Abstract
The chemotherapeutic drug Taxol is known to interact within a specific site on β-tubulin. Although the general location of the site has been defined by photoaffinity labeling and electron crystallography, the original data were insufficient to make an absolute determination of the bound conformation. We have now correlated the crystallographic density with analysis of Taxol conformations and have found the unique solution to be a T-shaped Taxol structure. This T-shaped or butterfly structure is optimized within the β-tubulin site and exhibits functional similarity to a portion of the B9-B10 loop in the α-tubulin subunit. The model provides structural rationalization for a sizeable body of Taxol structure–activity relationship data, including binding affinity, photoaffinity labeling, and acquired mutation in human cancer cells.
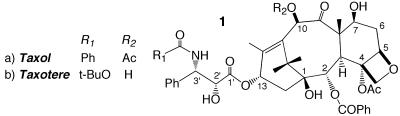
Taxol 1a [paclitaxel (PTX)] and Taxotere 1b (docetaxel) are important antitumor agents used clinically in the treatment of refractory ovarian cancer, small-cell lung cancer, metastatic breast disease, and other manifestations of the affliction (1). The drugs are believed to block cell-cycle progression during mitosis by binding to and stabilizing microtubules (MTs) (2, 3). One functional taxoid-binding site per tubulin (TB) dimer (4, 5) has been located on the β-subunit (6–11). The recently determined 3.7-Å structure of the αβ-TB-PTX complex (12), obtained by electron crystallography (EC) of zinc-induced TB sheets, clearly shows the location of the binding site but lacks the resolution to completely determine the ligand conformation. Therefore, in the original report, the x-ray crystal structure of Taxotere (13) served as a PTX surrogate. More recently, the EC crystal structure of TB has been docked into a 20-Å map of the MT to provide a high-resolution model of this polymer (14). The excellent fit indicates the similarity of the TB conformation in MTs and zinc-induced sheets. In the present work, we integrate the electron-density map of the refined complex and a three-step modeling strategy to define the PTX-binding conformation in the EC structure.
Methods
Conformational Selection and Fitting.
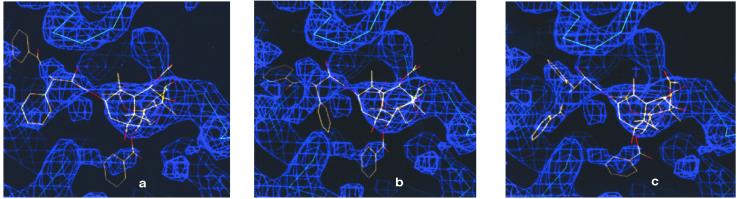
Taxol combines a polar and rigid tetracyclic core with an equally polar set of four flexible side chains, containing 10 single bonds that contribute to the conformational flexibility of the molecule. Here we have explicitly considered 26 PTX conformers derived from x-ray crystal structures and from NMR nuclear Overhauser effect data of PTX and its derivatives (15–24) and a large number of computer-generated conformers. The individual conformers were docked into the experimental density map of the TB-PTX complex. Examples are portrayed in Fig. 1. Most of the conformations leave the terminal C-13 phenyl rings in regions completely devoid of density. Two of these are shown in Fig. 1 a and b. On the other hand, three very similar conformations, two from the solid state (15, 16) and one from solution (24), gave reasonable fits. One of the former was used as a starting point for further refinement of the TB-PTX complex (Fig. 1c).
Figure 1.
Three PTX conformers docked into the ligand electron crystallographic density of β-TB. The first two conformers, a and b, were derived from PTX analogs determined by single-crystal x-ray crystallography. The best-fit conformer, shown in c, was obtained by NMR deconvolution.
Stepped Optimization.
Coordinates used for optimization were obtained by refinement¶ of those placed on deposit at the Brookhaven Protein Data Bank (1TUB.pdb). Results of restricted low-temperature dynamics and force-field optimization were monitored for fit within the experimental density. Details are published as supplemental data on the PNAS web site (www.pnas.org).
Automated Docking.
