Speed of spoligotyping could be a benefit in the clinical setting.
Keywords: Bacterial Typing, molecular epidemiology, multidrug resistance, tuberculosis
Abstract
We evaluated the clinical usefulness of spoligotyping, a polymerase chain reaction–based method for simultaneous detection and typing of Mycobacterium tuberculosis strains, with acid-fast bacilli–positive slides from clinical specimens or mycobacterial cultures. Overall sensitivity and specificity were 97% and 95% for the detection of M. tuberculosis and 98% and 96% when used with clinical specimens. Laboratory turnaround time of spoligotyping was less than that for culture identification by a median of 20 days. In comparison with IS6110-based restriction fragment length polymorphism typing, spoligotyping overestimated the number of isolates with identical DNA fingerprints by ≈50%, but showed a 100% negative predictive value. Spoligotyping resulted in the modification of ongoing antimycobacterial treatment in 40 cases and appropriate therapy in the absence of cultures in 11 cases. The rapidity of this method in detection and typing could make it useful in the management of tuberculosis in a clinical setting.
The last decade has seen a dramatic resurgence in the incidence of tuberculosis throughout the world and an increased need for more rapid methods to diagnose and prevent dissemination of this disease (1). Well-equipped clinical laboratories can detect tuberculosis cases within 14 to 21 days by using liquid culturing systems such as BACTEC (Becton Dickinson, Sparks, MD, USA). Moreover, several studies have verified the usefulness of nucleic acid amplification–based methods for diagnosis of Mycobacterium tuberculosis infections in <24 hours (2–4). Concomitantly, recently characterized molecular markers for typing mycobacterial strains have greatly facilitated and improved the study of tuberculosis epidemiology (5–8).
Restriction fragment length polymorphism (RFLP) typing with insertion element IS6110 as a probe has become the most widely used method for differentiating strains of M. tuberculosis isolates (7,8). However, because the application of RFLP typing is restricted to mycobacterial cultures, 20–40 days are required before sufficient mycobacteria are available to obtain sufficient DNA needed for this method. This time restriction limits the usefulness of RFLP typing, especially in studying possible nosocomial transmission of tuberculosis in a clinical setting.
Spoligotyping, a new method for simultaneous detection and typing of M. tuberculosis complex bacteria, has been recently developed (9–11). This method is based on polymerase chain reaction (PCR) amplification of a highly polymorphic direct repeat locus in the M. tuberculosis genome. Results can be obtained from a M. tuberculosis culture within 1 day. Thus, the clinical usefulness of spoligotyping is determined by its rapidity, both in detecting causative bacteria and in providing epidemiologic information on strain identities. Implementing such a method in clinic settings would be useful in surveillance of tuberculosis transmission and in interventions to prevent further spread of this disease.
The aims of this study were to evaluate 1) the reliability of spoligotyping when used with clinical specimens, 2) the potential usefulness of the method in distinguishing M. tuberculosis from other nontuberculous mycobacteria (primarily M. avium), and 3) the feasibility and impact of spoligotyping in managing tuberculosis in clinical settings.
Patients and Methods
Specimen Collection
We conducted a 2-year survey of suspected cases of tuberculosis with spoligotyping of acid-fast bacilli (AFB)–positive specimens collected consecutively from January 2000 to December 2001 in the Microbiology Laboratory at L. Sacco Hospital in Milan. Three types of specimens were included. The first was material scraped from slides prepared from all Ziehl-Neelsen–positive clinical specimens, which were obtained from patients admitted to the hospital. These clinical specimens included sputum, bronchoalveolar lavage, bone marrow aspirate, feces, cerebrospinal fluid, and urine. The second was samples obtained from liquid culture medium containing growing mycobacteria (BACTEC, Becton Dickinson). The third was a mycobacterial colony grown on solid medium (Lowenstein-Jensen). The hospital microbiology laboratory conducted isolation (both in solid and liquid media), identification, and antimicrobial susceptibility testing by using standard methods (12) on all specimens. The results of spoligotyping were immediately provided to the physicians treating the patients.
