Abstract
Clinical isolates of Staphylococcus aureus can express biofilm phenotypes promoted by the major cell wall autolysin and the fibronectin-binding proteins or the icaADBC-encoded polysaccharide intercellular adhesin/poly-N-acetylglucosamine (PIA/PNAG). Biofilm production in methicillin-susceptible S. aureus (MSSA) strains is typically dependent on PIA/PNAG whereas methicillin-resistant isolates express an Atl/FnBP-mediated biofilm phenotype suggesting a relationship between susceptibility to β-lactam antibiotics and biofilm. By introducing the methicillin resistance gene mecA into the PNAG-producing laboratory strain 8325-4 we generated a heterogeneously resistant (HeR) strain, from which a homogeneous, high-level resistant (HoR) derivative was isolated following exposure to oxacillin. The HoR phenotype was associated with a R602H substitution in the DHHA1 domain of GdpP, a recently identified c-di-AMP phosphodiesterase with roles in resistance/tolerance to β-lactam antibiotics and cell envelope stress. Transcription of icaADBC and PNAG production were impaired in the 8325-4 HoR derivative, which instead produced a proteinaceous biofilm that was significantly inhibited by antibodies against the mecA-encoded penicillin binding protein 2a (PBP2a). Conversely excision of the SCCmec element in the MRSA strain BH1CC resulted in oxacillin susceptibility and reduced biofilm production, both of which were complemented by mecA alone. Transcriptional activity of the accessory gene regulator locus was also repressed in the 8325-4 HoR strain, which in turn was accompanied by reduced protease production and significantly reduced virulence in a mouse model of device infection. Thus, homogeneous methicillin resistance has the potential to affect agr- and icaADBC-mediated phenotypes, including altered biofilm expression and virulence, which together are consistent with the adaptation of healthcare-associated MRSA strains to the antibiotic-rich hospital environment in which they are frequently responsible for device-related infections in immuno-compromised patients.
Author Summary
The acquisition of mecA, which encodes penicillin binding protein 2a (PBP2a) and methicillin resistance, by Staphylococcus aureus has added to an already impressive array of virulence mechanisms including enzyme and toxin production, biofilm forming capacity and immune evasion. And yet clinical data does not indicate that healthcare-associated methicillin resistant S. aureus (MRSA) strains are more virulent than their methicillin-susceptible counterparts. Here our findings suggest that MRSA sacrifices virulence potential for antibiotic resistance and that expression of methicillin resistance alters the biofilm phenotype but does not interfere with the colonization of implanted medical devices in vivo. High level expression of PBP2a, which was associated with a mutation in the c-di-AMP phosphodiesterase gene gdpP, resulted in these pleiotrophic effects by blocking icaADBC-dependent polysaccharide type biofilm development and promoting an alternative PBP2a-mediated biofilm, repressing the accessory gene regulator and extracellular protease production, and attenuating virulence in a mouse device-infection model. Thus the adaptation of MRSA to the hospital environment has apparently focused on the acquisition of antibiotic resistance and retention of biofilm forming capacity, which are likely to be more advantageous than metabolically-expensive enzyme and toxin production in immunocompromised patients with implanted medical devices offering a route to infection.
Introduction
Infections caused by healthcare-associated Staphylococcus aureus and methicillin resistant S. aureus (MRSA) pose a major threat to hospital patients. A significant risk factor for these healthcare-associated infections is the extensive use of implanted prosthetic biomaterials for diagnostic and therapeutic purposes, which can be colonized by staphylococci giving rise to device-related infections (DVIs) involving biofilms [1]. In addition to resistance to β-lactam antibiotics such as oxacillin, current chemotherapeutics for DVIs have limited effectiveness against biofilms.
The challenge of developing therapeutics to treat staphylococcal biofilm infections is compounded by the existence of multiple biofilm mechanisms in both S. aureus and S. epidermidis. Thus, although production of the exopolysaccharide polysaccharide intercellular adhesin (PIA) or polymeric N-acetyl-glucosamine (PNAG) synthesized and exported by proteins encoded by the icaADBC genes is common among clinical isolates of both species [2], [3], [4], [5], [6], ica-independent biofilm production has also been described under in vitro conditions [1]. Using clinical isolates of S. aureus, we reported that methicillin resistant S. aureus (MRSA) strains express an icaADBC-independent biofilm phenotype in vitro [3], [4], which is instead dependent on the fibronectin binding proteins (FnBPA and FnBPB) and the major autolysin (Atl) [6], [7]. Atl-dependent autolytic activity and extracellular DNA release are involved in the early stages of biofilm production by these MRSA isolates, whereas the FnBPs promote subsequent intercellular accumulation and biofilm maturation [6], [7]. Unlike MRSA, clinical isolates of methicillin susceptible S. aureus (MSSA) express a PNAG-dependent biofilm phenotype on hydrophilic surfaces and an Atl/PNAG-dependent biofilm on hydrophobic surfaces. Other staphylococcal surface proteins implicated in biofilm include the biofilm-associated protein (Bap, in bovine S. aureus isolates), accumulation-associated protein (Aap) of S. epidermidis and its S. aureus homologue SasG [8], [9], [10], [11], protein A [12], SasC [13] and the extracellular matrix binding protein (Embp) of S. epidermidis [14]. The growing number of bacterial factors involved in staphylococcal biofilm development underscores the importance of this phenotype to the pathogen and suggests that there may be redundancy between biofilm mechanisms in different clinical isolates or on different surfaces.
The methicillin resistance gene mecA encodes the low affinity penicillin binding protein 2a carried on a mobile staphylococcal cassette chromosomal mec element (SCCmec) [15] of which eight different types have been characterized to date [16]. Heterogeneity is a feature of S. aureus methicillin resistance [17], [18], [19]. Many S. aureus clinical isolates exhibit heterogeneous methicillin resistance (HeR) under laboratory growth conditions. In a HeR strain the majority of cells grown in the presence of a β-lactam antibiotic are susceptible to low concentrations of the drug, with only a subpopulation expressing higher-level resistance. However HeR strains become capable of expressing homogeneous resistance (HoR) after selection on elevated concentrations of β-lactam antibiotics or under specific growth conditions [20]. This transition from HeR to HoR is complex with mutations at the fem (factor essential for methicillin resistance), aux (auxiliary) and tagO loci all being implicated [21], [22], [23]. In addition, an oxacillin-induced increased SOS response was shown to increase the mutation rate during HeR to HoR selection in a mechanism dependent on the accessory gene regulator Agr [24], [25]. Nevertheless because HoR clinical isolates are not deficient in any of these accessory factors and because mutations at these loci alone are insufficient to explain HeR to HoR selection, the mechanism underpinning this phenomenon is clearly complex.