To certify that the T- or butterfly-binding motif is both unique and reproducible, PTX was removed from its protein complex, conformationally altered, and flexibly redocked into the binding pocket by using standard DOCK methodology (ref. 25; http://www.cmpharm.ucsf.edu/kuntz/kuntz.html). Although many partially docked PTX structures were generated, only two conformers were encased by the protein. The lowest energy form is identical in shape and location to that in Fig. 3. The other was scored 45 kcal/mol higher in energy and involves a torsional exchange of C-3′ phenyl and C-3′ NHCOPh. In this orientation, the side chains fall clearly outside the EC density, ruling out the alternative conformation. No other binding mode within the ligand groove was identified. A second docking experiment with FlexX, a fragment-based flexible docking algorithm (ref. 26; http://cartan.gmd.de/flexx/), generated 200 PTX conformers. T-Taxol was docked twice. The orientation is shown in Figs. 3 and 4a as the lowest energy form. Scoring data are given in the supplementary material.
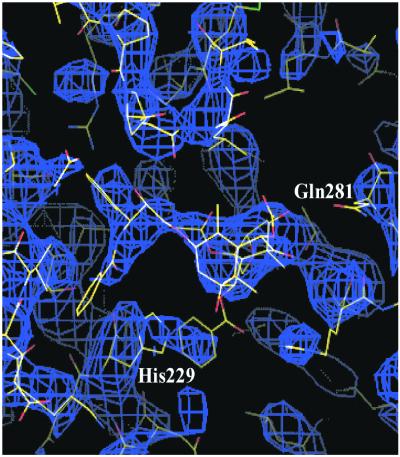
Figure 3.
The T-conformation of PTX sited within the experimental EC TB density following the refinement procedures described in the text. βHis-229 of the protein is stacked between the C-3′ benzamido and C-2 benzoyl phenyl rings of the PTX molecule.
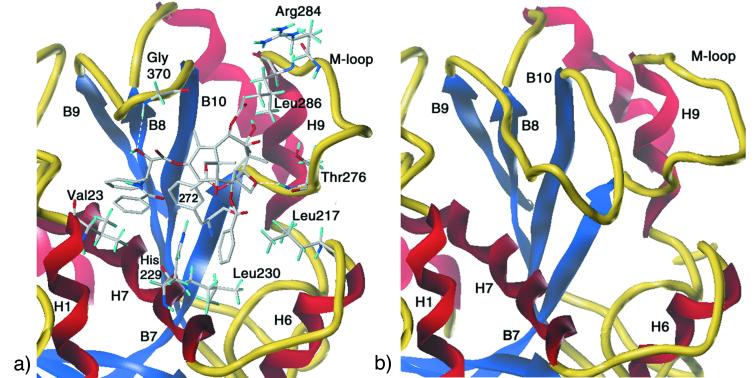
Figure 4.
Detail of important binding interactions illustrating similarities between PTX in β-TB (a) and the αB9-B10 loop extension in α-TB (b). The α and β subunits share both high-sequence homology and a common topology. (a) The PTX β-TB complex illustrating the ligand's pivotal role as a center of organization for α-helices H1, H6, and H7, the H6-H7 loop, β-strands B8-B10, and the B7-H9 M-loop. (b) The B9-B10 loop.
Discussion
Analysis of Taxol Conformation.
There have been many speculations as to the bioactive conformation of PTX complexed with MTs. Initially, the nonpolar conformer, with the C-2 and C-3′ phenyl side chains exhibiting hydrophobic collapse (Fig. 2a), was proposed as the bioactive one (17–19). Shortly thereafter, however, several laboratories argued in favor of the polar form (20–22) that juxtaposes the C-2 and C-3′ benzamido side chains (Fig. 2b) (27). More recently, during the processing of this manuscript, a pharmacophore model (28) and two studies using PTX docked into EC β-TB appeared, which also favor the polar structure (29, 30). The binding conformation derived from the present analysis belongs to neither extreme. As illustrated by Fig. 2c, PTX's bound state in our model is characterized by the C-2 benzoyl phenyl, nearly equidistant from both of the phenyl rings emanating from C-3′ in T-Taxol. A recent gas-phase ab initio conformational analysis of the PTX C-13 side chain led to a similar binding proposal (31), underscoring the notion that low-energy experimental conformational minima are prime candidates for protein-bound small molecules (32). The ligand structure corresponds to one member of the family of low population conformers found by NMR deconvolution analysis in chloroform (24) and DMSO/water (J.P.S., N. Nevins, J. Jiménez-Barbero, D. Cicero, and J. M. Jansen, unpublished work). With T-Taxol nestled in the EC density of the computationally refined PTX-TB complex, residue His-229 of the protein is interposed between the PTX C-3′ and C-2 phenyl rings, preventing their hydrophobic collapse (Figs. 3 and 4a). The same observation with respect to C-2 has been made in connection with 2-m-azido baccatin III (33). In association with β-TB, each of PTX's hydrophobic centers engages in complementary interactions with the protein (Fig. 4a) rather than self, as in the polar and nonpolar conformations. This picture predicts that taxoid design and synthesis based on a hydrophobically collapsed motif are likely to lead to inactive compounds. The point has been made decisively in one completely inactive series in which Georg, Himes, and coworkers tethered the C-3′ and C-2 phenyls with several two-atom spacers (34).
