Abstract
Neuroblastoma is a childhood cancer of the sympathetic nervous system that accounts for approximately 10% of all paediatric oncology deaths1,2. To identify genetic risk factors for neuroblastoma, we performed a genome-wide association study (GWAS) on 2,251 patients and 6,097 control subjects of European ancestry from four case series. Here we report a significant association within LIM domain only 1 (LMO1) at 11p15.4 (rs110419, combined P = 5.2 × 10−16, odds ratio of risk allele = 1.34 (95% confidence interval 1.25–1.44)). The signal was enriched in the subset of patients with the most aggressive form of the disease. LMO1 encodes a cysteine-rich transcriptional regulator, and its paralogues (LMO2, LMO3 and LMO4) have each been previously implicated in cancer. In parallel, we analysed genome-wide DNA copy number alterations in 701 primary tumours. We found that the LMO1 locus was aberrant in 12.4% through a duplication event, and that this event was associated with more advanced disease (P < 0.0001) and survival (P = 0.041). The germline single nucleotide polymorphism (SNP) risk alleles and somatic copy number gains were associated with increased LMO1 expression in neuroblastoma cell lines and primary tumours, consistent with a gain-of-function role in tumorigenesis. Short hairpin RNA (shRNA)-mediated depletion of LMO1 inhibited growth of neuroblastoma cells with high LMO1 expression, whereas forced expression of LMO1 in neuroblastoma cells with low LMO1 expression enhanced proliferation. These data show that common polymorphisms at the LMO1 locus are strongly associated with susceptibility to developing neuroblastoma, but also may influence the likelihood of further somatic alterations at this locus, leading to malignant progression.
Multiple somatically acquired chromosomal rearrangements, such as focal amplification of the MYCN oncogene or deletions at chromosome arms 1p or 11q, are each associated with an aggressive neuroblastoma phenotype2. Although these somatically acquired genomic alterations are of clinical use as prognostic biomarkers, until recently little was known about the constitutional genetic events that initiate tumorigenesis. Highly penetrant gain-of-function mutations in the anaplastic lymphoma kinase (ALK) tyrosine kinase domain were recently identified as the major cause of familial neuroblastoma, and somatic mutations in this gene implicate it as a target for therapeutic intervention3–6. In addition, a neuroblastoma GWAS identified common SNPs at 6p22 as being associated with susceptibility to aggressive neuroblastoma in sporadic cases7; follow-up association analysis on the clinically relevant group of patients with an aggressive tumour phenotype indicated that common SNPs within BARD1 also function as susceptibility variants8. Finally, our GWAS has also identified a common copy number variation at 1q21.1 being highly associated with neuroblastoma and probably playing a role in early tumorigenesis through disruption of a novel neuroblastoma breakpoint family gene (NBPF23)9. Taken together, it has become clear that the embryonal cancer neuroblastoma is genetically heterogeneous, and initiation of sporadically occurring disease requires multiple interacting genetic factors, including both sequence and copy number variants.
To identify additional genetic risk factors, we expanded our previous GWAS and analysed 1,627 neuroblastoma patients accrued through the North American-based Children’s Oncology Group with 3,254 genetically matched control subjects of European ancestry (see Supplementary Methods). All subjects were genotyped using the Illumina HumanHap550 BeadChip with over 550,000 SNP markers; the genomic control inflation factor was 1.08 (Supplementary Fig. 1). Clusters of SNPs from three genomic loci reached genome-wide significance (P < 5 × 10−8; Fig. 1a), including two SNPs within FLJ22536/FLJ44180 at the 6p22 locus (P values range from 2.46 × 10−14 to 3.25 × 10−13; Supplementary Table 1), nine SNPs within or nearby BARD1 at the 2q35 locus (P values range from 3.05 × 10−13 to 9.69 × 10−9; Supplementary Table 2), each previously reported, and two SNPs within LMO1 (LIM domain only 1), a newly identified neuroblastoma susceptibility locus at 11p15.4 (P values range from 5.12 × 10−10 to 2.83 × 10−8; Table 1 and Fig. 1b). Closer examination of the LMO1 locus identified a total of four SNPs that show strong association signals (P < 1 × 10−4) with neuroblastoma (Table 1), which are in a moderate degree of linkage disequilibrium (Supplementary Fig. 2). We then examined each of the most significant SNPs from the 2q35, 6p22, 11p15.4 susceptibility loci and the 1q21.1 copy number variation. However, we did not find evidence for epistasis (Supplementary Tables 3 and 4), indicating that these susceptibility loci increase disease risk independently.
