Abstract
Naturally emerging and deliberately released pathogens demand new detection strategies to allow early recognition and containment. We describe a diagnostic system for rapid, sensitive, multiplex discrimination of microbial gene sequences and report its application for detecting 22 respiratory pathogens in clinical samples.
Keywords: Mass spectroscopy, PCR, multiplex, differential diagnosis, infectious diseases
Efficient laboratory diagnosis of infectious diseases is increasingly important to clinical management and public health. Methods to directly detect nucleic acids of microbial pathogens in clinical specimens are rapid, sensitive, and may succeed when culturing the organism fails. Clinical syndromes are infrequently specific for single pathogens; thus, assays are needed that allow multiple agents to be simultaneously considered. Current multiplex assays employ gel-based formats in which products are distinguished by size, fluorescent reporter dyes that vary in color, or secondary enzyme hybridization assays. Gel-based assays are reported that detect 2–8 different targets with sensitivities of 2–100 PFU or <1–5 PFU, depending on whether amplification is carried out in a single or nested format, respectively (1–4). Fluorescence reporter systems achieve quantitative detection with sensitivity similar to that of nested amplification; however, their capacity to simultaneously query multiple targets is limited to the number of fluorescent emission peaks that can be unequivocally resolved. At present, up to 4 fluorescent reporter dyes can be detected simultaneously (5,6). Multiplex detection of up to 9 pathogens has been achieved in hybridization enzyme systems; however, the method requires cumbersome postamplification processing (7).
The Study
To address the need for sensitive multiplex assays in diagnostic molecular microbiology, we created a polymerase chain reaction (PCR) platform in which microbial gene targets are coded by a library of 64 distinct Masscode tags (Qiagen Masscode technology, Qiagen, Hilden, Germany). A schematic representation of this approach is shown in Figure 1. Microbial nucleic acids (RNA, DNA, or both) are amplified by multiplex reverse transcription (RT)-PCR using primers labeled by a photocleavable link to molecular tags of different molecular weight. After removing unincorporated primers, tags are released by UV irradiation and analyzed by mass spectrometry. The identity of the microbe in the clinical sample is determined by its cognate tags.
Figure 1.
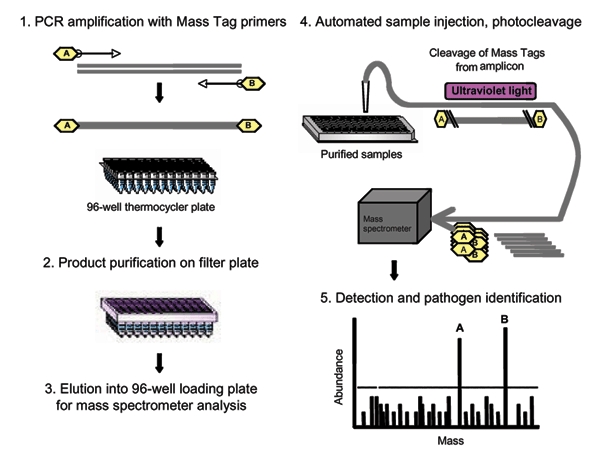
Schematic representation of Mass Tag polymerase chain reaction (PCR).
As a first test of this technology, we focused on respiratory disease because differential diagnosis is a common clinical challenge, with implications for outbreak control and individual case management. Multiplex primer sets were designed to identify up to 22 respiratory pathogens in a single Mass Tag PCR reaction; sensitivity was established by using synthetic DNA and RNA standards as well as titered viral stocks; the utility of Mass Tag PCR was determined in blinded analysis of previously diagnosed clinical specimens.