Clinical Characteristics
Demographic and epidemiologic data were obtained from the medical records of all patients with AFB-positive specimens, including medical history of mycobacteriosis, HIV status, dates and results of mycobacterial smears, signs, symptoms, radiographs of patients with tuberculosis, and CD4+ cell counts (for HIV-infected patients). Data regarding the response time of the method used, possible variation in treatments following spoligotyping results, and patient clinical responses were also obtained.
Isolation of DNA
DNA was isolated from AFB-positive slides as previously described (13). Briefly, stained microscopic preparations were washed in xylol and absolute ethanol, scraped with a sterile blade, and collected in a microcentrifuge tube in 1× phosphate buffer. The samples were centrifuged for 10 min at 13,000 rpm. The pellets were resuspended in 100 μL lysis buffer (10 mmol/L Tris-HCl, 50 mmol/L KCl, 2.5 mmol/L MgCl2, 0.45% Tween 20, 0.45% Nonidet P40, and 10 mg/mL proteinase K) and incubated for 3 h at 56°C or overnight at 37°C. The samples were then incubated for 15 min at 95°C and centrifuged for 15 min at 13,000 rpm, and the supernatants were transferred to a new microcentrifuge tube and used in PCR. Mycobacteria were grown in culture, and their DNA was isolated as previously described (6).
RFLP Fingerprinting Analysis and Spoligotyping
Isolates of M. tuberculosis were genotyped by RFLP using the IS6110 probe as a genetic marker, as previously described by van Embden et al. (6). Spoligotyping was performed on genomic DNA by using the standard method described by Kamerbeek et al. (9). Control samples were used in these procedures as previously described (14).
Computer-assisted Analysis of Typing Patterns
Gel Compar software version 4.1 (Applied Maths, Kortrijk, Belgium) was used to compare the hybridization patterns obtained by spoligotyping and RFLP fingerprinting. The software clustered strains with the same genotypic pattern and defined similarity dendrograms joining the obtained clusters. The results of the analysis were compared with our database containing all DNA patterns derived from tuberculosis cases analyzed (the database contains RFLP data from >4,500 different isolates in our region).
Statistical Analysis
The sensitivity and specificity of spoligotyping in distinguishing M. tuberculosis from nontuberculous mycobacteria were calculated in comparison with culture results that excluded analysis of patients without culture confirmation. The sensitivity and specificity of spoligotyping in typing M. tuberculosis isolates were calculated in comparison with IS6110 clustering.
Results
Patient Characteristics
Three hundred fifty AFB-positive slides from 164 episodes of suspected mycobacteriosis in 148 patients were analyzed. One hundred seven slides were obtained from fresh material: sputum (n = 65), stool (n = 19), lymph node aspirate (n = 12), bronchoaspirate (n = 4), urine (n = 3), skin biopsy (n = 2), biliar liquid (n =1), and pericardial fluid (n = 1). One hundred five samples were obtained after growth of mycobacteria from liquid medium, and 138 samples derived from culture of different materials (mainly blood, but also sputum, bone marrow aspirate, cerebral spinal fluid, and others) were obtained after growth on solid medium. The characteristics of the 148 patients are shown in Table 1.
Table 1. Characteristics of 148 patients with acid-fast bacilli in biologic specimens*.
| Age, y | |
|---|---|
| Median (range) | 34 (3–88) |
| Mean | 39.3 |
| Sex, no. (%) | |
| Male | 105 (70.9) |
| Female | 43 (29.1) |
| Type of patient, no. (%) | |
| Infectious diseases | 124 (83.8) |
| Pneumology | 10 (6.8) |
| Internal medicine | 10 (6.8) |
| Other | 4 (2.7) |
| HIV status, no. (%) | |
| Negative | 52 (35.1) |
| Positive | 96 (64.9) |
| CD4+ cell count/μL† | |
| Median | 47.5 |
| Mean (range) | 105 (1–589) |
| Previous tuberculosis, no. (%) | 20 (13.5) |
| Previous MAC infection, no. (%) |
10 (6.8) |
| No tuberculosis or MAC infection, no. (%) | 118 (79.7) |
*MAC, Mycobacterium avium complex. †Data available for 90 of 96 HIV-infected patients.