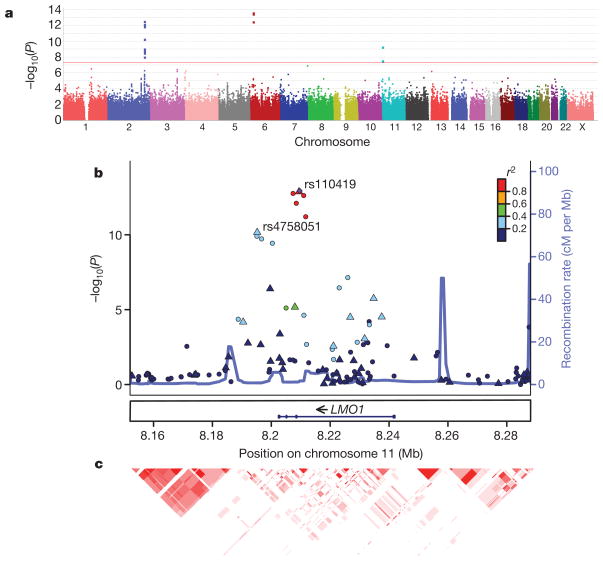
Figure 1. Discovery of LMO1 at 11p15.4 as a neuroblastoma susceptibility locus.
a, Manhattan plot of GWAS results from the discovery cases series, with the red horizontal line representing genome-wide significance threshold (P < 5 × 10−8). b, Genomic position (National Center for Biotechnology Information build 36) of genotyped (triangles) and imputed (circles) SNPs. The P values are calculated by combining discovery and replication case series with whole-genome genotypes, and SNPs are coloured based on their correlations with rs110419 (purple diamond). Estimated recombination rates from the HapMap data are overlaid. c, Degree of linkage disequilibrium between SNPs (as r2 values) is represented by red colour intensity in the corresponding cells.
Table 1.
Significantly associated SNPs at the LMO1 locus on 11p15.4
| SNP† | Risk/non-risk allele | Discovery (HumanHap550)* |
US replication (Human610)* |
UK replication (TaqMan)* |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency of cases (n = 1,627) | Frequency of controls (n = 3,254) | P‡ | Frequency of cases (n = 190) | Frequency of controls (n = 1,507) | P‡ | Frequency of cases (n = 253) | Frequency of controls (n = 845) | P‡ | ||
| rs4758051 | G/A | 0.51 | 0.45 | 2.8 × 10−8 | 0.55 | 0.45 | 2.1 × 10−4 | 0.51 | 0.45 | 0.039 |
| rs10840002 | A/G | 0.42 | 0.37 | 6.0 × 10−6 | 0.44 | 0.38 | 0.019 | 0.37 | 0.36 | 0.61 |
| rs110419 | A/G | 0.55 | 0.49 | 5.1 × 10−10 | 0.61 | 0.49 | 1.0 × 10−5 | 0.53 | 0.48 | 0.057 |
| rs204938 | C/T | 0.49 | 0.44 | 1.2 × 10−5 | 0.50 | 0.45 | 0.058 | 0.50 | 0.44 | 0.032 |
| SNP† | Risk/non-risk allele | Italian replication (TaqMan)* |
Combined
|
|||
|---|---|---|---|---|---|---|
| Frequency of cases (n = 181) | Frequency of controls (n = 491) | P‡ | CMH§P | CMH OR (95% confidence interval)|| | ||
| rs4758051 | G/A | 0.45 | 0.42 | 0.45 | 1.4 × 10−11 | 1.28 (1.19–1.37) |
| rs10840002 | A/G | — | — | — | 8.5 × 10−7 | 1.21 (1.12–1.30) |
| rs110419 | A/G | 0.49 | 0.41 | 0.004 | 5.2 × 10−16 | 1.34 (1.25–1.44) |
| rs204938 | C/T | — | — | — | 1.7 × 10−7 | 1.22 (1.13–1.31) |
No deviations from Hardy–Weinberg equilibrium were observed (P > 0.001) in all cohorts.