SCCmec elements can also carry resistance genes for other antibiotics and heavy metals as well as the psm-mec locus, which encodes a cytolysin termed phenol-soluble modulin-mec (PSM-mec) [26]. Carriage of the psm-mec locus from type II SCCmec elements attenuates virulence, suppresses colony spreading activity, reduces expression of the chromosomally encoded PSMα and promotes biofilm formation [26], [27], [28]. Furthermore both the psm-mec encoded RNA and the PSM-mec peptide contribute to the pleiotropic role of this locus [27], [28].
Our analysis of S. aureus clinical isolates identified a novel biofilm phenotype expressed by MRSA clinical isolates in which the major cell wall autolysin Atl and the fibronectin-binding proteins FnBPA and FnBPB have fundamental functions [3], [6], [7]. The Atl/FnBP biofilm phenotype appears to be absent or less prevalent among methicillin-susceptible S. aureus (MSSA) isolates, which produce PNAG-dependent biofilms in vitro. Interestingly the psm-mec locus from a type II SCCmec element increased expression of FnBPA in the MSSA strain Newman [28]. However because clinical MRSA isolates that produce an FnBP-dependent biofilm [4], [6] can contain either type II (psm-mec +) or type IV (psm-mec −) SCCmec elements [27], [28], it seems unlikely that carriage of the psm-mec locus alone can explain the expression of Atl/FnBP- or PNAG-dependent biofilm phenotypes by MRSA and MSSA clinical isolates, respectively. Furthermore when considered with earlier reports suggesting a correlation between β-lactam resistance and biofilm [29], [30], [31], [32] our data raise the question as to whether methicillin susceptibility itself influences biofilm and if so, how.
Here we investigated the impact of acquisition of PBP2a-induced homogeneous oxacillin resistance on biofilm and virulence in the laboratory MSSA strain 8325-4. Genetic changes associated with the HoR phenotype in 8325-4 were identified by whole genome sequencing. The biofilm phenotypes of 8325-4 and its HoR derivative were compared and the impact of HoR oxacillin resistance on transcription of the icaADBC and agr loci examined. The impact of loss of SCCmec and methicillin susceptibility on the biofilm phenotype of the MRSA strain BH1CC was also examined. Extracellular protease production by 8325-4 and 8325-4 HoR was measured and the virulence of both strains compared in a mouse model of device-related infection. Our data reveal that expression of homogeneous methicillin resistance in S. aureus influences the biofilm phenotype and attenuates virulence.
Results
Impact of induced oxacillin susceptibility on MRSA biofilm production
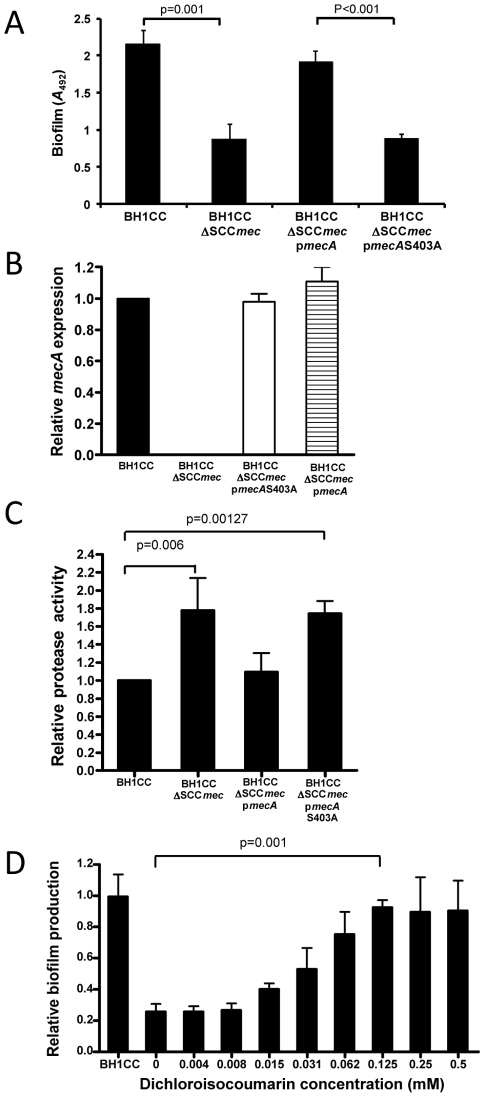
To investigate the relationship between susceptibility to β-lactam antibiotics and the biofilm phenotype we used plasmid pSR2 carrying the ccrAB recombinase genes to promote excision of SCCmec in the MRSA clinical isolate BH1CC, which contains a type II SCCmec element and produces an Atl/FnBP-dependent biofilm in growth media supplemented with glucose [6], [7]. Excision of the SCCmec element in BH1CC resulted in a reduction in the oxacillin MIC from >100 µg/ml to <1 µg/ml (data not shown). Biofilm assays revealed a significant reduction in biofilm production by BH1CC ΔSCCmec in BHI glucose compared to BH1CC (Figure 1A). Complementation of the ΔSCCmec mutant with pmecA restored both oxacillin resistance (data not shown) and biofilm production in BHI glucose to near wild type levels (Figure 1A). In contrast pmecAS403A, in which the serine residue in the PBP2a active site was replaced with alanine (as described in the Materials and Methods), failed to restore oxacillin resistance (data not shown) or biofilm production in the BH1CC ΔSCCmec mutant (Figure 1A). RT-PCR analysis demonstrated that mecA mRNA levels were similar in the BH1CC SCCmec mutant carrying pmecA and pmecAS403A (Figure 1B) indicating that functional PBP2a and not mecA mRNA was responsible for the observed phenotypes.
Figure 1. Impact of methicillin susceptibility on the MRSA biofilm phenotype.