Sensitivity and Specificity of Spoligotyping versus Culture
Culture confirmation was obtained in 317 (90.6%) of 350 AFB-positive slides from 138 of 164 episodes of suspected mycobacteriosis (Table 2). M. tuberculosis was isolated from 188 specimens from 77 patient episodes. Among these, isoniazid resistance was detected in 12 patients (15.6%), rifampin resistance in 11 patients (14.3%), streptomycin resistance in 4 patients (5.2%), and ethambutol resistance in 2 patients (2.6%). We also observed 6 patients with multidrug-resistant tuberculosis (resistance to at least isoniazid and rifampin). Fifty-six patients (72.7%) were infected with a strain susceptible to all 4 drugs.
Table 2. Comparison between spoligotyping and culture results in 350 acid-fast bacilli–positive samples.
| Mycobacteria grown in culture | No. episodes | Results of spoligotyping from clinical samples |
Results of spoligotyping from liquid medium |
Results of spoligotyping from solid medium |
Total |
||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | ||
| M. tuberculosis | 77 | 54 | 1 | 53 | 5 | 75 | 0 | 182 | 6 |
| M. avium | 28 | 0 | 15 | 0 | 20 | 2 | 31 | 2 | 66 |
| M. gordonae | 15 | 0 | 0 | 0 | 12 | 0 | 14 | 0 | 26 |
| M. xenopi | 8 | 0 | 2 | 1 | 5 | 1 | 6 | 2 | 13 |
| M. kansasii | 2 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 3 |
| M. chelonae | 2* | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 6 |
| Other | 6† | 1 | 1 | 0 | 4 | 1 | 4 | 2 | 9 |
| No growth | 26 | 9 | 21 | 1 | 2 | 0 | 0 | 10 | 23 |
| Subtotal | 64 | 43 | 55 | 50 | 79 | 59 | 198 | 152 | |
| Total | 164‡ | 107 | 105 | 138 | 350 | ||||
| Sensitivity, %§ | 98.2 (71.1–98.4) | 91.4 (75.7–90) | 100 | 96.8 (86.2–97) | |||||
| Specificity, %§ | 95.5 (67.7–97.7) | 97.7 (95.6–97.8) | 93.6 | 95.3 (88.4–96.1) | |||||
*Automated DNA sequencing rather than culture growth of M. chelonae confirmed one of these cases. †Other, 1 M. fortuitum, 1 M. asiaticum, and 4 nontyped mycobacteria. ‡16 patients had ≥2 episodes of mycobacterial infections. §Sensitivity and specificity were calculated without including the culture-negative specimens. Values in parentheses are ranges that include specimens with no growth.
One hundred ninety-eight of 350 AFB slides showed positive results by spoligotyping. Culture results confirmed the diagnosis of tuberculosis according to spoligotyping positivity in 182 (96.8%) of these 198 specimens. We did not observe definitive growth of mycobacteria in cultures from 10 patients. The PCR products obtained from 6 specimens of nontuberculous mycobacteria (2 M. xenopi, 2 M. fortuitum, and 2 M. avium) hybridized with M. tuberculosis–specific oligonucleotides, which indicated that that false-positive spoligotyping results were possible. However, mixed infections with 2 different mycobacteria cannot be ruled out.
The spoligotyping response was negative in specimens from 152 patients: 23 specimens showed no growth in culture, 123 were nontuberculous mycobacteria, and 6 showed growth characteristic of M. tuberculosis (Table 2). Three of these 6 false-negative spoligotyping results were from slides with very high concentrations of AFB, and a positive result was obtained when we repeated the test at a higher dilution (1:10).