SNP: r2 in controls between rs110419 and each of rs4758051, rs10840002 and rs204938 was 0.30, 0.17 and 0.29, respectively.
P values were calculated by allelic test.
CMH, Cochran–Mantel–Haenszel test.
OR, odds ratio of risk allele.
To replicate our findings, we examined the association results from an independent case series of 190 patients from the Children’s Oncology Group and 1,507 control subjects, all of whom were genotyped on the Human610-Quad arrays. All four LMO1 SNPs identified in the discovery effort showed the same direction of association in this replication cohort, with P values ranging from 1.01 × 10−5 to 0.058. To seek additional evidence of replication, we performed quantitative PCR-based genotyping of these four SNPs in a third independent case series from UK, as well as the two most significant SNPs in a fourth independent case series from Italy. Combined analysis by the Cochran–Mantel–Haenszel method demonstrated that two of the four SNPs had P values that extend well beyond the genome-wide significance threshold (Table 1). Additionally, using the two cohorts with whole-genome genotype data (discovery cohort and US replication cohort), we performed genotype imputation at 11p15.4 and identified six additional genome-wide significant markers, the most significant being rs110420 (P = 1.17 × 10−13), which is in complete linkage disequilibrium (r2 = 1 in HapMap CEU subjects (Utah residents with ancestry from northern and western Europe) with the genotyped marker rs110419 (Fig. 1c and Supplementary Table 5).
We next determined if the LMO1 genotypes were associated with a particular clinical phenotype and/or patient survival. Similar to the association pattern observed for the 6p22 and 2q35 (BARD1) loci7,8, the risk alleles of LMO1 were significantly associated with metastatic disease (P = 0.0040), advanced age (greater than 1 year, P < 0.0001) and a high-risk status by Children’s Oncology Group criteria for treatment stratification2 (P = 0.0010; Supplementary Tables 6 and 7). Consistent with this observation, the rs110419 risk allele was associated with decreased event-free survival (P = 0.0085; Supplementary Table 8 and Supplementary Fig. 3) and overall survival (P = 0.0217; Supplementary Fig. 4). Taken together, these data suggest that common germline variants at LMO1 are associated not only with predisposition to develop neuroblastoma, but also with a predilection to develop the more aggressive form of the disease. They emphasize that LMO1 genetic variations are associated with a particular neuroblastoma phenotype; however, this does not indicate that these variants have prognostic significance for an individual with neuroblastoma.
The LMO1 gene encodes a cysteine-rich transcriptional regulator with two LIM zinc-binding domains that is mainly expressed in the nervous system10. LMO1 belongs to a protein superfamily encoded by four genes, including LMO1, LMO2, LMO3 and LMO4. Multiple lines of evidence, including chromosomal translocation events and mouse models, strongly implicate this gene family in the aetiology of human cancer11–14. Most provocatively, retroviral insertion of the corrective gene for X-linked severe combined immunodeficiency into the LMO2 locus resulted in T-cell leukaemias in several participants in gene therapy trials15. LMO4 represses the transcription of BRCA1, and dys-regulation of LMO4 expression has been implicated in the breast car-cinogenesis16,17. Finally, LMO3 has been shown to act as an oncogene in neuroblastoma through the neuronal transcription factor HEN218. We therefore postulated that the common variants at the 11p15.4 locus discovered here may increase disease risk through a cis-acting effect on the regulation of expression or function of LMO1, but we cannot exclude the potential for trans-acting influences on loci distant from the discovered common variants.
We next examined tumour DNA genotyped on the Illumina SNP arrays for 701 neuroblastomas using a detection algorithm for copy number designed for tumour samples19. We detected relative segmental gain (copy number changes at a given locus relative to whole-genome copy number changes) at LMO1 in 87 out of 701 tumours (12.4%); this was particularly enriched in the high-risk group where the GWAS signal was most robust (Supplementary Fig. 5a). Most tumours with 11p gain showed a duplication of the entire chromosome p arm, but four tumours (approximately 5%) showed focal gain restricted to 11p15 including the LMO1 locus (Supplementary Fig 5b). These data demonstrate that LMO1 is one of many genes showing somatic copy number gain on 11p, and here we used the GWAS data to prioritize it as a potential target of this somatically acquired chromosomal rearrangement.