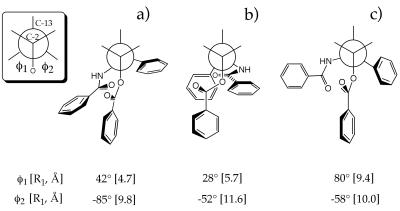
Figure 2.
Newman projections for PTX conformations in terms of the improper torsion angles O-C2-C3′-N(Bz) and O-C2-C3′-C(Ph) (φ1, φ2). The front atom is C-2, the unseen back atom, C-3′. R1 and R2 are the distances (in angstroms) from the center of the C-2 benzoyl phenyl ring and the center of the C-3′ phenyl ring corresponding to the appropriate φ1 and φ2. (a) The “nonpolar” conformation as derived by NMR in CDCl3. (b) The “polar” conformation as observed in the solid state (27). (c) Proposed T-Taxol conformation bound to β-TB.
Binding-Site Interactions.
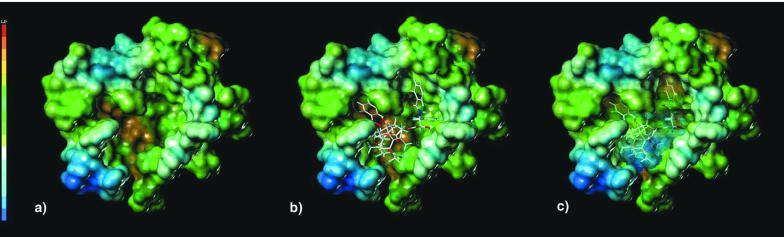
In our model, the binding pocket for PTX resides in a deep hydrophobic cleft near the surface of β-TB, where it interacts with the protein by means of three potential hydrogen bonds and multiple hydrophobic contacts. Importantly, the walls of the pocket are composed of several elements of secondary structure linked by the PTX molecule. Segments of helices H1, H6, H7, and the loop between H6 and H7 are anchored by hydrophobic interactions with PTX's 3′-benzamido phenyl, the 3′-phenyl, and the 2-benzoyl phenyl (Fig. 4a). In addition, the 3′-phenyl enjoys close contacts with β-sheet strands B8 and B10. The refined site model suggests that a backbone NH of the loop connecting strands B9 and B10 is hydrogen bonded to 2′-OH. The C-4 acetate is situated above a 10-residue hydrophobic basin [Leu-230, Ala-233 (H7); Phe-272, Pro-274, CH3 of Thr-276, Leu-286, Leu-291 (M-loop); Pro-360, Leu-371 (B9-B10 loop); and CH2 of Ser-374 (B10)], half of which derives from the N-terminal end of the long meandering loop that connects B7 and H9, the M-loop (14). The C-8 methyl is directed toward the M-loop, putting it in van der Waals contact with two residues near the C-terminal end, Thr-276 and Gln-281. The ligand's O-21 appears to experience a weak electrostatic interaction with the same loop via Thr-276 (35). Finally, the model places the taxoid's C-12 methyl in close proximity to Leu-371 on the B9-B10 loop. Thus, both side chains and baccatin core of the ligand contribute to the scaffold of interactions. Fig. 5 depicts β-TB with its surface represented in terms of hydrophobicity. The empty and Taxol-occupied binding pocket is presented in brown in Fig. 5 a and b. When the ligand–protein complex surface is recolored to include the ligand, it is clear that Taxol binding converts a hydrophobic cleft into a hydrophilic surface (Fig. 5c). The equivalent locus in α-TB is occupied by an eight-residue insertion in the loop connecting strands B9 and B10 (Thr-361–Leu-368; see Fig. 4b). Population of the β-TB cleft by Taxol or competitive biomimetics produces protofilaments with a homogeneous face that conceivably contributes to both the formation of MTs by lateral polymerization and to MT stability.‖ It likewise suggests the possibility that a properly constituted peptide, endogenous or synthetic, could operate in like manner in PTX's absence.
Figure 5.