In comparison with culture results, the sensitivity of spoligotyping was 98% for clinical specimens, 91% for slides obtained from liquid medium, and 100% for slides obtained directly from a mycobacterial colony on Lowenstein-Jensen solid medium. The corresponding specificities were 96% (clinical specimens), 98% (liquid medium), and 94% (solid medium), respectively (Table 2).
Clinical Application of Spoligotyping
Under optimal conditions, spoligotyping requires <24 hours for results. However, in the present study, response time was evaluated, taking into consideration routine processing time in the laboratory. The time from receipt of clinical specimens to obtaining spoligotyping results was 1–26 days (median 6 days). However, spoligotyping results from clinical specimens were obtained a median of 20 days (mean ± SD, 22.9 ± 18.6) sooner than those obtained by culture confirmation of tuberculosis and a median of 29 days (35.0 ± 25.2) sooner than those obtained by susceptibility testing. In contrast, RFLP typing results were obtained after a median of 75 days (range 24–160) (Table 3).
Table 3. Time required for obtaining results with clinical specimens by spoligotyping compared with 3 other methods*.
| Procedure | Days required, median (range) |
|---|---|
| Spoligotyping | 6 (1–26) |
| Culture confirmation | 28 (6–64) |
| Susceptibility testing | 37 (23–81) |
| RFLP typing | 75 (24–160) |
* RFLP, restriction fragment length polymorphism.
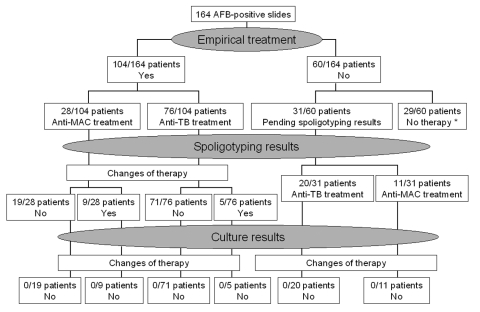
The use spoligotyping in determining treatment for 164 episodes of suspected mycobacteriosis was evaluated (Figure 1). In 25 episodes, patients did not begin antimycobacterial treatment because clinicians judged the AFB results to be not suggestive of true mycobacteriosis; none of these patients had clinical and radiologic features of tuberculosis (virtually all of these were infections with M. gordonae and M. xenopi isolates). Four patients died within a few days after admission without receiving any antimycobacterial drug.
Figure 1.
Clinical and therapeutic implications of spoligotyping results in treating suspected mycobacterial diseases. AFB, acid-fast bacilli; pts, patients; MAC, Mycobacterium avium complex; TB, tuberculosis. *Twenty-five patients did not begin treatment because they did not have clinical and radiologic features of tuberculosis. Four patients died within a few days after admission without receiving any antimycobacterial drug.
Clinicians waited until spoligotyping results were obtained before choosing the appropriate therapy for 31 other patients. Subsequently, 20 patients began standard antituberculosis regimens (isoniazid, rifampin, pyrazinamide, and/or ethambutol), whereas the 11 other patients began treatment for infection with M. avium (clarithromycin, ethambutol, rifabutin, and/or ciprofloxacin). In all cases, the choice of the treatment based on spoligotyping was not changed after culture and susceptibility test results were obtained.