(A) Biofilm phenotypes of BH1CC, BH1CC ΔSCCmec, BH1CC ΔSCCmec pmecA and BH1CC ΔSCCmec pmecAS403A grown for 24 h at 37°C in BHI glucose in hydrophilic 96-well polystyrene plates. (B) Comparison of relative mecA transcription by real time RT-PCR in BH1CC, BH1CC ΔSCCmec, BH1CC ΔSCCmec pmecA and BH1CC ΔSCCmec pmecAS403A. Total RNA was extracted from cultures grown at 37°C to A 600 = 1 (early exponential phase) in BHI glucose. Experiments were repeated at least three times and standard deviations are indicated. (C) Protease activity in culture supernatants of BH1CC, BH1CC ΔSCCmec and BH1CC ΔSCCmec pmecA grown for 8 h at 37°C in BHI glucose. (D) Relative biofilm production by BH1CC ΔSCCmec grown for 24 h at 37°C in BHI glucose media supplemented with the serine protease inhibitor dichloroisocoumarin (0–0.5 mM) in hydrophilic 96-well polystyrene plates compared to BH1CC (control). Experiments were repeated three times. Standard deviations and significance values are indicated.
Protease activity in the culture supernatant of BH1CC ΔSCCmec was reduced by approximately 50% compared to BH1CC and was restored to wild type levels by complementation with pmecA (Figure 1C). In addition the addition of the serine protease inhibitor dichloroisocoumarin to the growth media restored biofilm production by the BH1CC ΔSCCmec mutant strain in a dose-dependent manner (Figure 1D). Taken together these data indicate that oxacillin resistance promotes protein adhesin-dependent biofilm production in BH1CC at least in part by repressing extracellular protease production.
To determine if these findings could be extended to other clinical MRSA isolates, we deleted the SCCmec element from six clinical isolates in our collection that formed robust biofilm, were genetically amenable, had different SCCmec types and were from different clonal complexes. Furthermore we complemented all of the SCCmec mutants with the pmecA plasmid. Our data show that the impact of the SCCmec mutation on biofilm varied between the strains (Figure S1). In four strains biofilm production was significantly reduced whereas in the remaining two strains, biofilm was largely unaffected (Figure S1). However complementation with the pmecA plasmid increased biofilm production in all seven SCCmec mutants (Figure S1). The variable impact of the SCCmec deletions in different strains may reflect the complexity and multiple mechanisms of S. aureus biofilm production but nevertheless these data indicate that high level mecA/PBP2a expression always promoted biofilm production providing further evidence that methicillin resistance influences the biofilm phenotype in S. aureus clinical isolates.
Isolation of a homogeneously oxacillin resistant derivative of S. aureus 8325-4
To investigate the impact of oxacillin resistance on icaADBC/PNAG-dependent S. aureus biofilm production, we generated a methicillin (oxacillin) resistant derivative of the laboratory strain 8325-4. 8325-4 was chosen because is it amenable to genetic manipulation and exclusively produces icaADBC/PNAG-dependent biofilms [4]. The related laboratory strain SH1000 (an rsbU-repaired derivative of 8325-4 [33]) is capable of PNAG-independent biofilm production [34], [35], while HG003 [36] (an rsbU- and tcaR-repaired derivative of 8325) exhibited a smooth colony morphology on Congo red agar and did not produce detectable levels of PNAG in our experiments (Figure S2).
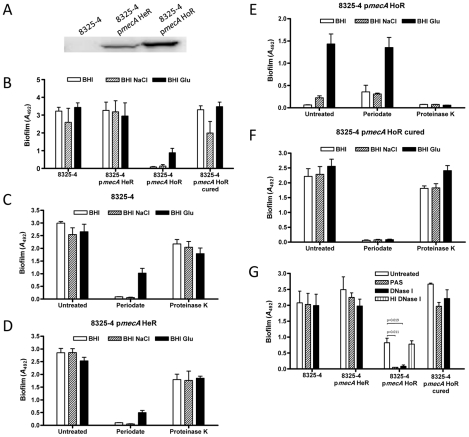
Transformation of 8325-4 with plasmid pRB474 carrying the mecA gene expressed from its own promoter (pmecA) was accompanied by heterogeneous resistance (HeR) to oxacillin (data not shown). A homogeneously, high-level oxacillin-resistant (HoR) derivative of 8325-4 pmecA was subsequently isolated on BHI media supplemented with 100 µg/ml oxacillin. Western blot analysis performed using commercial monoclonal antibody against PBP2a from Denka-Seiken (Japan) revealed substantially higher PBP2a levels in 8325-4 pmecA HoR than in 8325-4 pmecA HeR (Figure 2A). To investigate the genetic basis for the switch from expression of heterogeneous to homogeneous oxacillin resistance, the genomes of 8325-4 pmecA HeR and 8325-4 pmecA HoR were sequenced and aligned to the S. aureus NCTC8325 (CP000253) genome sequence as described in the Materials and Methods. Briefly one non-synonymous single nucleotide polymorphism (SNP) was identified in the gdpP (SAOUHSC_00015) gene of the HoR strain and confirmed by PCR amplification followed by capillary electrophoresis sequencing. This SNP results in a R602H substitution in GdpP, which has recently been identified as a c-di-AMP phosphodiesterase and implicated in resistance/tolerance to β-lactam antibiotics, biofilm formation and cell wall architecture [37], [38]. Subsequent characterization of nine independently isolated 8325-4 HoR strains identified a G308D substitution in seven strains, and Δ382–504 and Δ80–174 deletions in the other two strains. The GdpP R602 and G308 residues are highly conserved across multiple S. aureus species. These data revealed a strong correlation between high level PBP2a production and homogeneous oxacillin resistance in 8325-4 and suggest that gdpP mutations are also required for maximal PBP2a expression or stability in the 8325-4 pmecA HoR strain.
Figure 2. Impact of homogeneous oxacillin resistance on the biofilm phenotype of S. aureus 8325-4.