Shaded solvent accessible surface of Taxol-binding pocket of β-TB, colored according to degrees of hydrophobicity; color key is on left (maximum, red; minimum, dark blue). (a) The empty PTX-binding pocket (burnt orange) is highly hydrophobic. (b) The binding center is occupied by T-Taxol, showing excellent shape complementarity. (c) Surface recoloring illustrates that a hydrophobic depression has been converted to a hydrophilic surface on PTX binding.
Structure–Activity Relationships.
The model for the PTX–TB complex presented here is in excellent agreement with important elements of the structure–activity relationship data. First, the bioactivity of PTX is relatively insensitive to chemical manipulation from C-7 to C-10 (37). This section of the molecule is not engaged in binding the protein but is projected outward away from the surface of the macromolecule. Second, evidence has accumulated that the C-2′ hydroxyl is critical for biological activity (38, 39) and is a hydrogen bond donor (40). In agreement, a hydrogen bond from C-2′ OH to the backbone carbonyl of Arg-369 exists in the model. Third, replacement of phenyl groups with cyclohexyl moieties in the C-13 and C-2 side chains sustains the activity of PTX (41, 42). The location of each of the rings in a generous hydrophobic space is compliant. Fourth, extension of the C-4 acetate by longer alkyl chains without loss of activity (43) is explained by the location of the C-4 methyl at the top of a deep hydrophobic cleft. Fifth, meta substitution in the phenyl of the C-2 side-chain enhances activity, whereas para substitution reduces it (44). The hydrophobic subsite housing this phenyl moiety is relatively tight on three sides of the ring but open at one of the meta positions. The elegant deduction by Horwitz, Kingston, and colleagues that appropriate m-substitution permits the elimination of the C-13 side chain without sacrificing potency or efficacy amplifies the point (33). Confirming the structural aspects of their work, our model positions the carboxylate of Asp-226 directly above the central cationic nitrogen of the azide in 2-m-azido baccatin. Sixth, C-6 nor-Taxol derivatives drop activity by 10–20 times (45, 46). Contraction of the C-ring from six members to five pulls the oxetane ring away from Thr-276 and reduces the effectiveness of the O-21/HN-Thr interaction.
Acquired Resistance.
PTX- and epothilone-resistant human ovarian cancer cell lines have been isolated by Giannakakou et al. and shown to express mutant β-TB with diminished capacity for TB polymerization by the drugs (47, 48). The acquired mutations are Phe-270→Val, Ala-364→Thr, Thr-274→Ile, and Arg-282→Gln [corresponding to positions 272, Ser-374, 276, and 284 in the structure-based sequence alignment of α- and β-TB] (12)].** The first two residues, conferring PTX resistance, line the floor of the deep hydrophobic pocket surrounding the PTX ligand. In the model of Fig. 4a, Phe-272 resides at one end of β-sheet strand B7 in van der Waals contact with the methyl group of PTX's C-4 acetate. Ser-374, although not in direct contact with the ligand, is part of a larger hydrophobic cluster that includes Ala-273 and Phe-272. Its replacement by threonine can be viewed as causing a reorganization of the cluster with concomitant adjustment in the position of Phe-272. In this interpretation, both mutations modify the character of the floor of the pocket in a similar manner to effectively block PTX binding. A smaller taxoid surrogate might well escape these changes in protein sequence and stimulate other mutations, as appears to be the case for epothilone. The third and fourth mutations, affording epothilone resistance, are found on the M-loop contiguous with the PTX-binding site (Fig. 4a). They cause resistance to both PTX and epothilone-driven TB polymerization. From a structural point of view, Thr-276→Ile will influence any hydrogen bond between O-21 and threonine, a phenomenon consistent with the resistance shown by a carcinoma subline expressing this β-TB mutant (48). The local electrostatic reorganization does not, however, capture what must be a more subtle effect. Although it has been generally believed that the taxane oxetane ring is “absolutely required” for activity, our recent analysis suggests this not to be the case. Both a minireceptor treatment and the complex of Fig. 4a were used to predict that the oxetane ring could be replaced, among others, with a cyclopropane ring without loss in activity (35). The prediction has been borne out in the Taxotere series in the work of Dubois et al. (49). Given the uncertainties in the conformation of the M-loop, our model does not provide a specific rationale for the Arg-284→Gln substitution apart from proximity. However, an interesting proposal regarding the disruption of a hydrogen-bonding network mediated by water molecules has been advanced (48).
Photoaffinity Labeling.