Oligonucleotide primers were designed in conserved genomic regions to detect the broadest number of members for a given pathogen species by efficiently amplifying a 50- to 300-bp product. In some instances, we selected established primer sets; in others, we used a software program designed to cull sequence information from GenBank, perform multiple alignments, and maximize multiplex performance by selecting primers with uniform melting temperatures and minimal cross-hybridization potential (Table A1). Primers, synthesized with a 5´ C6 spacer and aminohexyl modification, were covalently conjugated by a photocleavable link to Masscode tags (Qiagen Masscode technology) (8,9). Masscode tags have a modular structure, including a tetrafluorophenyl ester for tag conjugation to primary amines; an o-nitrobenzyl photolabile linker for photoredox cleavage of the tag from the analyte; a mass spectrometry sensitivity enhancer, which improves the efficiency of atmospheric pressure chemical ionization of the cleaved tag; and a variable mass unit for variation of the cleaved tag mass (8,10–12). A library of 64 different tags has been established. Forward and reverse primers in individual primer sets are labeled with distinct molecular weight tags. Thus, amplification of a microbial gene target produces a dual signal that allows assessment of specificity.
Gene target standards were cloned by PCR into pCR2.1-TOPO (Invitrogen, Carlsbad, CA, USA) by using DNA template (bacterial and DNA viral targets) or cDNA template (RNA viral targets) obtained by reverse transcription of extracts from infected cultured cells or by assembly of overlapping synthetic polynucleotides. Assays were initially established by using plasmid standards diluted in 2.5-µg/mL human placenta DNA (Sigma, St. Louis, MO, USA) and subjected to PCR amplification with a multiplex PCR kit (Qiagen), primers at 0.5 µmol/L each, and the following cycling protocol: an annealing step with a temperature reduction in 1°C increments from 65°C to 51°C during the first 15 cycles and then continuing with a cycling profile of 94°C for 20 s, 50°C for 20 s, and 72°C for 30 s in an MJ PTC200 thermal cycler (MJ Research, Waltham, MA, USA). Amplification products were separated from unused primers by using QIAquick 96 PCR purification cartridges (Qiagen, with modified binding and wash buffers). Masscode tags were decoupled from amplified products through UV light-induced photolysis in a flow cell and analyzed in a single quadrapole mass spectrometer using positive-mode atmospheric pressure chemical ionization (Agilent Technologies, Palo Alto, CA, USA). A detection threshold of 100 DNA copies was determined for 19 of 22 cloned targets by using a 22-plex assay (Table 1).
Table 1. Sensitivity of pathogen detection by Mass Tag polymerase chain reaction determined by using plasmid and synthetic RNA standards*.
| Pathogen or protein | Detection threshold (DNA copies/RNA copies) |
|---|---|
| Influenza A matrix | 100/1,000 |
| Influenza A N1 | 100/NA |
| Influenza A N2 | 100/NA |
| Influenza A H1 | 100/NA |
| Influenza A H2 | 100/NA |
| Influenza A H3 | 100/NA |
| Influenza A H5 | 100/NA |
| Influenza B H | 500/1,000 |
| RSV group A | 100/1,000 |
| RSV group B | 100/500 |
| Metapneumovirus | 100/1,000 |
| CoV-SARS | 100/500 |
| CoV-OC43 | 100/500 |
| CoV-229E | 100/500 |
| HPIV-1 | 100/1,000 |
| HPIV-2 | 100/1,000 |
| HPIV-3 | 100/500 |
| Chlamydia pneumoniae | 100/NA |
| Mycoplasma pneumoniae | 100/NA |
| Legionella pneumophila | 100/NA |
| Enterovirus (genus) | 500/1,000 |
| Adenovirus (genus) | 5,000/NA |
*NA, not assessed; RSV, respiratory syncytial virus; CoV, coronavirus; SARS, severe acute respiratory syndrome; HPIV, human parainfluenza virus.