(A) Western blot analysis of PBP2a levels in 8325-4, 8325-4 pmecA HeR and 8325-4 pmecA HoR. (B) Biofilm phenotypes of 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured) grown for 24 h at 37°C in BHI, BHI NaCl and BHI glucose in hydrophilic 96-well polystyrene plates. (C–F) Dispersal of 8325-4 (C), 8325-4 pmecA HeR (D), 8325-4 pmecA HoR (E), 8325-4 pmecA HoR (cured) (F) biofilms grown for 24 h in BHI, BHI NaCl and BHI glucose by sodium metaperiodate and proteinase K. (G) Biofilm formation by 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured) grown for 24 h in BHI glucose in the absence or presence of 500 µg/ml PAS, 0.25 mg/ml DNase I and heat-inactivated (HI) 0.25 mg/ml DNase I. Biofilm assays were repeated at least three times and standard deviations are indicated. * indicates a statistically significant difference p<0.01.
Impact of mecA-induced oxacillin resistance on biofilm development
Comparison of the biofilm phenotypes of 8325-4, 8325-4 pmecA HeR and 8325-4 pmecA HoR under different growth conditions revealed that homogeneous oxacillin resistance was associated with a substantial reduction in biofilm forming capacity in BHI and BHI NaCl (P<0.0001) but that 8325-4 pmecA HoR retained the capacity to form reduced levels of biofilm in BHI glucose (Figure 2B). Our previous studies have shown that biofilm production by 8325-4 is dependent on PNAG under all growth conditions, whereas glucose-induced biofilm by MRSA isolates is mediated by protein adhesins [4], [6], [7]. Thus the loss of NaCl-induced biofilm was suggestive of impaired PNAG production in 8325-4 pmecA HoR. The reduced levels of biofilm produced by 8325-4 pmecA HoR compared to 8325-4 pmecA HeR in BHI glucose can be attributed to the altered biofilm phenotype expressed by this strain, but as described below, both strains formed similar levels of biofilm under flow conditions in BHI glucose. The biofilm phenotypes of 8325-4 and 8325-4 pmecA HeR were similar (Figure 2B) indicating the heterogeneous oxacillin resistance phenotype was not associated with a change in the biofilm phenotype. Curing the pmecA plasmid from 8325-4 pmecA HoR was associated with a return to oxacillin susceptibility and a restoration of the wild type biofilm phenotype (Figure 2B).
Sodium metaperiodate, which is known to break down PNAG-dependent biofilms, degraded 8325-4 and 8325-4 pmecA HeR biofilms grown in BHI, BHI NaCl and BHI glucose, whereas proteinase K had no significant effect (Figures 2C and D). In contrast 8325-4 pmecA HoR biofilms were completely dispersed by proteinase K (P<0.0001) but not by sodium metaperiodate (Figure 2E). Biofilms produced by 8325-4 pmecA HoR (cured) were dispersed by sodium metaperiodate but not proteinase K (Figure 2F). 8325-4 pmecA HoR biofilms were also inhibited by growth in the presence of DNase I whereas 8325-4 and 8325-4 pmecA HeR were unaffected (Figure 2G). Furthermore polyanethole sodium sulfonate (PAS), which blocks autolysis (and consequently eDNA release) [6], [7], completely blocked biofilm production by 8325-4 pmecA HoR but not 8325-4 and 8325-4 pmecA HeR (Figure 2G). Finally, independently isolated 8325-4 pmecA HoR isolates all exhibited the same biofilm phenotype (data not shown). Taken together these data indicate that acquisition of homogeneous oxacillin resistance is apparently accompanied by a switch from PNAG- to protein-mediated biofilm formation and that in contrast to 8325-4, the 8325-4 pmecA HoR biofilm phenotype is dependent on autolytic activity and eDNA release similar to that observed among clinical MRSA isolates expressing an Atl/FnBP-dependent biofilm [6], [7].
A BioFlux system was used to compare biofilm production by 8325-4 pmecA HeR and 8325-4 pmecA HoR grown in BHI NaCl and BHI glucose under flow conditions. These data showed that both strains formed abundant and similar levels of biofilm in BHI glucose (Figure S3). However neither strain was capable of biofilm production in BHI NaCl media (Figure S3) making it impossible to determine the contribution of PNAG to the biofilm phenotypes of both strains using the BioFlux system. The biofilm negative phenotype in BHI NaCl is likely due to the more hydrophobic surface of the BioFlux flow cell compared to the very hydrophilic, tissue-culture treated polystyrene used in our microtitre plate assay. We have previously reported that 8325-4 is incapable of producing a PNAG-type biofilm on hydrophobic polystyrene when grown in BHI NaCl, but can produce a complex biofilm dependent not only on PNAG but also protein adhesin(s) and eDNA on hydrophobic polystyrene in BHI glucose media [7].
Contribution of the icaADBC, fnbAB, atl and srtA loci to 8325-4 pmecA HoR biofilm phenotype
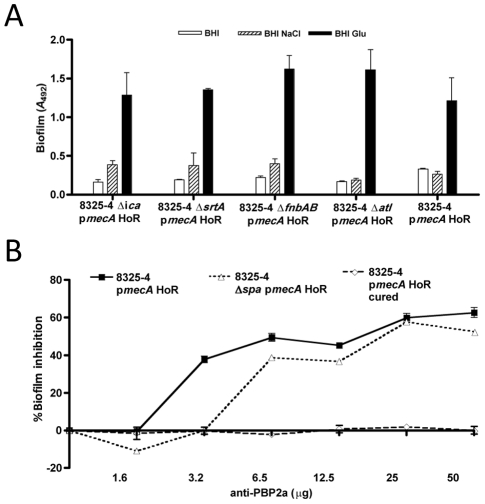
To investigate the genetic basis for protein-mediated biofilm in 8325-4 pmecA HoR, the pmecA plasmid was transformed into fnbAB::Tcr, atl::Cmr and srtA::Tcr derivatives of 8325-4 and homogeneous oxacillin resistant variants were isolated as described above. Biofilm assays revealed that HoR derivatives of 8325-4 icaADBC::Tcr, 8325-4 fnbAB::Tcr, 8325-4 atl::Cmr and 8325-4 srtA::Tcr exhibited a similar biofilm phenotype to 8325-4 pmecA HoR (Figure 3A). These data strongly suggest that the biofilm phenotypic switch associated with acquisition of oxacillin resistance is not dependent on PNAG, LPXTG cell wall anchored proteins (including FnBPA, FnBPB and Protein A) or the major autolysin.
Figure 3. Contribution of PBP2a to the S. aureus biofilm phenotype.