Empiric antimycobacterial therapy was given to the remaining 104 patients before spoligotyping results were obtained. Seventy-six of these patients received antituberculosis treatment against M. tuberculosis infections, and 28 received therapy for infection with M. avium based on clinical presentation. Therapy was subsequently modified as a result of the spoligotyping results in 14 of these 104 patients. In 5 patients in whom spoligotyping results were negative and subsequent cultures showed nontuberculous mycobacteria, antituberculosis treatment was replaced with treatment for infection with M. avium. In contrast, in 8 patients in whom spoligotyping results were positive and subsequent cultures were positive for M. tuberculosis, treatment for infection with M. avium was replaced by antituberculosis treatment. In 1 patient, the homology of the spoligotyping pattern with patterns of other patients included in our data bank demonstrated that the isolate originated from an outbreak caused by multidrug-resistant strains. Consequently, therapy was modified to include second-line antituberculosis drugs, which previous in vitro susceptibility data had shown were active against the specific strains. Subsequent in vitro susceptibility data confirmed the spoligotyping results. False-positive spoligotyping results in 9 patients did not result in errors in prescribing treatment. These patients continued ongoing treatment, primarily on the basis of previous spoligotyping results or because of a positive response to treatment.
Analysis of Different Isolates from the Same Patient
We analyzed 34 spoligotyping-positive specimens from 6 patients with successive episodes of culture-confirmed tuberculosis (≥3 months apart). In these patients, spoligotyping was modified to distinguish relapses from new infections. After the samples were decoded, all episodes were classified as true relapses, and no new infections were detected. The banding patterns of the successive specimens matched those of their corresponding initial isolates. The subsequent RFLP results confirmed those obtained with spoligotyping.
Genotyping
The reproducibility of spoligotyping was demonstrated by the identity of results obtained with clinical samples and corresponding cultures from different anatomic sites in the same episode (18 patients), and from episodes of recurrent tuberculosis in the same person (5 patients). Thirty-nine distinct spoligotyping patterns were observed; 55% of the specimens were grouped into 10 clusters, and the others had unique spoligotypes. Sixty-one different RFLP patterns were seen in 64 isolates. Of these 61 patterns, 3 were shared by 2 isolates, while the remaining 58 patterns (95%) were observed in only 1 isolate.
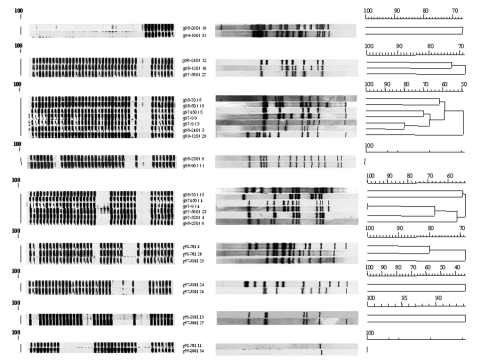
One of the 3 clusters identified by RFLP was a false cluster because it showed a 1-band pattern that correctly matched 2 different spoligotypes. In the remaining 2 RFLP clusters, complete concordance with spoligotyping was observed. The remaining 8 clusters detected by spoligotyping were not confirmed by RFLP analysis. Therefore, although it demonstrated 100% sensitivity, spoligotyping overestimated the number of clustered isolates by ≈50% (specificity 47.5%). Conversely, RFLP analysis had 100% specificity, but lower sensitivity. However, most of the isolates classified as clustered by spoligotyping but not by RFLP showed >50% similarity in their IS6110 patterns (Figure 2). Using spoligotyping of clinical samples without culture confirmation, we were able to diagnose an M. bovis infection and rapidly identify 2 cases of recurrent tuberculosis in patients with the same spoligotyping pattern in both followup specimens and the initial M. tuberculosis strain isolated several months earlier.
Figure 2.
Spoligotype dendrograms generated by clustered Mycobacterium tuberculosis strains after computer analysis compared with the corresponding dendrograms of IS6110 DNA fingerprints.
Discussion
Recent characterization of biologic markers for typing M. tuberculosis strains has greatly facilitated and improved tuberculosis epidemiology. IS6110 RFLP typing is the most widely used method for differentiating M. tuberculosis strains (15–19). However, RFLP analysis requires growth of mycobacterial colonies, which involves a consistent lag time between epidemiologic events and typing results. Thus, time remains a limiting factor in obtaining a highly effective method for epidemiologic surveys and preventing the spread of disease.