We next examined whether somatic LMO1 alterations were associated with neuroblastoma clinical phenotype and survival of patients (Supplementary Table 9). Gain of LMO1 was significantly more common in tumours from patients with metastatic disease (P < 0.0001), advanced age (greater than 1 year, P < 0.0001), unfavourable pathological grade (P = 0.0013) and Children’s Oncology Group high-risk classification (P < 0.0001). Gain of 11p was rarely observed in the MYCN amplified cases (Supplementary Table 9). Despite the strong association of 11p gain in cases without MYCN amplification, a known powerful adverse prognostic factor1, LMO1 gain was associated with decreased overall survival of patients (P = 0.041) (Supplementary Table 10 and Supplementary Figs 6 and 7).
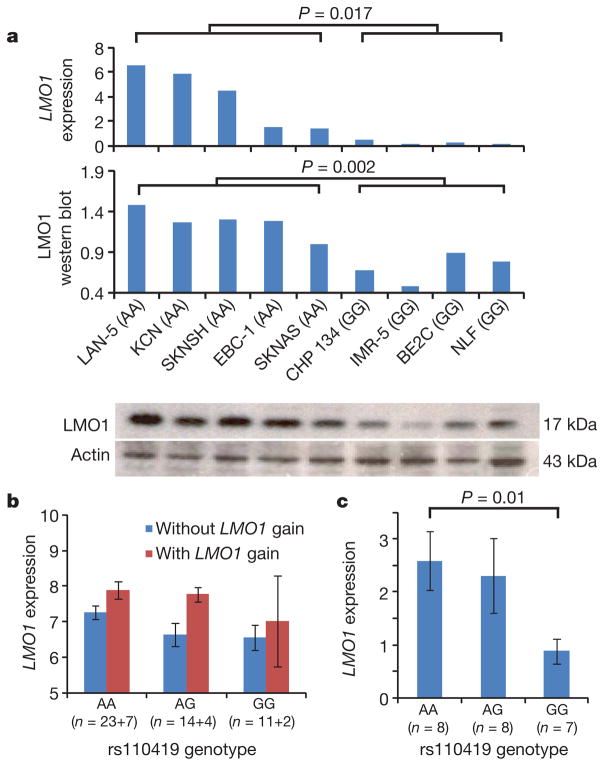
To investigate how the neuroblastoma-associated LMO1 alleles may contribute to tumour initiation and/or clinical phenotype, we next genotyped a set of human neuroblastoma-derived cell lines with Illumina SNP arrays, and measured messenger RNA (mRNA) and protein expression levels on the subset of lines without copy number changes at 11p to avoid the influence of somatic DNA alterations on gene expression. Cell lines with diploid 11p status and harbouring homozygous risk alleles showed significantly higher LMO1 mRNA and protein expression than those with homozygous non-risk alleles (Fig. 2a and Supplementary Table 11). This trend held in an expanded set of 25 neuroblastoma cell lines with variable 11p status (Supplementary Fig. 8). To determine if this correlation existed in diagnostic tumour tissues, we next examined mRNA expression levels on a whole-genome Affymetrix expression microarray20 in a subset of 61 neuroblastoma primary tumours from patients whose blood samples and primary tumours had both been genotyped on the Illumina SNP arrays. Among these 61 tumours, 13 harboured somatic gain of 11p. Considering both somatic and germline genotypes in the same linear regression model, we detected an association between LMO1 copy number gains and increased LMO1 expression (P = 0.02; Fig. 2b and Supplementary Table 12), as well as an association between rs110419 risk alleles and increased LMO1 expression (P = 0.022; Fig. 2b). To refine the genotype-expression relationships further, we subsequently used quantitative PCR to measure LMO1 expression in an additional set of 23 tumours without LMO1 gain. We confirmed that the rs110419 risk allele is associated with LMO1 expression (P = 0.01), independent of copy number changes (Fig. 2c). To determine whether a regulatory variant exists at a narrow promoter region of LMO1, we performed Sanger sequencing in 20 neuroblastoma cell lines but did not detect any potential causal variant (Supplementary Table 13). Examination of the 1000 Genomes Project data identified over 300 SNPs within or surrounding LMO1 that are in moderate to strong linkage disequilibrium (D′ > 0.5) with rs110419 (Supplementary Table 14); however, fine mapping of this region through resequencing will be required to identify whether any are causal cis-regulatory variants. Subsequent experimentation will be required to determine if causal DNA variations directly impact LMO1 expression, and if somatic copy-number gain indeed is targeting LMO1 for further increased expression in tumour cells.