(A) Biofilm phenotypes of 8325-4 ΔicaADBC::Tcr pmecA HoR, 8325-4 ΔsrtA::Emr pmecA HoR, 8325-4 ΔfnbAB::Tcr pmecA HoR, 8325-4 Δatl::Cmr pmecA HoR and 8325-4 pmecA HoR (control) grown for 24 h at 37°C in BHI, BHI NaCl and BHI glucose in hydrophilic 96-well polystyrene plates. (B) Biofilm production by 8325-4 pmecA HoR, 8325-4 pmecA HoR (cured) and 8325-4 Δspa pmecA HoR (control) grown in the presence of monoclonal anti-PBP2a antibody. Biofilms were grown at 37°C for 24 h in BHI glucose in hydrophilic 96-well polystyrene plates using monoclonal anti-PBP2a antibody. Experiments were repeated three times and average data are shown.
This raised the possibility that overexpression of the PBP2a protein itself may promote biofilm development in BHI glucose. Consistent with this, commercial monoclonal antibodies (Calbiochem) to PBP2a reduced biofilm production by 8325-4 pmecA HoR by up to 50% (P<0.05)(Figure 3B). Biofilm production by 8325-4 Δspa pmecA HoR was also inhibited by the PBP2a antibody in a concentration dependent manner (Figure 3B), ruling out any non-specific interference due to antibody binding to Protein A. In contrast the PBP2a monoclonal antibody had no significant effect on PNAG-dependent biofilm production by the plasmid-cured 8325-4 pmecA HoR strain (Figure 3B). These data directly implicate the PBP2a protein in 8325-4 pmecA HoR biofilm development. However it is important to note that the failure of the pmecAS403A plasmid to complement biofilm production in the BH1CC SCCmec strain (Figure 1A) also suggests that PBP2a-induced oxacillin resistance is required for the PBP2a-mediated biofilm phenotype
Repression of PNAG production in 8325-4 pmecA HoR
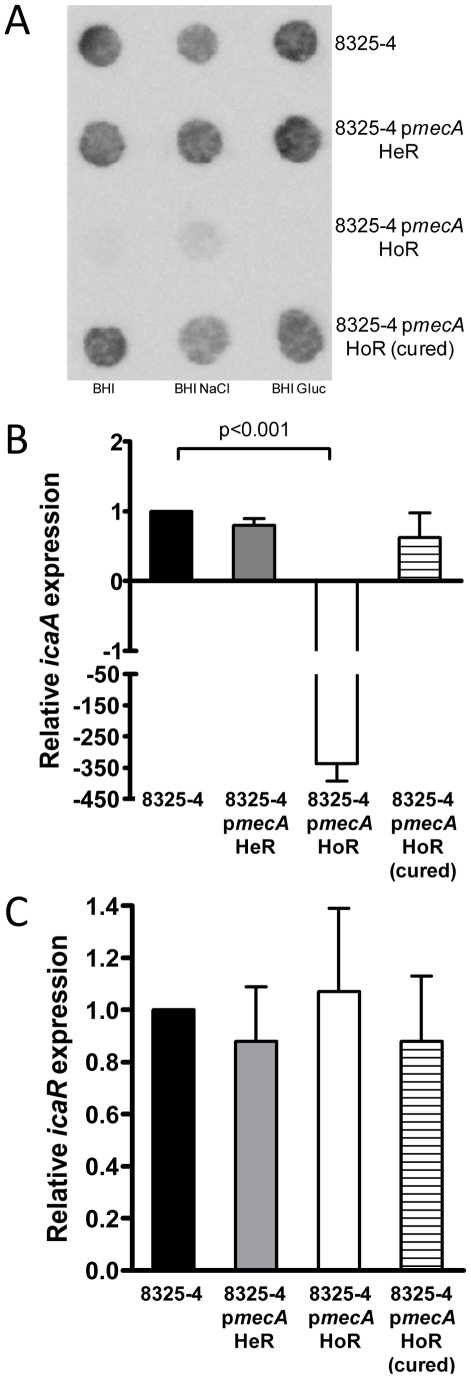
Comparison of 8325-4, 8325-4 pmecA HeR and 8325-4 pmecA HoR on Congo red agar revealed that the acquisition of homogeneous resistance was associated with a switch from a crusty to a smooth colony morphology, which is indicative of reduced PNAG production (data not shown). Accordingly immunoassays demonstrated that, unlike 8325-4 and 8325-4 pmecA HeR, PNAG was not produced by 8325-4 pmecA HoR (Figure 4A). Interestingly PNAG production was restored in the cured 8325-4 HoR strain (Figure 4A). Using real time RT-PCR, a >300-fold repression of icaADBC transcription was measured in 8325-4 pmecA HoR compared to 8325-4 (Figure 4B). Furthermore wild type levels of icaADBC transcription were restored in the plasmid-cured 8325-4 pmecA HoR strain (Figure 4B). Transcriptional activity of icaR was similar in both strains (Figure 4C). In addition genome sequencing of 8325-4 pmecA HoR confirmed the absence of any mutations in any known ica operon transcriptional regulator. Thus homogeneous oxacillin resistance alters the biofilm phenotype by repressing icaADBC transcription and PNAG production.
Figure 4. Impact of homogeneous oxacillin resistance on PNAG production in 8325-4.
(A) Immunoblot analysis of PNAG production in whole cell extracts of 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured) grown overnight at 37°C in BHI, BHI NaCl and BHI glucose. (B and C) Comparison of relative icaA (B) and icaR (C) transcription by real time RT-PCR in 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured). Total RNA was extracted from cultures grown at 37°C to A 600 = 1 (early exponential phase) in BHI glucose. Experiments were repeated at least three times and standard deviations are indicated. Statistical significance (p value) is indicated.