Many respiratory pathogens have RNA genomes; thus, where indicated, assay sensitivity was determined by using synthetic RNA standards or RNA extracts of viral stocks. Synthetic RNA standards were generated by using T7 polymerase and linearized plasmid DNA. After quantitation by UV spectrometry, RNA was serially diluted in 2.5-µg/mL yeast tRNA (Sigma), reverse transcribed with random hexamers by using Superscript II (Invitrogen, Carlsbad, CA, USA), and used as template for Mass Tag PCR. As anticipated, sensitivity was reduced by the use of RNA instead of DNA templates (Table 1). The sensitivity of Mass Tag PCR to detect live virus was tested by using RNA extracted from serial dilutions of titered stocks of coronaviruses (severe acute respiratory syndrome [SARS] and OC43) and parainfluenzaviruses (HPIV 2 and 3). A 100-µL volume of each dilution was analyzed. RNA extracted from a 1-TCID50/mL dilution, representing 0.025 TCID50 per PCR reaction, was consistently positive in Mass Tag PCR.
RNA extracted from banked sputum, nasal swabs, and pulmonary washes of persons with respiratory infection was tested by using an assay panel comprising 30 gene targets that represented 22 respiratory pathogens. Infection in each of these persons had been previously diagnosed through virus isolation, conventional nested RT-PCR, or both. Reverse transcription was performed using random hexamers, and Mass Tag PCR results were consistent in all cases with the established diagnosis. Infections with respiratory syncytial virus, human parainfluenza virus, SARS coronavirus, adenovirus, enterovirus, metapneumovirus, and influenza virus were correctly identified (Table 2 and Figure 2). A panel comprising gene targets representing 17 pathogens related to central nervous system infectious disease (influenza A virus matrix gene; influenza B virus; human coronaviruses 229E, OC43, and SARS; enterovirus; adenovirus; human herpesvirus-1 and -3; West Nile virus; St. Louis encephalitis virus; measles virus; HIV-1 and -2; and Streptococcus pneumoniae, Haemophilus influenzae, and Nesseria meningitidis) was applied to RNA obtained from banked samples of cerebrospinal fluid and brain tissue that had been previously characterized by conventional diagnostic RT-PCR. Two of 3 cases of West Nile virus encephalitis were correctly identified. Eleven of 12 cases of enteroviral meningitis were detected representing serotypes CV-B2, CV-B3, CV-B5, E-6, E-11, E-13, E-18, and E-30 (data not shown).
Table 2. Multiplex pathogen detection by Mass Tag polymerase chain reaction using Masscode-labeled primers in a 30-plex assay with clinical specimens with previously identified pathogens*.
| Pathogen | No. positive/no. tested† |
|---|---|
| RSV A | 2/2 |
| RSV B | 3/3 |
| HPIV-1 | 1/1 |
| HPIV-3 | 2/2 |
| HPIV-4 | 2/2 |
| CoV-SARS | 4/4 |
| Metapneumovirus | 2/3 |
| Influenza B | 1/3 |
| Influenza A | 2/6 |
| Adenovirus | 2/2 |
| Enterovirus | 2/2 |
*RSV, respiratory syncytial virus; HPIV, human parainfluenza virus; CoV, coronavirus; SARS, severe acute respiratory syndrome. †No. positive and consistent with previous diagnosis/number tested (with respective previous diagnosis).
Figure 2.
