentrations of 75 mol% MPTMS/TEOS particles in TSB at 37 °C for 24 h (1010 cfu mL-1 starting bacterial concentration) and spontaneous mutation parameters before and after exposure to inhibitory concentrations of NO.
| MIC24h (mg mL-1) | ||||
|---|---|---|---|---|
| species | ATCC# | Day 1 | Day 20 | ΔMIC |
| S. aureus | 29213 | 3.13 | 3.13 | 0 |
| MRSA | 33591 | 3.13 | 1.65 | −50% |
| S. epidermidis | 35983 | 6.25 | 3.13 | −50% |
| E. coli (0157:H7) | 35150 | 6.25 | 6.25 | 0 |
| P. aeruginosa | 19143 | 6.25 | 6.25 | 0 |
1.3.2 Spontaneous mutagenesis assay
Even after exposure to bactericidal doses of an antimicrobial, some microbes may survive depending on the antimicrobial concentration, environmental conditions, and microbial species.[58] In the case of antibiotics, some of the surviving microbes are the result of a spontaneous mutation that confers greater resistance to future treatment.[58] Thus, the rate of spontaneous mutations occurring at inhibitory NO concentrations was evaluated for each bacterial species to address the possibility of NO-resistance. Nitric oxide-releasing particle concentrations ranging from 2 to 8 times the MIC were utilized to provide adequate selective pressure against NO-susceptible bacteria. Surviving colonies were isolated and propagated in TSB, and the MIC assays were repeated to observe if the microbes were more or less susceptible to NO treatment. Exposure of E. coli to NO-releasing particles at 2 times the MIC resulted in 19 surviving colonies in 1 mL. Each colony was reinoculated in TSB, and all resulted in a cloudy suspension after overnight incubation. An MIC assay was performed individually on each colony and the susceptibility of all 19 colonies was unchanged from the parent strain (6.25 mg mL−1). Nanoparticle exposure to MRSA at 7 times the MIC resulted in one surviving colony in 1 mL. Although the MIC of this survivor was increased by 2 times to 6.25 mg mL−1, this increase is considered to be within the experimental variation and is thus not significant. After exposure of P. aeruginosa to 2 times the MIC, 7 surviving colonies were isolated and propagated successfully in TSB. The MIC of all P. aeruginosa survivors was increased two fold to 12.5 mg mL−1, but again this increase is within the experimental variation and not significant.
Some colonies that were able to survive NO treatment were not able to grow successfully in TSB. These colonies were instead propagated three times on TSA to assess if the mutation that limited growth in broth was stable. After exposure of S. epidermidis to 4 times the MIC, two of the three surviving colonies could not be propagated further in TSB, even after three successful passages on TSA. Regrowth of the bacterial precipitate was possible on TSA. However, the inability of the S. epidermidis to grow to turbidity in fresh TSB prevented the determination of the MIC. The spontaneous mutation that resulted in this NO tolerance seemed to have prevented regrowth in nutrient broth. Others have observed similar behavior where mutations conferring resistance to a therapeutic also result in a fitness cost to the bacteria, sometimes preventing further propagation.[62,63] A third colony of S. epidermidis was successfully regrown in TSB, but viability was not evident following NO exposure even at 1/8 of the MIC. To assess the fitness of this colony, it was propagated three times in succession on TSA and then inoculated in TSB. The solution again grew to a cloudy suspension overnight. An MIC assay was successfully completed and found to be identical to the parent strain (~6.25 mg mL−1). These results indicate that for one mutant, the growth defect preventing propagation in TSB in the absence of NO was resolved, possibly due to a second mutation that also abolished the observed increase in NO resistance. Methicillin-susceptible S. aureus exposed to 8 times the MIC resulted in 12 surviving colonies on TSA. Reinoculation of each colony in TSB produced a precipitate similar to that of the S. epidermidis colonies described above. Similarly, the colony failed to successfully grow to a cloudy suspension in TSB after regrowth on TSA three times in succession. Therefore, no MIC assay was performed. However, propagation of the aggregated bacteria on TSA was successful. A comparison of all parameters (initial and final MIC, survivors, and colonies propagated in TSB) is shown in Table 1.