Repression of extracellular protease production and Agr activity in 8325-4 pmecA HoR
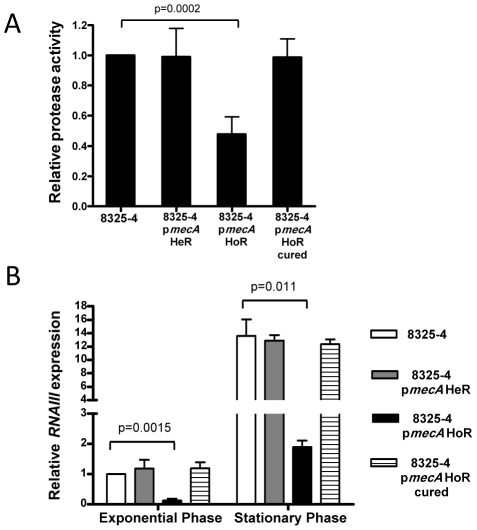
Assays of protease activity in culture supernatants revealed an approximately 2-fold reduction in protease levels in 8325-4 pmecA HoR compared to 8325-4, 8325-4 pmecA HeR and the cured 8325-4 HoR strain (Figure 5A). Because extracellular protease activity is subject to regulation by the accessory gene regulator (Agr) system, we used real time RT-PCR to compare RNAIII transcript levels in these strains. These data revealed that RNAIII expression was significantly repressed in 8325-4 pmecA HoR grown to both the exponential and stationary phases of growth (Figure 5B). Genome sequence analysis confirmed the absence of any mutations in the agr locus of 8325-4 pmecA HoR. Thus the homogeneous oxacillin resistance phenotype in 8325-4 is associated with repression of the Agr system and extracellular protease production, both of which are consistent with PBP2a-mediated biofilm development by 8325-4 pmecA HoR. Furthermore these data correlate with our earlier observation that protease activity was increased in BH1CC ΔSCCmec culture supernatants (Figure 1B).
Figure 5. Impact of homogeneous oxacillin resistance on protease activity and expression of the accessory gene regulator locus in 8325-4.
(A) Protease activity in culture supernatants of 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured) grown for 8 h at 37°C in BHI glucose. (B) Comparison of relative RNAIII transcription by real time RT-PCR in 8325-4, 8325-4 pmecA HeR, 8325-4 pmecA HoR and 8325-4 pmecA HoR (cured). Total RNA was extracted from cultures grown at 37°C to A 600 = 1 (early exponential phase) and for 8 hours (late exponential/early stationary phase; A 600≈8) in BHI glucose. Experiments were repeated at least three times and standard deviations are indicated. Statistical significance (p value) is indicated.
Homogeneous methicillin resistance attenuates virulence in 8325-4
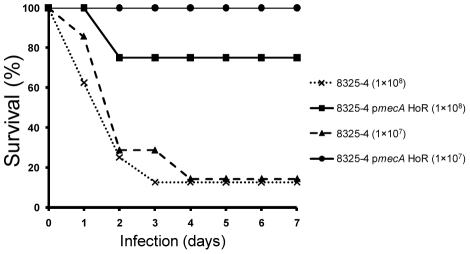
Because 8325-4 and 8325-4 pmecA HoR express PNAG-dependent and PNAG-independent biofilm phenotypes, respectively, their virulence was compared using an established mouse model of device-related infection [39], [40]. Briefly two 1-cm segments of 14-gauge polyethylene intravenous catheter were implanted subcutaneously per mouse. In total four groups of eight mice were inoculated with bacterial cell suspensions of 1×107 or 1×108 of either 8325-4 or 8325-4 pmecA HoR, injected directly into the catheter. These experiments revealed a significantly higher mortality rate among mice inoculated with 8325-4 compared to 8325-4 pmecA HoR (Figure 6). Seven of the eight mice inoculated with 1×108 8325-4 died or were euthanized by Day 4. Similarly only one of seven mice (one mouse failed to recover from the anesthetic) inoculated with 1×107 8325-4 survived beyond Day 4 (Figure 6). In contrast all eight animals inoculated with 1×107 8325-4 pmecA HoR survived to Day 7 and six of the eight animals inoculated with 1×108 8325-4 pmecA HoR survived to Day 7 (Figure 6).
Figure 6. Impact of homogeneous oxacillin resistance on survival in a mouse device-related S. aureus infection model.
1-cm segments of 14-gauge polyethylene intravenous catheter were implanted into male 6-week old C57BL/6 mice (2 per mouse) and injected with 1×107 or 1×108 8325-4 and 8325-4 pmecA HoR. Survival of the animals over seven days is plotted as a percentage of total numbers (n = 8 mice per group except for 8325-4 (1×107) where n = 7 mice).
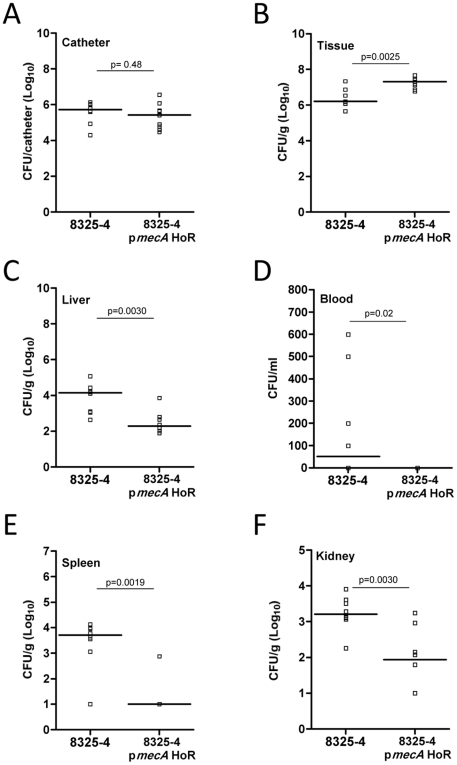
The high mortality rate associated with infection of 8325-4 made it impossible for us to compare the invasiveness and dissemination of 8325-4 and 8325-4 pmecA HoR. Thus, the above experiment was repeated over an 18-hour time period with an inoculum of 1×107 bacteria after which all animals were sacrificed. The implanted catheter sections were aseptically removed and bacteria associated with the implanted biomaterial were quantitatively cultured on tryptone soya agar (TSA). In addition, peri-catheter tissue, liver, kidneys and spleen were dissected, weighed, homogenized and quantitatively cultured on TSA, as were bacteria present in blood. These data revealed that the numbers of adherent bacteria on implanted catheters were similar for 8325-4 and 8325-4 pmecA HoR (Figure 7A) indicating that the altered biofilm phenotype expressed by 8325-4 pmecA HoR does not diminish its ability to colonize implanted devices. There were significantly more 8325-4 pmecA HoR bacteria in the peri-catheter tissue than 8325-4 bacteria (Figure 7B). However significantly fewer 8325-4 pmecA HoR bacteria were recovered from the liver, blood, spleen and kidneys than 8325-4 bacteria (Figure 7 C–F). Thus, these findings are consistent with the mortality data and indicate that 8325-4 is significantly more invasive than 8325-4 pmecA HoR, which accumulated in the peri-catheter tissue and was less capable of disseminating to other organs.