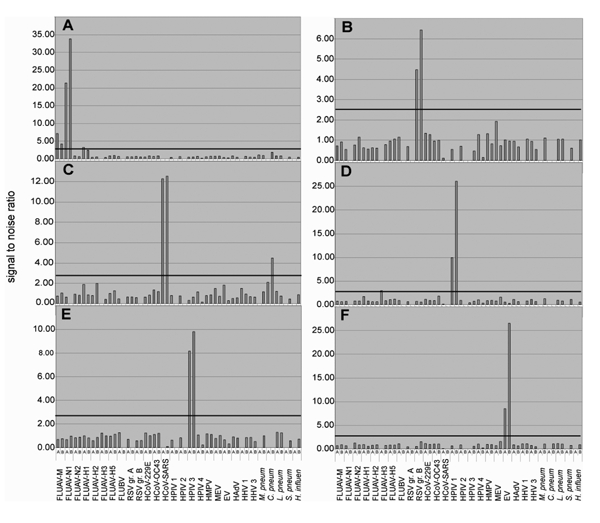
Analysis of clinical specimens. RNA extracts from clinical specimens containing known pathogens were reverse transcribed into cDNA (Superscript RT system, Invitrogen, Carlsbad, CA; 20-µL volume). Five microliters of the reaction were subjected to Mass Tag PCR by using primers coupled to Masscode tags (Qiagen Masscode technology, Qiagen, Hilden, Germany). Detection of (A) influenza virus A (H1N1), (B) respiratory syncytial virus (RSV) group B, (C) human coronavirus SARS (HCoV-SARS), (D) human parainfluenza virus (HPIV) types 1 and (E) 3, and (F) enterovirus (EV) by using a 30-plex assay, including 60 primers targeting influenza A virus matrix gene (FLUAV-M), and for typing N1, N2, H1, H2, H3, and H5 sequences, as well as influenza B virus (FLUBV), RSV groups A and B, HCoV-229E, -OC43, and -SARS, HPIV types 1, 2, 3, and 4 (groups A and B combined; 4 primers), human metapneumovirus (HMPV, 4 primers), measles virus (MEV), EV (degenerate primer pair targeting all serogroups), human adenoviruses (HAdV, degenerate primer pair targeting all serogroups), human herpesvirus 1 (HHV-1, herpes simplex virus), human herpesvirus 3 (HHV-3; varicella-zoster virus), Mycoplasma pneumoniae, Chlamydia pneumoniae, Legionella pneumophila, Streptococcus pneumoniae, Haemophilus influenzae. The bar indicates an arbitrary cut-off threshold of 2.7 (4 times average background determined with random human DNA).
Conclusions
Our results indicate that Mass Tag PCR is a sensitive and specific tool for molecular characterization of microflora. The advantage of Mass Tag PCR is its capacity for multiplex analysis. Although the use of degenerate primers (e.g., enteroviruses and adenoviruses, Table A1 and Table 1) may reduce sensitivity, the limit of multiplexing to detect specific targets will likely be defined by the maximal primer concentration that can be accommodated in a PCR mix. Analysis requires the purification of product from unincorporated primers and mass spectroscopy. Although these steps are now performed manually, and mass spectrometers are not yet widely distributed in clinical laboratories, the increasing popularity of mass spectrometry in biomedical sciences and the advent of smaller, lower-cost instruments could facilitate wider use and integrated instrumentation. In addition to developing additional pathogen panels, our continuing work is focused on optimizing multiplexing, sensitivity, and throughput. Potential applications include differential diagnosis of infectious diseases, blood product surveillance, forensic microbiology, and biodefense.
Acknowledgments
We are grateful to Cinnia Huang, Jill Taylor, and Tony Mazzulli for providing sample materials with previously identified pathogens for analysis.
This work was supported by National Institutes of Health awards AI51292, AI056118, AI55466, U54AI057158 (Northeast Biodefense Center-Lipkin) and the Ellison Medical Foundation. M. Kokoris was a consultant for Qiagen GmbH while the work reported in this manuscript was pursued. He is currently a consultant for Operon Biotechnologies, Inc., which holds the rights on Masscode technology.
Biography
Dr. Briese is associate professor of epidemiology at the Columbia University Mailman School of Public Health and associate director of the Jerome L. and Dawn Greene Infectious Disease Laboratory. His research interests include the molecular epidemiology of emerging viral diseases, virus-host interactions, and novel techniques for pathogen detection and discovery.
Table A1. Respiratory panel Mass Tag primers.