The development of new PCR-based typing methods (4,5,9,10,20–23) has allowed rapid mycobacterial identification to be combined with epidemiologic typing results. Thus, molecular epidemiologic information can be combined in the context of epidemic events and tuberculosis transmission. Spoligotyping appears to have the specific characteristics needed to satisfy these issues of epidemics and tuberculosis transmission (24–28). This method permits the concomitant identification and differentiation of M. tuberculosis strains and avoids the timing problems associated with the slow growth of these bacteria (10,11). However, although spoligotyping can be used with clinical specimens (9), the usefulness of any PCR-based typing method in the clinical setting has yet to be demonstrated. In addition, although previous studies have investigated the effect of specific methodologic issues on the performance of several typing techniques (20–22,24–30), we have demonstrated the usefulness of a PCR–based technique in the clinical setting for diagnostic purposes, as well as for epidemiologic studies of tuberculosis transmission.
We have also shown that spoligotyping can be satisfactorily used with clinical samples. The performance of spoligotyping was satisfactory with all clinical specimens used, and its specificity and sensitivity were 98% and 96%, respectively. These features are comparable with those of commercial PCR methods used for detecting M. tuberculosis from clinical samples (19). Moreover, the opportunity to combine rapid diagnostic information and molecular epidemiologic data represents an important advance in the epidemiologic control of tuberculosis.
Several molecular typing studies have compared different methods for M. tuberculosis typing. IS6110-based RFLP has been found to be more discriminative than direct repeat–based spoligotyping (20–31). Although our data confirm that spoligotyping vastly overestimates the number of clustered isolates, this method has a lower discriminatory power than IS6110-RFLP. However, spoligotyping also has a higher negative predictive value, thus enabling the clinician to exclude a particular clustered strain as a cause of infection, if known drug-resistant variants are present.
This study showed that spoligotyping can provide useful data to clinicians in different settings. Although co-infection with M. tuberculosis and nontuberculous mycobacteria cannot be ruled out, the ability to differentiate between M. tuberculosis and other mycobacteria was demonstrated in 27% of the patients. Some patients began therapy after evaluation of spoligotyping results, while others changed treatments because spoligotyping did not confirm the initial diagnosis. Time of response for spoligotyping was shorter than that of culture confirmation of tuberculosis by a median of 20 days and that of susceptibility results by a median of 29 days. In addition, a median of 6 days was needed for obtaining spoligotyping results, compared with 75 days for RFLP typing results.
Comparison of molecular typing patterns identified 6 patients with reactivation of tuberculosis caused by the same strain of M. tuberculosis, a finding that can differentiate between relapse and new infection in a new episode of tuberculosis. In 1 patient, we found that the spoligotyping pattern was identical to that of other strains that belonged to a cluster of multidrug-resistant tuberculosis. This observation resulted in the modification of antituberculosis treatment 34 days before susceptibility data were available. Moreover, information obtained by spoligotyping was relevant and useful in therapeutic management of ≈33% of the patients.
The clinical utility of spoligotyping may not be fully apparent by analyzing the results of this study, primarily because of the lack of clustered episodes of tuberculosis during the study period. However, spoligotyping would have been useful during a period or in a setting characterized by the emergence of M. tuberculosis outbreaks (8,32,33). We have also shown that the usefulness of spoligotyping is increased when results are compared with data on other tuberculosis patients and a DNA database on M. tuberculosis strains.
In conclusion, this study underscores the need to implement rapid molecular epidemiologic methods in managing tuberculosis epidemics. We have shown that spoligotyping is a useful method for screening and epidemiologic control of tuberculosis dissemination, particularly when results are required quickly, such as in outbreaks, or in the management of transmission of multidrug-resistant tuberculosis, especially in restricted high-risk situations such as prisons, schools, and hospitals.