Photoaffinity-labeling studies distributing photoactive moieties at distant atoms around PTX (i.e., groups at C-2, C-7, and C-3′) have successfully identified specific residues in the β-TB dimer in close proximity to the binding pocket. Thus, [3H]2-(m-aziodbenzoyl) PTX (AzBo-PTX) photolabels residues 217–231 on and adjacent to helix H7 in β-TB (10). Providing perfect consistency, the model nestles the C-2 OCO-Ph ring in a hydrophobic subsite composed of Leu-217, Leu-219, a CH2 of Asp-226, and His-229 (Fig. 4a). A second probe, [3H]3′-(p-azidobenzamido) PTX (AzBa-PTX), competes with PTX binding and labels the N-terminal 31 amino acids of β-TB (6–8). In complete accord, the binding model of Fig. 4a juxtaposes the phenyl ring of C-3′ NHCO-Ph and the isopropyl group of Val-23 on helix H1. Additionally, the same ring enjoys short contacts with methylene groups (CH2) of Lys-19, Glu-22, and Asp-26. Most recently, a compound that stabilizes MTs in the presence of GTP but does not promote TB polymerization, [3H]7-(benzoyldihydro-cinnamoyl) PTX (BzDC-PTX), has been shown to crosslink to βArg-284 (29). This M-loop residue is the same amino acid that arises from one of the epothilone-resistant cell lines discussed above. Flexible docking of the BzDc-PTX label in the ligand pocket sketched in Fig. 4a effectively superimposes the baccatin core with that of PTX while siting the C-7 benzophenone end-group near βArg-284, as deduced by Ojima, Horwitz, and coworkers (29). However, the intrusive C-7 side chain has the potential to induce an unnatural M-loop conformation. Thus, our docked BzDC-PTX not only accounts for the labeling of Arg-284 but also suggests a reason for the failure of the photolabel to promote TB polymerization.
Comparison with Other Models.
Three recent reports have used the 3.7-Å resolution EC coordinates of αβ-TB to derive a protein-bound conformation for either PTX or epothilone (29, 30, 48). It is important to recall that, whereas the actual TB sheets examined by EC were stabilized by PTX, the deposited protein coordinates contain the single-crystal coordinates for Taxotere in place of the actual ligand (12) [Brookhaven Protein Data Bank (1TUB.pdb)]. All three studies began the PTX-TB model building with the isolated small molecule crystal structure coordinates of either Taxotere or PTX as the starting point, and each ended with essentially the same conformation. By contrast, the PTX model portrayed by Fig. 4a was derived by fitting numerous conformations followed by a treatment that remains fully faithful to the EC density of both the protein and the ligand. Of particular note is the Bane, Kingston, and Schaefer model (30) derived from a combination of solid-state rotational echo double resonance (REDOR) NMR and fluorescence spectroscopy [fluorescence resonance energy transfer (FRET)]. REDOR, performed on labeled and MT-bound PTX, is a conformational determinant permitting measurement of the distance between 19F at the para-position of the C-2 benzoyl substituent and 13C at the 3′-amide carbonyl and the C-3′ methine carbons. The values of 9.8 ± 0.5 Å and 10.3 ± 0.5 Å, respectively, led to the choice of the hydrophobically collapsed polar conformation b in Fig. 2 (rcalc 9.6 and 10.4 Å, respectively). In good agreement with this measurement, T-Taxol (Fig. 2c) sustains the corresponding distances at 9.1 and 9.9 Å, respectively. Consequently, both structures are viable REDOR candidates, although the intervention of βHis-229 between the C-2 and C-13 PTX side chains (Figs. 3 and 4a) favors the T-form in the pictured binding mode. FRET, on the other hand, is a ligand orientation probe that depends on the incorporation of both colchicine (COL) and PTX in β-TB. To fit the FRET-derived COL-to-PTX distances in the context of the collapsed conformer b, the data were interpreted in terms of an alternative binding mode in which PTX is rotated 180° in the binding pocket relative to the present model. Two factors argue against this binding orientation. First, the polar form in this locus matches the ligand density much less well than that shown in Fig. 1 a and b. Second, the requirement that COL and PTX must simultaneously bind to β-TB to derive the PTX-binding mode adds a new variable to the problem. The two antimitotic drugs influence the polymerization of TB in a qualitatively different manner, one destabilizing, the other stabilizing MTs, respectively. Consequently, there is a reasonable possibility that the altered placement of PTX in TB doped by COL results from reorganization of the binding site in the presence of the second drug. As a result, we favor the PTX-only solution in the refined TB coordinates as shown in Figs. 3 and 4a.