Figure 7. Catheter colonisation and dissemination of 8325-4 and 8325-4 pmecA HoR in a mouse device-related infection model.
Implanted catheter segments were injected with 1×107 8325-4 and 8325-4 pmecA HoR and animals (n = 8 mice per group) were sacrificed after 18 hours. Colony forming units (CFU) per catheter (A), per g peri-catheter tissue (B), per g liver (C), per ml blood (D), per g spleen (E) and per g kidney (F) recovered from animals infected with 8325-4 and 8325-4 pmecA HoR. Statistical significance (p values) is indicated.
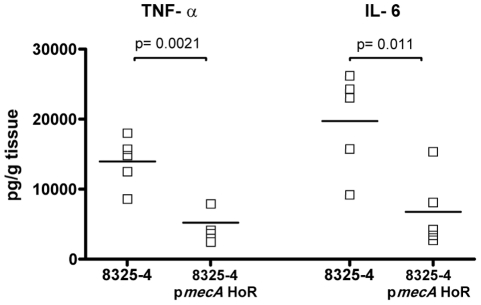
The immune response of the mice to 8325-4 and 8325-4 pmecA HoR was measured by assaying levels of TNF-α and IL-6 in the peri-catheter tissue. Levels of both pro-inflammatory cytokines were significantly increased in mice inoculated with 8325-4 compared to 8325-4 pmecA HoR (Figure 8), despite the fact that there were more 8325-4 pmecA HoR cells recovered from the tissue (Figure 7B). Taken together these data reveal that homogeneous, high-level resistance to oxacillin significantly attenuated the virulence of 8325-4.
Figure 8. Enzyme-linked immunosorbent assay (ELISA) of TNF-α and IL-6 in peri-catheter tissue recovered from mice (n = 7 mice per group) infected for 18 h with 8325-4 and 8325-4 pmecA HoR.
Statistical significance (p values) is indicated.
Impact of homogeneous methicillin resistance on biofilm production and δ-hemolytic activity in clinical isolates of S. aureus
To investigate the impact of homogenous methicillin resistance on biofilm production and Agr activity in clinical S. aureus isolates, we first introduced pmecA into the MSSA strains MSSA476 and 15981 and isolated HoR derivatives on media containing oxacillin 100 µg/ml. As observed in 8325-4, acquisition of the HoR phenotype in MSSA476 and 15981 was associated with repression of polysaccharide-type biofilm production and expression of a protein adhesin-mediated biofilm phenotype (Figure S4). Furthermore the MSSA476 and 15981 HoR strains exhibited reduced δ-hemolytic activity on sheep blood BHI agar (Figure S4). Given that the δ-hemolysin is encoded by the hld gene within the agr locus RNAIII transcript [41], these data indicate that the HoR phenotype in MSSA476 and 15981 is accompanied by repression of Agr activity.
The impact of the HoR phenotype on biofilm production by the CA-MRSA USA300 strain LAC was also examined. In our experiments this strain formed a protein-adhesin type biofilm in BHI glucose media (Figure S5A). A LAC HoR derivative isolated on media containing oxacillin 100 µg/ml produced significantly more biofilm (Figure S5A) and exhibited reduced δ-hemolytic activity on sheep blood BHI agar (Figure S5B). Finally we examined the impact of the HoR phenotype on three CC5 strains with different SCCmec elements exhibiting a heterogeneous pattern of oxacillin resistance, namely DAR173, DAR26 and DAR9. These strains produced a polysaccharide-type biofilm, whereas their HoR derivates isolated on media containing oxacillin 100 µg/ml produced protein adhesin-type biofilms and exhibited reduced δ-hemolytic activity (Figure S5C–K).
In the laboratory strain 8325-4, non-synonymous SNPs in gdpP were associated with the HoR phenotype. Similarly, DNA sequencing of the gdpP gene from the USA300 LAC HoR derivative identified an R450STOP mutation. GdpP amino acid substitutions were also identified in HoR derivatives of the clinical isolates 15981 (P392S, D105N), MSSA476 (V52I, D105N, P392S) and DAR26 (N105D, S392P). However these latter substitutions occur in GdpP residues that are not conserved in multiple S. aureus species and so their significance is unclear. No gdpP mutations were identified in HoR derivatives of the clinical isolates DAR176 and DAR9 suggesting that the HoR phenotypes of these strains may be independent of c-di-AMP or that these strains contain unidentified mutations in c-di-AMP target genes.
Discussion
Antibiotic resistance, enzyme and toxin production, biofilm forming capacity and immune evasion capability have combined to accelerate the emergence of S. aureus as a globally important human pathogen. Our recent research has focused on the relationship between two of these virulence determinants, antibiotic resistance and biofilm-forming capacity, in clinical S. aureus isolates. Our data show that MSSA clinical isolates are more likely to produce a PNAG-dependent biofilm than MRSA isolates which produce an Atl/FnBP-dependent biofilm [3], [4], [6], [7], suggesting that methicillin susceptibility influences biofilm expression. MRSA strains contain the Staphylococcus cassette chromosome mec (SCCmec) including the mecA gene, which encodes the PBP2a protein that confers resistance to β-lactam antibiotics. To investigate how methicillin resistance influences the biofilm phenotype we introduced a plasmid expressing the mecA gene from its own promoter (pmecA) into the PNAG-producing laboratory strain 8325-4. 8325-4 pmecA exhibited heterogeneous resistance to oxacillin (designated 8325-4 pmecA HeR) from which an homogeneous, high-level oxacillin resistant derivative was isolated, designated 8325-4 pmecA HoR. Western blots show that expression of homogeneous resistance was associated with substantially increased expression of PBP2a. Genome re-sequencing identified a number of amino acid substitutions and small deletions in the gdpP gene of independent 8325-4 HoR strains and in HoR derivatives of the clinical isolates USA300 strain LAC, MSSA476, 15981 and DAR26. GdpP has recently been identified as a phosphodiesterase controlling intracellular levels of the secondary messenger c-di-AMP, which in turn influences phenotypes such as cell wall architecture, biofilm formation and, most relevant to this study, resistance/tolerance to β-lactam antibiotics [37], [38]. Thus c-di-AMP may be involved in the multiple phenotypic changes associated with homogeneous oxacillin resistance. However, given that the GdpP amino acid sequence is identical in 8325-4 (MSSA), USA300 strain LAC (HeR MRSA) and BH1CC (HoR MRSA), mutations in gdpP and/or altered c-di-AMP signaling alone may not be sufficient to explain the HeR to HoR transition. Furthermore no gdpP SNPs were identified in HoR derivatives of two clinical isolates (DAR9 and DAR176) suggesting that either these strains contain mutations in c-di-AMP target genes or that c-di-AMP-independent mechanisms may also be involved in the HoR phenotype. In this context our recent finding that loss of methicillin resistance in the HoR MRSA strain BH1CC reduced biofilm forming capacity and increased virulence in a mouse sepsis model [42], also indicates that altered c-di-AMP levels alone are unlikely to account for the pleiotropic effects of the HoR phenotype. Clearly homogenous methicillin resistance remains a complex phenotype and future research to identify c-di-AMP targets is necessary to gain mechanistic insights into how this cyclic dinucleotide contributes to the co-regulation of antibiotic resistance, biofilm and virulence.