1.3.3 Serial passage assay
Repeated exposure to sub-therapeutic concentrations of antibiotics often hastens the development of antibiotic-resistant bacteria.[64,65] Genetic mutations may result, leading to an increased resistance to the antibiotic that the microbes were exposed to at sub-cidal or sub-inhibitory doses. Repeated or prolonged exposure to sub-therapeutic antibiotic concentrations would further enrich the resistant strain. To investigate possible resistance, the susceptibility to NO treatment following exposure to sub-inhibitory NO doses was examined using the NO-releasing nitrosothiol-modified particles. Bacterial cultures were treated with a range of concentrations both above and below the MIC for 24 h in nutrient growth conditions employing a serial passage mutagenesis assay described previously.[60,61] The assay was repeated by propagating the bacteria exposed to the highest concentration of particles that did not inhibit growth. After the completion of 20 passages of NO exposure in this manner, no sustained increases in the MIC for any of the bacterial species were observed versus the parent strains (Table 2). The two-fold increase in susceptibility observed for S. aureus and S. epidermidis was not significant and is considered normal inter-experimental variation.
Table 2.
Minimum inhibitory concentrations of 75 mol% MTPMS/TEOS particles after 1 and 20 exposure passages (106 cfu mL-1 starting bacterial concentration).
| species | ATCC #a | MIC24h (mg mL-1) | Exposure concentration (mg mL-1)b | Colonies after >MIC NO exposure | Colonies propagated in TSB | Final MIC24h (mg mL-1) |
|---|---|---|---|---|---|---|
| S. aureus | 29213 | 3.13 | 25.0 | 12 | 0 | N/Ac |
| MRSA | 33591 | 3.13 | 21.9 | 1 | 1 | 6.25 |
| S. epidermidis | 35983 | 6.25 | 25.0 | 3 | 1 | 6.25 |
| E.coli (0157:H7) | 35150 | 6.25 | 12.5 | 19 | 19 | 6.25 |
| P. aeruginosa | 19143 | 6.25 | 12.0 | 7 | 7 | 12.5 |
ATCC, American Type Culture Collection.
Starting bacterial concentrations were ~109 cfu mL-1.
Regrowth in broth did not result in turbidity, thus an MIC could not be performed.
1.4 Conclusions
The inability of bacteria to develop resistance to exogenous NO delivered from a silica vehicle was not surprising primarily because of the multiple mechanisms by which NO presents toxicity towards microbes.[2,11,12,13,16,51] The hydrophobicity and small size of NO allows it to rapidly migrate across bacterial lipid membranes where a number of nitrosative and oxidative reactions may occur.[13] The diversity of NO's antimicrobial mechanisms thus would require multiple mutations to occur simultaneously for microbial survival, hindering resistance development. Nevertheless, it would be naïve to conclude that bacteria absolutely cannot develop increased resistance to exogenous NO. Spellberg et al. points out the fallacy of assuming that we (humans) can win a war against bacteria that have been “creating and defeating antibiotics for 20 million times longer than Homo sapiens have known that antibiotics existed.”[65] It is likely that the emergence of resistance to exogenous NO will depend heavily on environmental conditions such as nutrient availability, temperature, exposure duration/intensity, the presence of other bacterial species, and infection location (i.e., in vivo vs. in vitro). Clearly, it is imperative that future studies examining the efficacy of NO-releasing therapeutics also consider the ability of bacteria to develop resistance, especially as such therapeutics are applied clinically.
We evaluate the ability of bacteria to develop a resistance to exogenous NO.
Nitrosothiol nanoparticles were utilized as an exogenous antimicrobial model.
Prolonged exposure to sub-therapeutic NO did not result in increased resistance.
Single-dose exposure to bactericidal NO did not result in increased resistance.
These assays may serve as a blueprint for future NO resistance studies.