Summary and Conclusions
A model of the bioactive conformation of PTX in its β-TB-binding site depicted in Figs. 3 and 4a has been constructed by making maximum use of empirical information and refinement of the EC structure. In short, two dozen conformers of PTX derived from various structural studies were fitted into the partial EC density associated with the ligand in β-TB. One of the best solutions was optimized in the binding site by applying computational tools in a way that kept the evolving model fully consistent with the EC density of the protein complex. The final construct, Figs. 3 and 4a, portrays a T-shaped or butterfly Taxol conformation operating as a center of organization for a diversity of secondary structures. Unlike the commonly proposed polar and nonpolar conformations of the drug, which experience intramolecular hydrophobic collapse, T-PTX opens up to permit intermolecular hydrophobic association as seen for the irregularly stacked C-3′ benzamido, His-229, and C-2 benzoyl moieties. The ligand–protein construct rationalizes a range of structure–activity data involving synthetic substitution at all parts of the PTX molecule. It addresses the issue of drug resistance for several acquired TB mutants in human cancer cells. The new model is in complete harmony with three photoaffinity-labeling studies focused on β-TB. By comparing the nearly identical α and β subunits of the αβ-TB dimer, we note that PTX in β-TB appears to serve a purpose similar to an extended loop in the same location in α-TB. The two centers can be viewed as acting in tandem to promote lateral aggregation of TB protofilaments to give mature MTs. Finally, we review the proposals for PTX conformation at the ligand-binding site and argue that the present model incorporates numerous advantages that should prove useful in evaluating and predicting the behavior of taxanes and taxane-mimetic drugs. We also anticipate that the strategy of combining diversity-rich, small-molecule, empirical conformational analysis with 3–4 Å resolution electron crystallography will assist the identification of ligand conformation where partial EC density alone is insufficient to reconcile ambiguities.
Supplementary Material
Scheme 1.
Acknowledgments
We are grateful to Dr. Qi Gao (Bristol-Myers Squibb) for providing unpublished x-ray crystal coordinates of several PTX analogs and to Prof. Dennis Liotta (Department of Chemistry, Emory University) for encouragement and support. Prof. Gunda Georg (Department of Medicinal Chemistry, University of Kansas) generously provided access to unpublished results. Support for part of this work was received from the National Institutes of Health and the Department of Energy.
Abbreviations
- MT
microtubule
- TB
tubulin
- EC
electron crystallography
- PTX
paclitaxel
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
The structure of the αβ-tubulin dimer refined to 3.5 Å was obtained by using simulated annealing torsion angle refinement and phase information from experimental images; R factor 0.23 and free R factor 0.30; J. Lowe., T. Li, K.H.D., and E.N., unpublished data.
A protofilament consists of a longitudinal head-to-tail stacking of αβ-tubulin dimers. These extended and observable macrostructures are capable of assembling laterally to give cylindrical MTs; see ref. 36.
The EC structure determination used wild-type pig-brain tubulin with β-Ser. Formally, the β-Ala-364→Thr mutation is β-Ser-364→Thr here.
References
- 1.Holmes F A, Kudelka A P, Kavanagh J J, Huber M H, Ajani J A, Valero V. In: Taxane Anticancer Agents. Georg G I, Chen T T, Ojima I, Vyas D M, editors. Washington, DC: Am. Chem. Soc.; 1995. , ACS Symposium Series 583, p. 31. [Google Scholar]