Figure 2. LMO1 germline genotypes and somatic copy number gains are associated with mRNA and protein expression.
a, LMO1 mRNA and protein expression in nine human neuroblastoma-derived cell lines are highly correlated with rs110419 genotype. b, Microarray-based expression profiling on 61 primary tumours confirms that LMO1 gene expression is associated with both LMO1 gain (P = 0.02, t-test) and risk genotypes (P = 0.022, linear regression). c, Quantitative PCR-based expression profiling of an independent set of primary neuroblastomas without LMO1 gain confirms the same association. Error bars, s.e.m.
As our germline and somatic genomic analyses implicated LMO1 as a neuroblastoma oncogene, we next sought to determine the functional consequences of LMO1 depletion or overexpression in a genotype- and expression-specific manner. First, after lentiviral-based shRNA infection of neuroblastoma cell lines, we were able to recover stable clones with 45–63% depletion of LMO1 mRNA and protein (Fig. 3e). Cells with the homozygous neuroblastoma-associated genotype and high LMO1 expression showed significantly decreased proliferation compared with mock-infected controls (Fig. 3a, b), whereas cells with homozygous non-risk alleles showed little phenotypic effect (Fig. 3c, d). Finally, to determine the cellular phenotypes of forced overexpression of LMO1, we stably overexpressed LMO1 with approximately fourfold higher levels in the SK-N-BE2C cell line with low de novo LMO1 expression, and detected significantly enhanced proliferation (Fig. 3f). Therefore it appears that inhibition of LMO1 in cells expressing high levels of LMO1 or activation of LMO1 in cells with low levels of LMO1 leads to pronounced phenotypes. Taken together, these data suggest that LMO1 may function as an oncogene in a subset of human neuroblastomas.
Figure 3. Genetic manipulation of LMO1 expression in neuroblastoma cell line models influences proliferative phenotype in an expression-specific manner.
a–d, In cells with neuroblastoma risk alleles and higher LMO1 expression levels, LMO1 knockdown leads to inhibition of cellular proliferation. e, LMO1 knockdown as measured by quantitative reverse-transcription PCR and western blot for experiments a–d. f, In SK-N-BE2C cells with non-risk alleles and low LMO1 expression levels, forced overexpression of LMO1 leads to enhanced cellular proliferation. Approximate fourfold overexpression of LMO1 RNA and protein are shown. Error bars, s.e.m.
In conclusion, here we have identified germline sequence variants at the LMO1 locus that are robustly associated with neuroblastoma. We have applied an integrative genomics approach to demonstrate that common genetic polymorphisms associated with cancer predisposition may also mark regions of the genome prone to somatic alterations influencing tumour progression. Our data suggest that GWAS studies can identify previously undiscovered oncogenic drivers of a malignant phenotype, especially when they occur in a region of the genome involved in large segmental rearrangements impacting hundreds of genes. In paediatric cancers such as neuroblastoma, the real translational potential of GWAS efforts may be in discovering therapeutic targets and predictive biomarkers of tumour aggressiveness.
METHODS SUMMARY
All genome-wide SNP genotyping for the discovery cohorts was performed using the Illumina HumanHap550 BeadChip at the Center for Applied Genomics at the Children’s Hospital of Philadelphia. Multi-dimensional scaling was performed using PLINK version 1.06 on a subset of SNPs not in linkage disequilibrium to identify subjects of European ancestry, and all control subjects were genetically matched to patients. The first replication case series was genotyped by Illumina Human610 BeadChip, yet two additional replication case series were genotyped by TaqMan. Genotype imputation was performed by MACH (http://www.sph.umich.edu/csg/abecasis/MaCH/) on discovery and replication case series with whole-genome genotypes. Alteration calls in tumour copy number were generated from data of SNP signal intensity by the OverUnder19. Survival analyses used the methods of Kaplan and Meier, with standard errors following the methods of Peto et al.21. For gene expression profiling by Affymetrix U95Av2 microarrays, the expression measures for each probe set was extracted and normalized using robust multi-array average protocols from raw CEL files. Association tests on genotype and expression were performed on log-transformed expression values by linear regression or t-test. For quantitative PCR on LMO1, TaqMan probes were purchased from Applied Biosystems with assay identity Hs00231133_m1. Relative expression of the target gene was determined by normalization to HPRT1 using a standard curve method with ten serial dilutions according to the manufacturer’s instructions. All quantitative PCR reactions were performed in triplicate with an ABI PrismTM 7900HT Sequence Detection System (Applied Biosystems). For the LMO1 knockdown experiments, the lentiviral particles for shRNA knockdown were purchased from Santa Cruz, including copGFP Control Lentiviral Particles (catalogue number sc-108084) and LMO1 shRNA(h) Lentiviral Particles (catalogue number sc-38025-v). Pooled clones of SK-N-BE2C cells with LMO1 overexpression were created through stable transfection of full-length LMO1 complementary DNA in pCDNA3.1 as previously described22.