1.5 Acknowledgements
This research was supported by the National Institutes of Health (EB000708).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1.6 References
- [1].Wink DA, Mitchell JB. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. and Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- [2].Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- [3].Fang FC. Host/pathogen interactions: Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Foley E, O'Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 2003;17:115–125. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- [6].Mannick JB. Immunoregulatory and antimicrobial effects of nitrogen oxides. Proc. Am. Thorac. Soc. 2006;3:161–165. doi: 10.1513/pats.200505-048BG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Moller MN, Li Q, Lancaster JRJ, Denicola A. Acceleration of nitric oxide autooxidation and nitrosation by membranes. Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- [8].Zaki MH, Akuta T, Akaike T. Nitric oxide-induced nitrative stress involved in microbial pathogenesis. J. Pharmacol. Sci. 2005;98:117–129. doi: 10.1254/jphs.crj05004x. [DOI] [PubMed] [Google Scholar]
- [9].Keefer LK. Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates. Annu. Rev. Pharmacol. Toxicol. 2003;43:585–607. doi: 10.1146/annurev.pharmtox.43.100901.135831. [DOI] [PubMed] [Google Scholar]
- [10].Nablo BJ, Rothrock AR, Schoenfisch MH. Nitric oxide-releasing sol-gels as antibacterial coatings for orthopedic implants. Biomaterials. 2005;26:917–924. doi: 10.1016/j.biomaterials.2004.03.031. [DOI] [PubMed] [Google Scholar]
- [11].Hetrick EM, Schoenfisch MH. Antibacterial nitric oxide-releasing xerogels: Cell viability and parallel plate flow cell adhesion studies. Biomaterials. 2007;28:1948–1956. doi: 10.1016/j.biomaterials.2007.01.006. [DOI] [PubMed] [Google Scholar]
- [12].Hetrick EM, Shin JH, Paul HS, Schoenfisch MH. Anti-biofilm efficacy of nitric oxide-releasing silica nanoparticles. Biomaterials. 2009;30:2782–2789. doi: 10.1016/j.biomaterials.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hetrick EM, Shin JH, Stasko NA, Johnson CB, Wespe DA, Holmuhamedov E, Schoenfisch MH. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2008;2:235–246. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shin JH, Metzger SK, Schoenfisch MH. Synthesis of nitric oxide-releasing silica nanoparticles. J. Am. Chem. Soc. 2007;129:4612–4619. doi: 10.1021/ja0674338. [DOI] [PubMed] [Google Scholar]
- [15].Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide-Biol. Chem. 2006;14:21–29. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- [16].Feelisch M. The use of nitric oxide donors in pharmacological studies. Naunyn-Schmiedebergs Arch. Pharmacol. 1998;358:113–122. doi: 10.1007/pl00005231. [DOI] [PubMed] [Google Scholar]
- [17].Muscará MN, Wallace JL. V. Therapeutic potential of nitric oxide donors and inhibitors. Am. J. Physiol. - Gastrointestinal and Liver Physiology. 1999;276:G1313–G1316. doi: 10.1152/ajpgi.1999.276.6.G1313. [DOI] [PubMed] [Google Scholar]
- [18].Artz JD, Toader V, Zavorin SI, Bennett BM, Thatcher GRJ. In vitro activation of soluble guanylyl cyclase and nitric oxide release: A comparison of NO donors and NO mimetics. Biochemistry. 2001;40:9256–9264. doi: 10.1021/bi002885x. [DOI] [PubMed] [Google Scholar]
- [19].Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hetrick EM, Shin JH, Stasko NA, Johnson BA, Wespe DA, Holmuhamedov E, Schoenfisch MH. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2007;2 doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Riccio DA, Nugent JL, Schoenfisch MH. Stober synthesis of nitric oxide-releasing S-nitrosothiol-modified silica particles. Chem. Mater. 2011;23:1727–1735. doi: 10.1021/cm102510q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stasko NA, Fischer TH, Schoenfisch MH. S-nitrosothiol-modified dendrimers as nitric oxide delivery vehicles. Biomacromolecules. 2008;9:834–841. doi: 10.1021/bm7011746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stasko NA, Schoenfisch MH. Dendrimers as a scaffold for nitric oxide release. J. Am. Chem. Soc. 2006;128:8265–8271. doi: 10.1021/ja060875z. [DOI] [PubMed] [Google Scholar]