Acknowledgments
We thank the patients of the infectious diseases ward and outpatient clinic at L. Sacco Hospital for their participation; the staff of the Department of Infectious Diseases, L. Sacco Hospital, and the Institute of Infectious Disease, University of Milan, including Cecilia Paoli, Patrizia Franza, and Cristina Bessi, for their assistance; Carmela Perna for her contribution in coordinating the study; Michael Alan Rosen and Shane Fogden for their assistance in the English revision of the text; and Annalisa Angelantoni for her continuous support.
This work was supported by grants from the National Institute of Health, Minister of Health, 1% 2000, project 0AL/F and RF/101, Rome, Italy; and the AHSI Company, Milan, Italy.
Biography
Dr. Gori is a senior physician at the Institute of Infectious Diseases and Tropical Medicine, L. Sacco Hospital, University of Milan. His primary research interests include molecular diagnostics and molecular epidemiology of tuberculosis for clinical applications.
Footnotes
Suggested citation for this article: Gori A, Bandera A, Marchetti G, Esposti AD, Catozzi L, Nardi GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis [serial on the Internet]. 2005 Aug [date cited]. http://dx.doi.org/10.3201/eid1108.040982
References
- 1.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, et al. The growing burned of tuberculosis. Global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–21. 10.1001/archinte.163.9.1009 [DOI] [PubMed] [Google Scholar]
- 2.Brisson-Noël A, Gicquel B, Lecossier D, Lévy-Frébault V, Nassif X, Hance AJ. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet. 1989;2:1069–71. 10.1016/S0140-6736(89)91082-9 [DOI] [PubMed] [Google Scholar]
- 3.Clarridge JE III, Shawar RM, Shinnick TM, Plikaytis BB. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J Clin Microbiol. 1993;31:2049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenach KD, Cave MD, Bates JH, Crawford JT. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–81. 10.1093/infdis/161.5.977 [DOI] [PubMed] [Google Scholar]
- 5.van Soolingen D, de Haas PEW, Hermans PWM, Groenen PMA, van Embden JDA. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behr MA, Small PM. Molecular fingerprinting of Mycobacterium tuberculosis: how can it help the clinician? Clin Infect Dis. 1997;25:806–10. 10.1086/515550 [DOI] [PubMed] [Google Scholar]
- 8.Franzetti F, Gori A, Iemoli E, Meraviglia P, Mainini F, Quirino T, et al. Outcome of multidrug resistant tuberculosis in HIV-infected patients. Clin Infect Dis. 1999;29:553–60. 10.1086/598633 [DOI] [PubMed] [Google Scholar]
- 9.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenen PMA, Bunschoten AE, van Soolingen D, van Embden JDA. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–65. 10.1111/j.1365-2958.1993.tb00976.x [DOI] [PubMed] [Google Scholar]
- 11.Hermans PWM, van Soolingen D, Bik EM, de Haas PEW, Dale JW, van Embden JDA. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metchock BG, Nolte FS, Wallance RJ Jr. Mycobacterium. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. 7th ed. Washington: American Society for Microbiology; 1999. p. 399–437. [Google Scholar]
- 13.Rossi MC, Gori A, Zehender G, Marchetti G, Ferrario G, de Maddalena C, et al. A PCR-colorimetric microwell plate hybridization assay for detection of M. tuberculosis and M. avium from culture samples and Ziehl-Neelsen positive smears. J Clin Microbiol. 2000;38:1772–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrario G, Gori A, Rossi A, Catozzi L, Molteni C, Marchetti G, et al. PCR-hybridization assay for Mycobacterium avium complex: optimization of detection in peripheral blood from humans. J Clin Microbiol. 2001;39:1638–43. 10.1128/JCM.39.4.1638-1643.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck-Sague C, Dooley SW, Hutton MD, Otten J, Breeden A, Crawford JT, et al. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. JAMA. 1992;268:1280–6. 10.1001/jama.1992.03490100078031 [DOI] [PubMed] [Google Scholar]
