Abstract
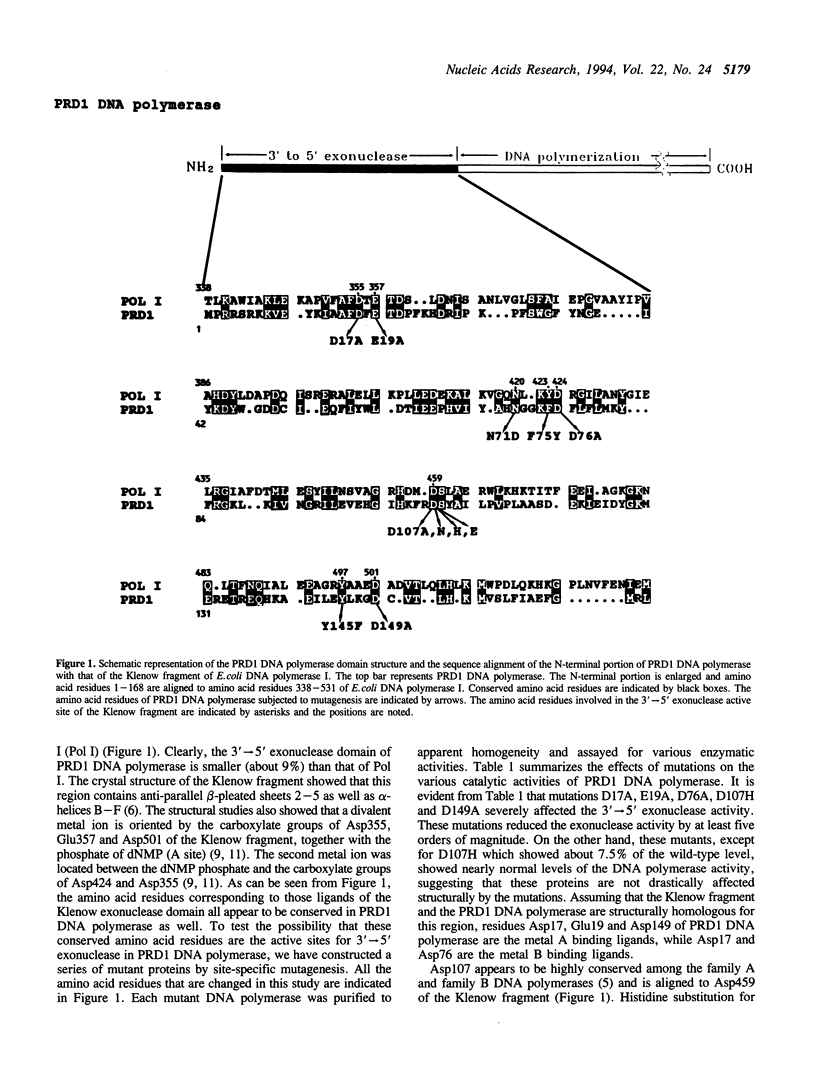
In order to establish the evolutionary relationship between the family A and B DNA polymerases, we have closely compared the 3'-->5' exonuclease domains between the Klenow fragment of E.coli DNA polymerase I (a family A DNA polymerase) and the bacteriophage PRD1 DNA polymerase, the smallest member of the DNA polymerase family B. Although the PRD1 DNA polymerase has a smaller 3'-->5' exonuclease domain, its active sites appear to be very similar to those of the Klenow fragment. Site-directed mutagenesis studies revealed that the residues important for the 3'-->5' exonuclease activity, particularly metal binding ligands for the Klenow fragment, are all conserved in the PRD1 DNA polymerase as well. The metal binding ligands are also essential for the strand-displacement activity of the PRD1 DNA polymerase. Based on these results and the studies by others in various systems, we conclude that family A and B DNA polymerases, at least in the 3'-->5' exonuclease domain, are structurally as well as evolutionarily related.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beese L. S., Derbyshire V., Steitz T. A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993 Apr 16;260(5106):352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Blasco M. A., Salas M. A general structure for DNA-dependent DNA polymerases. Gene. 1991 Apr;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Lázaro J. M., Martín G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989 May 25;264(15):8935–8940. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braithwaite D. K., Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993 Feb 25;21(4):787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Almassy R. J., Hostomska Z., Ferre R. A., Hostomsky Z. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994 Mar 25;76(6):1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Foury F., Vanderstraeten S. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 1992 Jul;11(7):2717–2726. doi: 10.1002/j.1460-2075.1992.tb05337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. W., Nossal N. G., Capson T. L., Benkovic S. J. Construction and characterization of a bacteriophage T4 DNA polymerase deficient in 3'-->5' exonuclease activity. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura R. K., Roop B. C. Characterization of DNA polymerase induced by bacteriophage T5 with DNA containing single strand breaks. J Biol Chem. 1976 Apr 10;251(7):2168–2174. [PubMed] [Google Scholar]
- Gibbs J. S., Weisshart K., Digard P., deBruynKops A., Knipe D. M., Coen D. M. Polymerization activity of an alpha-like DNA polymerase requires a conserved 3'-5' exonuclease active site. Mol Cell Biol. 1991 Sep;11(9):4786–4795. doi: 10.1128/mcb.11.9.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz G. S., Dean F. B., Hurwitz J., Matson S. W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988 Jan 5;263(1):383–392. [PubMed] [Google Scholar]
- Ito J. Bacteriophage phi29 terminal protein: its association with the 5' termini of the phi29 genome. J Virol. 1978 Dec;28(3):895–904. doi: 10.1128/jvi.28.3.895-904.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991 Aug 11;19(15):4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis T. C., Newport J. W., von Hippel P. H. Stimulation of the processivity of the DNA polymerase of bacteriophage T4 by the polymerase accessory proteins. The role of ATP hydrolysis. J Biol Chem. 1991 Jan 25;266(3):1830–1840. [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Hsieh J. C., Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8287–8291. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Schultz M., Ito J. Site-specific mutagenesis of PRD1 DNA polymerase: mutations in highly conserved regions of the family B DNA polymerase. Biochem Biophys Res Commun. 1990 Aug 16;170(3):1294–1300. doi: 10.1016/0006-291x(90)90534-t. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leavitt M. C., Ito J. T5 DNA polymerase: structural--functional relationships to other DNA polymerases. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4465–4469. doi: 10.1073/pnas.86.12.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner R. L., Engler M. J., Richardson C. C. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983 Sep 25;258(18):11174–11184. [PubMed] [Google Scholar]