- [24].Rothrock AR, Donkers RL, Schoenfisch MH. Synthesis of nitric oxide-releasing gold nanoparticles. J. Am. Chem. Soc. 2005;127:9362–9363. doi: 10.1021/ja052027u. [DOI] [PubMed] [Google Scholar]
- [25].Hetrick EM, Schoenfisch MH. Antibacterial nitric oxide-releasing xerogels: Cell viability and parallel plate flow cell adhesion studies. Biomaterials. 2007;28:1948–1956. doi: 10.1016/j.biomaterials.2007.01.006. [DOI] [PubMed] [Google Scholar]
- [26].Marxer SM, Rothrock AR, Nablo BJ, Robbins ME, Schoenfisch MH. Preparation of nitric oxide (NO)-releasing sol-gels for biomaterial applications. Chem. Mater. 2003;15:4193–4199. [Google Scholar]
- [27].Coneski PN, Schoenfisch MH. Synthesis of nitric oxide-releasing polyurethanes with S-nitrosothiol-containing hard and soft segments. Polym. Chem. 2011;2:906–913. doi: 10.1039/C0PY00269K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Coneski PN, Nash JA, Schoenfisch MH. Nitric oxide-releasing electrospun polymer microfibers. ACS Appl. Mater. Interfaces. 2011;3:426–432. doi: 10.1021/am101010e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Privett BJ, Nutz ST, Schoenfisch MH. Efficacy of surface-generated nitric oxide against Candida albicans adhesion and biofilm formation. Biofouling. 2010;26:973–983. doi: 10.1080/08927014.2010.534552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Coneski PN, Rao KS, Schoenfisch MH. Degradable nitric oxide-releasing biomaterials via post-polymerization functionalization of cross-linked polyesters. Biomacromolecules. 2010;11:3208–3215. doi: 10.1021/bm1006823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Riccio DA, Dobmeier KP, Hetrick EM, Privett BJ, Paul HS, Schoenfisch MH. Nitric oxide-releasing S-nitrosothiol-modified xerogels. Biomaterials. 2009;30:4494–4502. doi: 10.1016/j.biomaterials.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nablo BJ, Schoenfisch MH. Antibacterial properties of nitric oxide-releasing sol-gels. J. Biomed. Mater. Res., Part A. 2003;67A:1276–1283. doi: 10.1002/jbm.a.20030. [DOI] [PubMed] [Google Scholar]
- [33].Nablo BJ, Chen TY, Schoenfisch MH. Sol-gel derived nitric-oxide releasing materials that reduce bacterial adhesion. J. Am. Chem. Soc. 2001;123:9712–9713. doi: 10.1021/ja0165077. [DOI] [PubMed] [Google Scholar]
- [34].Akimitsu N, Hamamoto H, Inoue R, Shoji M, Akamine A, Takemori K, Hamasaki N, Sekimizu K. Increase in resistance of methicillin-resistant Staphylococcus aureus to β-lactams caused by mutations conferring resistance to benzalkonium chloride, a disinfectant widely used in hospitals. Antimicrob. Agents Chemother. 1999;43:3042–3043. doi: 10.1128/aac.43.12.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hiramatsu K. Vancomycin resistance in Staphylococci. Drug Res. Updates. 1998;1:135–150. doi: 10.1016/s1368-7646(98)80029-0. [DOI] [PubMed] [Google Scholar]
- [36].Levy SB. Multidrug resistance: a sign of the times. New Eng. J. Med. 1998;338:1376–1378. doi: 10.1056/NEJM199805073381909. [DOI] [PubMed] [Google Scholar]
- [37].Percival SL, Bowler PG, Russell D. Bacterial resistance to silver in wound care. J. Hosp. Infect. 2005;60:1–7. doi: 10.1016/j.jhin.2004.11.014. [DOI] [PubMed] [Google Scholar]
- [38].Silver S, Phung LT, Silver G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 2006;33:627–634. doi: 10.1007/s10295-006-0139-7. [DOI] [PubMed] [Google Scholar]
- [39].Suller MTE, Russell AD. Antibiotic and biocide resistance in methicillin-resistant staphylococcus aureus and vancomycin-resistant enterococcus. J. Hosp. Infect. 1999;43:281–291. doi: 10.1016/s0195-6701(99)90424-3. [DOI] [PubMed] [Google Scholar]
- [40].Delcour AH. Outer membrane permeability and antibiotic resistance. BBA-Protiens Proteom. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Charrel RN, Pages JM, DeMicco P, Mallea M. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob. Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nunoshiba T, Derojaswalker T, Tannenbaum SR, Demple B. Roles of nitric-oxide in inducible resistance of Escherichia-coli to activated murine macrophages. Infect. Immun. 1995;63:794–798. doi: 10.1128/iai.63.3.794-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nunoshiba T, Derojaswalker T, Wishnok JS, Tannenbaum SR, Demple B. Activation by nitric-oxide of an oxidative-stress response that defends Escherichia-coli against activated macrophages. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- [46].Tillmann A, Gow NAR, Brown AJP. Nitric oxide and nitrosative stress tolerance in yeast. Biochem. Soc. Trans. 2011;39:219–223. doi: 10.1042/BST0390219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Husain M, Bourret TJ, McCollister BD, Jones-Carson J, Laughlin J, Vazquez-Torres A. Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J. Biol. Chem. 2008;283:7682–7689. doi: 10.1074/jbc.M708845200. [DOI] [PubMed] [Google Scholar]