Supplementary Material
Acknowledgments
We acknowledge the Children’s Oncology Group for providing most blood and tumour specimens and clinical and outcome data (U10-CA98543 and U10-CA98413) from neuroblastoma patients. We thank G. P. Tonini for providing neuroblastoma DNA samples in the Italian replication cohort. This work was supported in part by National Institutes of Health grant R01-CA124709 (to J.M.M.), the Giulio D’Angio Endowed Chair (J.M.M.), the Alex’s Lemonade Stand Foundation (J.M.M.), the Evan Dunbar Foundation (J.M.M.), the Rally Foundation (J.M.M.), Andrew’s Army Foundation (J.M.M.), the Abramson Family Cancer Research Institute (J.M.M.), a Howard Hughes Medical Institute Research Training Fellowship (K.B.), a fellowship from Associazione Oncologia Pediatrica e Neuroblastoma (M.C.), a Research Development Award from the Cotswold Foundation (H.H.), UL1-RR024134-03 (H.H.) and an Institutional Development Award to the Center for Applied Genomics from the Children’s Hospital of Philadelphia (H.H.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.H. and J.M.M. conceived the study, guided interpretation of results and helped preparation of the manuscript. K.W., H.Z. and C.H. performed SNP association analysis. K.W., S.J.D., E.F.A. and J.J. performed gene expression and copy number analysis. C.W. and K.B. performed PCR validation of gene expression data. C.W., R.W.S., K.B., P.A.M., S.J.D. and K.A.C. performed and/or analysed shRNA transfection and LMO1 overexpression experiments. N.S. and H.S. generated viral construct for human LMO1 complementary DNA. M.C. and A.I. performed the replication study on the Italian case series, and N.R. performed the replication study on the UK case series. P.W.M. and W.B.L. performed outcome and clinical covariate analyses on the Children’s Oncology Group samples. C.H., C.K., E.F., M.G., W.G. and R.C. generated the genotyping data. L.N. and M.D. helped with data analysis. S.F.A.G., Y.P.M., H.L. and M.D. advised on data interpretation. K.W. drafted the manuscript; H.H., J.M.M. and other authors edited it.
Author Information Microarray data are deposited in the GEO database under accession number GSE3960. The genotypic and phenotypic information from this study is deposited in dbGaP (www.ncbi.nlm.gov/gap) under accession number phs000124.v2.p1. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 6.George RE, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maris JM, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capasso M, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nature Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabbitts TH. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 1998;12:2651–2657. doi: 10.1101/gad.12.17.2651. [DOI] [PubMed] [Google Scholar]
- 12.Rabbitts TH, et al. The effect of chromosomal translocations in acute leukemias: the LMO2 paradigm in transcription and development. Cancer Res. 1999;59:1794s–1798s. [PubMed] [Google Scholar]
- 13.Fisch P, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 14.Neale GA, Rehg JE, Goorha RM. Disruption of T-cell differentiation precedes T-cell tumor formation in LMO-2 (rhombotin-2) transgenic mice. Leukemia. 1997;11(suppl 3):289–290. [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 16.Sum EY, et al. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- 17.Visvader JE, et al. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoyama M, et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 19.Attiyeh EF, et al. Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy. Genome Res. 2009;19:276–283. doi: 10.1101/gr.075671.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 21.Peto R, et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. analysis and examples. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeki N, et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene. 2007;26:6488–6498. doi: 10.1038/sj.onc.1210475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.