- Lechner R. L., Richardson C. C. A preformed, topologically stable replication fork. Characterization of leading strand DNA synthesis catalyzed by T7 DNA polymerase and T7 gene 4 protein. J Biol Chem. 1983 Sep 25;258(18):11185–11196. [PubMed] [Google Scholar]
- Masamune Y., Richardson C. C. Strand displacement during deoxyribonucleic acid synthesis at single strand breaks. J Biol Chem. 1971 Apr 25;246(8):2692–2701. [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Mindich L., McGraw T. Molecular cloning of bacteriophage PRD1 genomic fragments. Mol Gen Genet. 1983;190(2):233–236. doi: 10.1007/BF00330645. [DOI] [PubMed] [Google Scholar]
- Morrison A., Bell J. B., Kunkel T. A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Patel S. S., Wong I., Johnson K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991 Jan 15;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Reha-Krantz L. J. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J Mol Biol. 1988 Aug 20;202(4):711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Liesner E. M., Parmaksizoglu S., Stocki S. Isolation of bacteriophage T4 DNA polymerase mutator mutants. J Mol Biol. 1986 May 20;189(2):261–272. doi: 10.1016/0022-2836(86)90508-5. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Nonay R. L. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3'-->5'-exonuclease activity. J Biol Chem. 1993 Dec 25;268(36):27100–27108. [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., RICHARDSON C. C., KORNBERG A. ENZYMIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID. XVII. SOME UNUSUAL PHYSICAL PROPERTIES OF THE PRODUCT PRIMED BY NATIVE DNA TEMPLATES. J Mol Biol. 1964 Jul;9:24–45. doi: 10.1016/s0022-2836(64)80089-9. [DOI] [PubMed] [Google Scholar]
- Savilahti H., Caldentey J., Lundström K., Syväoja J. E., Bamford D. H. Overexpression, purification, and characterization of Escherichia coli bacteriophage PRD1 DNA polymerase. In vitro synthesis of full-length PRD1 DNA with purified proteins. J Biol Chem. 1991 Oct 5;266(28):18737–18744. [PubMed] [Google Scholar]
- Sawaya M. R., Pelletier H., Kumar A., Wilson S. H., Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994 Jun 24;264(5167):1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- Shiue S. Y., Hsieh J. C., Ito J. Mapping of the DNA linking tyrosine residue of the PRD1 terminal protein. Nucleic Acids Res. 1991 Jul 25;19(14):3805–3810. doi: 10.1093/nar/19.14.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Giot L., Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991 Aug;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas M. S., Esteban J. A., Lázaro J. M., Bernad A., Blasco M. A., Salas M., Blanco L. Site-directed mutagenesis at the Exo III motif of phi 29 DNA polymerase; overlapping structural domains for the 3'-5' exonuclease and strand-displacement activities. EMBO J. 1992 Nov;11(11):4227–4237. doi: 10.1002/j.1460-2075.1992.tb05517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989 Apr 15;264(11):6447–6458. [PubMed] [Google Scholar]
- Wang Y. S., Woodward S., Hall J. D. Use of suppressor analysis to identify DNA polymerase mutations in herpes simplex virus which affect deoxynucleoside triphosphate substrate specificity. J Virol. 1992 Mar;66(3):1814–1816. doi: 10.1128/jvi.66.3.1814-1816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe K., Leusch M., Ito J. Replication of bacteriophage phi 29 DNA in vitro: the roles of terminal protein and DNA polymerase. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5374–5378. doi: 10.1073/pnas.81.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M., Schwarz M. W., Stahl H. Simian virus 40 large T antigen DNA helicase. Characterization of the ATPase-dependent DNA unwinding activity and its substrate requirements. J Biol Chem. 1988 Jan 5;263(1):436–442. [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Ito J. Protein-primed replication of bacteriophage PRD1 genome in vitro. Virology. 1989 Jun;170(2):442–449. doi: 10.1016/0042-6822(89)90435-2. [DOI] [PubMed] [Google Scholar]
- Zhu W., Ito J. Purification and characterization of PRD1 DNA polymerase. Biochim Biophys Acta. 1994 Oct 18;1219(2):267–276. doi: 10.1016/0167-4781(94)90048-5. [DOI] [PubMed] [Google Scholar]