- 2.Jordon M A, Wilson L. In: Taxane Anticancer Agents. Georg G I, Chen T T, Ojima I, Vyas D M, editors. Washington, DC: Am. Chem. Soc.; 1995. , ACS Symposium Series 583, pp. 138–153. [Google Scholar]
- 3.Nicolaou K C, Dai W-M, Guy R K. Angew Chem Int Ed Engl. 1994;33:15–44. [Google Scholar]
- 4.Parness J, Horwitz S B. J Cell Biol. 1981;91:479–587. doi: 10.1083/jcb.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz J F, Andreu J M. Biochemistry. 1993;32:2747–2755. doi: 10.1021/bi00062a003. [DOI] [PubMed] [Google Scholar]
- 6.Rao S, Horwitz S B, Ringel I. J Natl Cancer Inst. 1992;84:785–788. doi: 10.1093/jnci/84.10.785. [DOI] [PubMed] [Google Scholar]
- 7.Rao S, Krauss N E, Heerding J M, Swindell C S, Ringel I, Orr G A, Horwitz S B. J Biol Chem. 1994;269:3132–3134. [PubMed] [Google Scholar]
- 8.Dasagupta D, Park H, Harriman G C B, Georg G I, Himes R. J Med Chem. 1994;37:2976–2980. doi: 10.1021/jm00044a019. [DOI] [PubMed] [Google Scholar]
- 9.Combeau C, Commerçon A, Mioskowski C, Rousseau B, Aubert F, Goeldner M. Biochemistry. 1994;33:6676–6683. doi: 10.1021/bi00187a038. [DOI] [PubMed] [Google Scholar]
- 10.Rao S, Orr G A, Chaudhary A G, Kingston D G I, Horwitz S B. J Biol Chem. 1995;270:20235–20238. doi: 10.1074/jbc.270.35.20235. [DOI] [PubMed] [Google Scholar]
- 11.Loeb C, Combeau C, Shret-Sabatier L, Breton-Gilet A, Faucher D, Rousseau B, Commerçon A, Goeldner M. Biochemistry. 1997;36:3820–3825. doi: 10.1021/bi961602r. [DOI] [PubMed] [Google Scholar]
- 12.Nogales E, Wolf S G, Downing K H. Nature (London) 1998;391:199–202. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 13.Guéritte-Voegelein F, Guenard D, Mangatal L, Potier P, Guilhem J, Cesario M. Acta Crystallogr C. 1990;46:781–784. [Google Scholar]
- 14.Nogales E, Whittaker M, Milligan R A, Downing K H. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Wei J-M, Chen S-H. Pharm Res. 1995;12:337–341. doi: 10.1023/a:1016236014766. [DOI] [PubMed] [Google Scholar]
- 16.Gao Q, Chen S-H. Tetrahedron Lett. 1996;37:3425–3428. [Google Scholar]
- 17.Dubois J, Guénard D, Guéritte-Voegelein F, Guedira N, Potier P, Gillet B, Beloeil J-C. Tetrahedron. 1993;49:6533–6544. [Google Scholar]
- 18.Williams H J, Scott A I, Dieden R A, Swindell C S, Chirlian L E, Francl M M, Heerding J M, Krauss N E. Tetrahedron. 1993;49:6545–6560. [Google Scholar]
- 19.Cachau R E, Gussio R, Beutler J A, Chmurny G N, Hilton B D, Muschik G M, Erickson J W. Supercomput Appl High Perform Comput. 1994;6:24–34. [Google Scholar]
- 20.Vander Velde D G, Georg G I, Grunewald G L, Gunn C W, Mitscher L A. J Am Chem Soc. 1993;113:11650–11651. [Google Scholar]
- 21.Paloma L G, Guy R K, Wrasidlo W, Nicolaou K C. Chem Biol. 1994;1:107–112. doi: 10.1016/1074-5521(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 22.Ojima I, Kuduk S D, Chakravarty S, Ourevitch M, Bégue J-P. J Am Chem Soc. 1997;119:5519–5527. [Google Scholar]
- 23.Jiménez-Barbero J, Souto A A, Abal M, Barasoain I, Evangelio J A, Acuña A U, Andreu J M, Amat-Guerri F. Biorg Med Chem. 1998;6:1857–1863. doi: 10.1016/s0968-0896(98)00149-7. [DOI] [PubMed] [Google Scholar]
- 24.Snyder J P, Nevins N, Cicero D O, Jansen J J. J Am Chem Soc. 2000;172:724–725. [Google Scholar]
- 25.Ewing T J A, Kuntz E D. J Comp Chem. 1997;18:1175–1189. [Google Scholar]