| Target – Masscode FWD/REV | Name FWD | Sequence | Name REV | Sequence | Ref. |
|---|---|---|---|---|---|
| FLUAV-M 618/690 | AM-U151 | CATGGAATGGCTAAAGACAAGACC | AM-L397 | AAGTGCACCAGCAGAATAACTGAG | (8) |
| FLUAV-N1 499/439 | NA1-U1078 | ATGGTAATGGTGTTTGGATAGGAAG | NA1-L1352 | AATGCTGCTCCCACTAGTCCAG | (8) |
| FLUAV-N2 658/730 | NA2-U560 | AAGCATGGCTGCATGTTTGTG | NA2-L858 | ACCAGGATATCGAGGATAACAGGA | (8) |
| FLUAV-HA1 650/590 | HA1-U583 | GGTGTTCATCACCCGTCTAACAT | HA1-L895 | GTGTTTGACACTTCGCGTCACAT | (8) |
| FLUAV-HA2 662/539 | H2A208U27 | GCTATGCAAACTAAACGGAATYCCTCC | H2A559L26 | TATTGTTGTACGATCCTTTGGCAACC | |
| FLUAV-HA3-1 586/475 | HA3-U115 | GCTACTGAGCTGGTTCAGAGTTC | HA3-L375 | GAAGTCTTCATTGATAAACTCCAG | (8) |
| FLUAV-HA5 646/395 | HA5hum-u71 | TTACTGTTACACATGCCCAAGACA | HA5hum-L147 | AGGYTTCACTCCATTTAGATCGCA | |
| FLUBV 698/598 |
BHA-U188 |
AGACCAGAGGGAAACTATGCCC |
BHA-L347 |
CTGTCGTGCATTATAGGAAAGCAC |
(8) |
| HCoV-SARS 527/666 | CIID-28891F | AAGCCTCGCCAAAAACGTAC | CIID-29100R | AAGTCAGCCATGTTCCCGAA | |
| HCoV-229E 670/558 | Taq-Co22-418F | GGCGCAAGAATTCAGAACCA | Taq-Co22-636R | TAAGAGCCGCAGCAACTGC | |
| HCoV-OC43 686/548 |
Taq-Co43-270F |
TGTGCCTATTGCACCAGGAGT |
Taq-Co43-508R |
CCCGATCGACAATGTCAGC |
|
| RSV A 467/455 | RSA-U1137 | AGATCAACTTCTGTCATCCAGCAA | RSV-L1192 | GCACATCATAATTAGGAGTATCAAT | (9) |
| RSV B 483/479 |
RSB-U1248 |
AAGATGCAAATCATAAATTCACAGGA |
RSV-1318 |
TGATATCCAGCATCTTTAAGTATCTTTATAGTG |
(9) |
| HMPV-1 718/654 | MPV01.2 | AACCGTGTACTAAGTGATGCACTC | MPV02.2 | CATTGTTTGACCGGCCCCATAA | (10) |
| HMPV-2 718/654 |
MV-Can-U918 |
AAGTCCAAAGGCAGGRCTGTTATC |
MV-Can-L992 |
CCTGAAGCATTRCCAAGAACAACAC |
|
| HPIV-1 566/357 | HPIV1-U82 | TACTTTTGACACATTTAGTTCCAGGAG | HPIV1-L167 | CGGTACTTCTTTGACCAGGTATAATTG | |
| HPIV-2 463/590 | HPIV2-U908 | GGACTTGGAACAAGATGGCCT | HPIV2-L984 | AGCATGAGAGCYTTTAATTTCTGGA | |
| HPIV-3 423/539 | HPIV3-U590 | GCTTTCAGACAAGATGGAACAGTG | HPIV3-L668 | GCATKATTGACCCAATCTGATCC | |
| HPIV 4A 622/606 | HPIV4A-U191 | AACAGAAGGAAATGATGGTGGAAC | HPIV4A-L269 | TGCTGTGGATGTATGGGCAG | |
| HPIV 4B 622/606 |
HPIV4B-U194 |
AGAAGAAAACAACGATGAGACAAGG |
HPIV4B-L306 |
GTTTCCCTGGTTCACTCTCTTCA |
|
| Measles 578/562 | MEA-U1103 | CAAGCATCATGATYGCCATTCCTGG | MEA-L1183 | CCTGAATCYCTGCCTATGATGGGTTT | |
| Adenovirus 503/630 | ADV2HEX2F | CCCMTTYAACCACCACCG | ADVHEX1R | ACATCCTTBCKGAAGTTCCA | (11) |
| Enterovirus 702/495 | 5UTR-U447 | TCCTCCGGCCCCTGAATGCGGCTAATCC | 5UTR-L541 | GAAACACGGWCACCCAAAGTASTCG | |
| HHV-1 443/706 | HSV-U27 | CCCGGATGCGGTCCAGACGATTAT | HSV-L121 | CCCGCGGAGGTTGTACAAAAAGCT | |
| HHV-3 515/471 | VZV-U138 | ACGTGGATCGTCGGATCAGTTGT | VZV-L196 | TCGCTATGTGCTAAAACACGCGG | |
| Legionella pneumophila 678/582 | LegPneu-U149 | GCATWGATGTTARTCCGGAAGCA | LegPneu-L223 | CGGTTAAAGCCAATTGAGCG | |