- [48].Miller CC, Rawat M, Johnson T, Av-Gay Y. Innate Protection of Mycobacterium smegmatis against the Antimicrobial Activity of Nitric Oxide Is Provided by Mycothiol. Antimicrob. Agents Chemother. 2007;51:3364–3366. doi: 10.1128/AAC.00347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Newton GL, Av-Gay Y, Fahey RC. A Novel Mycothiol-Dependent Detoxification Pathway in Mycobacteria Involving Mycothiol S-Conjugate Amidaseâ€. Biochemistry. 2000;39:10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- [50].Nunoshiba T, Derojaswalker T, Wishnok JS, Tannenbaum SR, Demple B. Activation by Nitric-Oxide of an Oxidative-Stress Response That Defends Escherichia-Coli against Activated Macrophages. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Deupree SM, Schoenfisch MH. Morphological analysis of the antimicrobial action of nitric oxide on Gram-negative pathogens using atomic force microscopy. Acta Biomaterialia. 2009;5:1405–1415. doi: 10.1016/j.actbio.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dobmeier KP, Schoenfisch MH. Antibacterial properties of nitric oxide-releasing sol-gel microarrays. Biomacromolecules. 2004;5:2493–2495. doi: 10.1021/bm049632u. [DOI] [PubMed] [Google Scholar]
- [53].Privett BJ, Deupree SM, Backlund CJ, Rao KS, Johnson CB, Coneski PN, Schoenfisch MH. Synergy of nitric oxide and silver sulfadiazine against gram-negative, gram-positive, and antibiotic-resistant pathogens. Mol. Pharmaceutics. 2010;7:2289–2296. doi: 10.1021/mp100248e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- [55].Miller C, McMullin B, Ghaffari A, Stenzler A, Pick N, Roscoe D, Ghahary A, Road J, Av-Gay Y. Gaseous nitric oxide bactericidal activity retained during intermittent high-dose short duration exposure. Nitric Oxide-Biol. Chem. 2009;20:16–23. doi: 10.1016/j.niox.2008.08.002. [DOI] [PubMed] [Google Scholar]
- [56].Ghaffari A, Jalili R, Ghaffari M, Miller C, Ghahary A. Efficacy of gaseous nitric oxide in the treatment of skin and soft tissue infections. Wound Rep. Regen. 2007;15:368–377. doi: 10.1111/j.1524-475X.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- [57].Miller CC, Miller MK, Ghaffari A, Kunimoto B. Treatment of chronic nonhealing leg ulceration with gaseous nitric oxide: A case study. J. Cutan. Med. Surg. 2004;8:233–238. doi: 10.1007/s10227-004-0106-8. [DOI] [PubMed] [Google Scholar]
- [58].Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vasquez JE, Walker ES, Franzus BW, Overbay BK, Reagan DR, Sarubbi FA. The epidemiology of mupirocin resistance among methicillin-resistant Staphylococcus aureus at a veterans' affairs hospital. Infect. Control Hosp. Epidemiol. 2000;21:459–464. doi: 10.1086/501788. [DOI] [PubMed] [Google Scholar]
- [60].Suller MTE, Russell AD. Antibiotic and biocide resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococcus. J. Hosp. Inf. 1999;43:281–291. doi: 10.1016/s0195-6701(99)90424-3. [DOI] [PubMed] [Google Scholar]
- [61].Suller MTE, Russell AD. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 2000;46:11–18. doi: 10.1093/jac/46.1.11. [DOI] [PubMed] [Google Scholar]
- [62].Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- [63].Normark BH, Normark S. Evolution and spread of antibiotic resistance. J. Intern. Med. 2002;252:91–106. doi: 10.1046/j.1365-2796.2002.01026.x. [DOI] [PubMed] [Google Scholar]
- [64].Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the infectious diseases society of america. Clin. Infect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- [65].Ghosh S, LaPara TM. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007;1:191–203. doi: 10.1038/ismej.2007.31. [DOI] [PubMed] [Google Scholar]