- 26.Rarey M, Kramer B, Lengauer T, Klebe G. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 27.Mastropaolo D, Camerman A, Luo Y, Brayer G D, Camerman N. Proc Natl Acad Sci USA. 1995;92:6920–6924. doi: 10.1073/pnas.92.15.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojima I, Chakravarty S, Inoue T, Lin L, He L, Horwitz S B, Kuduk S D, Danishefsky S J. Proc Natl Acad Sci USA. 1999;96:4256–4261. doi: 10.1073/pnas.96.8.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao S, He L, Chakravarty S, Ojima I, Orr G A, Horwitz S B. J Biol Chem. 1999;274:37990–37994. doi: 10.1074/jbc.274.53.37990. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Poliks B, Cegelski L, Poliks M, Gryczynski Z, Piszczek G, Jagtap P G, Studelska D R, Kingston D G I, Schaefer J, Bane S. Biochemistry. 2000;39:281–291. doi: 10.1021/bi991936r. [DOI] [PubMed] [Google Scholar]
- 31.Milanesio M, Ugliengo P, Viterbo D, Appendino G. J Med Chem. 1999;42:291–299. doi: 10.1021/jm981082t. [DOI] [PubMed] [Google Scholar]
- 32.Klebe G. Perspect Drug Discovery Des. 1995;3:85–105. [Google Scholar]
- 33.He L, Jagtap P G, Kingston D G I, Shen H-J, Orr G A, Horwitz S B. Biochemistry. 2000;39:3972–3978. doi: 10.1021/bi992518p. [DOI] [PubMed] [Google Scholar]
- 34.Boge T C, Wu Z-J, Himes R H, Vander Velde D G, Georg G I. Biorg Med Chem Lett. 1999;9:3047–3052. doi: 10.1016/s0960-894x(99)00522-3. [DOI] [PubMed] [Google Scholar]
- 35.Wang M, Cornett B, Nettles J, Liotta D C, Snyder J P. J Org Chem. 2000;65:1059–1068. doi: 10.1021/jo9916075. [DOI] [PubMed] [Google Scholar]
- 36.Garret R H, Grisham C M. Biochemistry. 2nd Ed. New York: Sanders College Pub.; 1999. pp. 534–535. [Google Scholar]
- 37.Kingston D G I. In: Taxane Anticancer Agents. Georg G I, Chen T T, Ojima I, Vyas D M, editors. Washington, DC: Am. Chem. Soc.; 1995. , ACS Symposium Series 583, pp. 203–216. [Google Scholar]
- 38.Guéritte-Voegelein F, Guénard D, Lavell F, LeGoff M-T, Mangatal L, Potier P. J Med Chem. 1991;34:992–998. doi: 10.1021/jm00107a017. [DOI] [PubMed] [Google Scholar]
- 39.Swindell C S, Krauss N E, Horwitz S B, Ringel I. J Med Chem. 1991;34:1176–1184. doi: 10.1021/jm00107a042. [DOI] [PubMed] [Google Scholar]
- 40.Williams H J, Moyna G, Scott A I, Swindell C S, Chirlian L E, Heerding J M, Williams D K. J Med Chem. 1996;39:1555–1559. doi: 10.1021/jm950796q. [DOI] [PubMed] [Google Scholar]
- 41.Ojima I, Duclos O, Zucco M, Bissery M-C, Combeau C, Vrignaud P, Riou J F, Lavelle F. J Med Chem. 1994;37:2602–2608. doi: 10.1021/jm00042a013. [DOI] [PubMed] [Google Scholar]
- 42.Boge T C, Himes R H, Vander Velde D G, Georg G I. J Med Chem. 1994;37:3337–3343. doi: 10.1021/jm00046a018. [DOI] [PubMed] [Google Scholar]
- 43.Chen S-H, Wei J-M, Long B H, Fairchild C A, Carboni J, Mamber W W, Rose W C, Johnston K, Casazza A M, Kadow J F, et al. Bioorg Med Chem Lett. 1995;5:2741–2746. [Google Scholar]
- 44.Chaudhary A G, Gharpure M M, Rimoldi J M, Chordia M D, Gunatilaka A A L, Kingston D G I, Grover S, Lin C, Hamel E. J Am Chem Soc. 1994;116:4097–4098. [Google Scholar]
- 45.Wender P A, Lee D, Lal T K. Bioorg Med Chem Lett. 1997;7:1941–1944. [Google Scholar]
- 46.Liang X, Kingston D G I, Long B H, Fairchild C A, Johnston K A. Tetrahedron. 1997;53:3441–3456. [Google Scholar]
- 47.Giannakakou P, Sackett D L, Kang Y-K, Shan Z, Buters J T M, Fojo T, Poruchynsky M S. J Biol Chem. 1997;272:17118–17125. doi: 10.1074/jbc.272.27.17118. [DOI] [PubMed] [Google Scholar]
- 48.Giannakakou P, Gussio R, Nogales E, Downing K H, Zaharevitz D, Bollbuck B, Poy G, Sackett D, Nicolaou K C, Fojo T. Proc Natl Acad Sci USA. 2000;97:2904–2909. doi: 10.1073/pnas.040546297. . (First Published February 25, 2000; 10.1073/pnas.040546297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubois J, Thoret S, Guéritte F, Guénard D. Tetrahedron Lett. 2000;41:3331–3334. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.