| Chlamydia pneumoniae 519/383 | CLPM1 | CATGGTGTCATTCGCCAAGT | CLPM2 | CGTGTCGTCCAGCCATTTTA | (12) |
| Streptococcus pneumonia 714/694 | SPPLY-U532 | AGCGATAGCTTTCTCCAAGTGG | SPPLY-L606 | CTTAGCCAACAAATCGTTTACCG | |
| Haemophilus influenzae 734/726 | HINF-U82 | AAGCTCCTTGMATTTTTTGTATTAGAA | Hinf-L158 | GCTGAATTGGCTTRGATACCGAG | |
| Mycoplasma pneumoniae 602/614 | MTPM1 | CCAACCAAACAACAACGTTCA | MTPM2 | ACCTTGACTGGAGGCCGTTA | (12) |
Footnotes
Suggested citation for this article: Briese T, Palacios G, Kokoris M, Jabado O, Liu Z, Renwick N, et al. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis [serial on the Internet]. 2005 Feb [date cited].http://dx.doi.org/10.3201/eid1102.040492
These authors contributed equally to this study.
References
- 1.Fan J, Henrickson KJ, Savatski LL. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin Infect Dis. 1998;26:1397–402. 10.1086/516357 [DOI] [PubMed] [Google Scholar]
- 2.Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998;36:2990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis JS, Zambon MC. Molecular diagnosis of influenza. Rev Med Virol. 2002;12:375–89. 10.1002/rmv.370 [DOI] [PubMed] [Google Scholar]
- 4.Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol. 2004;72:484–95. 10.1002/jmv.20008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vet JA, Majithia AR, Marras SA, Tyagi S, Dube S, Poiesz BJ, et al. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci U S A. 1999;96:6394–9. 10.1073/pnas.96.11.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–3. 10.1128/JCM.42.3.1220-1223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl JA, Schmitt HJ. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kokoris M, Dix K, Moynihan K, Mathis J, Erwin B, Grass P, et al. High-throughput SNP genotyping with the Masscode system. Mol Diagn. 2000;5:329–40. [DOI] [PubMed] [Google Scholar]
- 9.Lukhtanov EA, Kutyavin IV, Gamper HB, Meyer RB Jr. Oligodeoxyribonucleotides with conjugated dihydropyrroloindole oligopeptides: preparation and hybridization properties. Bioconjug Chem. 1995;6:418–26. 10.1021/bc00034a012 [DOI] [PubMed] [Google Scholar]
- 10.Venkatesan H, Greenberg MM. Improved utility of photolabile solid phase synthesis supports for the synthesis of oligonucleotides containing 3´-hydroxyl termini. J Org Chem. 1996;61:525–9. 10.1021/jo951550w [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Baky S, Allam K, Giese RW. Detection of electrophore-labeled DNA and albumin by gas chromatography: labile amide electrophoric release tags. Anal Chem. 1993;65:498–9. 10.1021/ac00052a031 [DOI] [PubMed] [Google Scholar]
- 12.Saha M, Saha J, Giese RW. 4-(Trifluoromethyl)-2,3,5,6-tetrafluorobenzyl bromide as a new electrophoric derivatizing reagent. J Chromatogr A. 1993;641:400–4. 10.1016/0021-9673(93)80158-5 [DOI] [PubMed] [Google Scholar]