- 16.Pearson ML, Jereb JA, Frieden TR, Crawford JT, Davis BJ, Dooley SW, et al. Nosocomial transmission of multidrug-resistant Mycobacterium tuberculosis. Ann Intern Med. 1992;117:191–6. [DOI] [PubMed] [Google Scholar]
- 17.Edlin BR, Tokars JL, Grieco MH, Crawford JT, Williams J, Sordillo EM, et al. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N Engl J Med. 1992;326:1514–21. 10.1056/NEJM199206043262302 [DOI] [PubMed] [Google Scholar]
- 18.Coninx R, Mathieu C, Debacker M, Mirzoev F, Ismaelov A, de Haller R, et al. First-line tuberculosis therapy and drug-resistant Mycobacterium tuberculosis in prisons. Lancet. 1999;353:969–73. 10.1016/S0140-6736(98)08341-X [DOI] [PubMed] [Google Scholar]
- 19.Drobniewski FA, Gibson A, Ruddy M, Yates MD. Evaluation and utilization as a public health tool of a national molecular epidemiological tuberculosis outbreak database within the United Kingdom from 1997 to 2001. J Clin Microbiol. 2003;41:1861–8. 10.1128/JCM.41.5.1861-1868.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz R, Kremer K, de Has PE, Gomez RI, Marrero A, Valdivia JA, et al. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994–June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis. 1998;2:743–50. [PubMed] [Google Scholar]
- 21.Sola C, Horgen L, Maisetti J, Devallois A, Goh KS, Rastogi N. Spoligotyping followed by double-repetitive-element PCR as rapid alternative to IS6110 fingerprinting for epidemiological studies of tuberculosis. J Clin Microbiol. 1998;36:1122–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goyal M, Saunders NA, van Embden JDA, Young DB, Shaw RJ. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci U S A. 2001;98:1901–6. 10.1073/pnas.98.4.1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goguet de la Salmoniére YO, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranaz A, Liebana E, Mateos A, Dominguez L, Cousins D. Restriction fragment length polymorphism and spacer oligonucleotide typing: a comparative analysis of fingerprinting strategies for Mycobacterium bovis. Vet Microbiol. 1998;61:311–24. 10.1016/S0378-1135(98)00192-8 [DOI] [PubMed] [Google Scholar]
- 26.Roring S, Brittain D, Bunschoten AE, Hughes MS, Skuce RA, van Embden JD, et al. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet Microbiol. 1998;61:111–20. 10.1016/S0378-1135(98)00178-3 [DOI] [PubMed] [Google Scholar]
- 27.Bauer J, Andersen AB, Kremer K, Miorner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML, et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–7. 10.1001/jama.1996.03530300036037 [DOI] [PubMed] [Google Scholar]
- 29.Moss AR, Alland D, Telzak E, Hewlett D Jr, Sharp V, Chiliade P, et al. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuberc Lung Dis. 1997;1:115–21. [PubMed] [Google Scholar]
- 30.Alito A, Morcillo N, Scipioni S, Dolmann A, Romano MI, Cataldi A, et al. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J Clin Microbiol. 1999;37:788–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soini H, Pan X, Amin A, Graviss EA, Siddiqui A, Musser JM. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moro ML, Gori A, Errante I, Infuso A, Franzetti F, Sodano L, et al. An outbreak of multidrug resistant tuberculosis involving HIV-infected patients of two hospitals in Milan Italy. AIDS. 1998;12:1095–102. 10.1097/00002030-199809000-00018 [DOI] [PubMed] [Google Scholar]
- 33.Moro ML, Salamina G, Gori A, Penati V, Sacchetti R, Mezzetti F, et al. Two-year population-based molecular epidemiological study of tuberculosis transmission in the metropolitan area of Milan, Italy. Eur J Clin Microbiol Infect Dis. 2002;21:114–22. 10.1007/s10096-001-0664-2 [DOI] [PubMed] [Google Scholar]