The biofilm phenotype of 8325-4 pmecA HoR was dramatically altered and characterized by the loss of PNAG production and expression of a protein-adhesin mediated biofilm phenotype. Using a series of surface protein mutants, we were able to show that the PBP2a-induced biofilm phenotype was independent of PNAG, Atl, and any of the LPXTG-anchored surface proteins (including the FnBPs and protein A). Furthermore, a monoclonal antibody against PBP2a reduced 8325-4 pmecA HoR biofilm production by approximately 50% implicating PBP2a itself in intercellular accumulation. The inability of the PBP2a antibody to fully inhibit 8325-4 pmecA HoR biofilm may be explained, at least in part, by our observation that this biofilm phenotype was also dependent on autolytic activity and extracellular DNA. We have previously implicated autolytic activity and eDNA in the early stages of PNAG-independent biofilm phenotypes expressed by S. aureus clinical isolates [6], [7]. In contrast biofilm expression by 8325-4 pmecA HeR was similar to wild type, indicating that high level PBP2a expression is required to change the biofilm phenotype. In the MRSA strain BH1CC, excision of SCCmec and induced methicillin susceptibility also resulted in an impaired biofilm phenotype that could be complemented by the wild type mecA gene but not a mecA allele expressing a PBP2a mutant with an S403A substitution in the active site of the enzyme. Taken together, these data suggest that both high level PBP2a expression and methicillin resistance are required for the PBP2a-induced biofilm phenotype. In addition, because Atl/FnBP-dependent biofilm development by BH1CC was unaffected by PBP2a antibody (data not shown), our data also reveal redundancy among S. aureus surface proteins that can promote biofilm development.
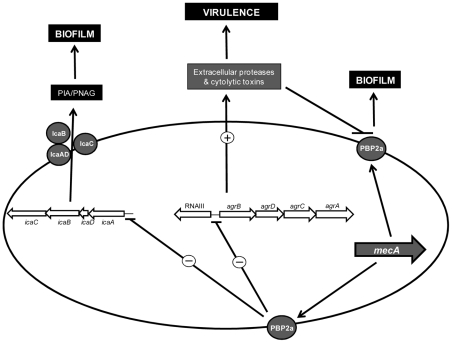
How PBP2a promotes biofilm is uncertain but possibly the altered cell wall architecture in methicillin resistant strains expressing high levels of PBP2a may facilitate PBP2a-promoted cell-cell interactions that are not possible between MSSA cells. What is clear, however, is that high level PBP2a expression resulted in significant repression of icaADBC transcription and PNAG production in 8325-4 pmecA HoR (Figure 9). Comparison of acetic acid levels in 8325-4 and 8325-4 pmecA HoR culture supernatants revealed no significant difference (data not shown) suggesting that altered TCA cycle activity, which is known to regulate PIA/PNAG production [43], [44], may not be involved in this phenotype. PBP2a overexpression also resulted in significant repression of the agr locus. These data are supported by our recent findings that excision of the SCCmec element from the MRSA strain BH1CC and loss of oxacillin resistance had the opposite effect and was associated with increased agr transcription [42]. Furthermore our previous data showed that MRSA cells are unable to detect the Agr-encoded auto-inducing peptide (AIP), thus preventing normal activation of the Agr system and concomitant virulence gene regulation [42]. However this does not explain why the icaADBC locus is repressed in strains expressing high levels of PBP2a. Given that both the agr and ica loci are repressed by PBP2a expression, it seems possible that a transcriptional regulator(s) involved in regulating both operons may also play a role in this phenotype. For instance preliminary data indicate that the activity of the virulence and carbon metabolism regulator CcpA, which is known to increase oxacillin resistance levels [45], [46], was activated approx 4-fold in 8325-4 pmecA HoR (unpublished findings). On the other hand CcpA positively regulates both icaADBC and agr expression [45], [46], suggesting that the impact of PBP2a overexpression on biofilm and virulence is complex and experiments are underway to investigate the potential involvement of other transcription factors.
Figure 9. Model of PBP2a-mediated modulation of biofilm expression and virulence in S. aureus.
High level PBP2a expression and homogeneous methicillin resistance results in repression of the icaADBC and agr loci blocking PNAG production and reducing expression of extracellular proteases and cytolytic toxins. Reduced levels of extracellular protease and toxin production in turn correlate with PBP2a-promoted biofilm development and attenuated virulence.
Repression of the Agr locus in response to homogenous methicillin resistance was accompanied by down-regulation of extracellular protease production [35], [47] (Figure 9). Protease activity has previously been shown to alter murein hydrolase activity and in turn biofilm [35], which appears to correlate with our finding implicating autolytic activity in the 8325-4 pmecA HoR biofilm phenotype. Thus repression of extracellular proteases combined with high level PBP2a expression and methicillin resistance appears to be required for PBP2a-mediated biofilm development in 8325-4. Consistent with this, activation of agr following excision of SCCmec from BH1CC increased production of extracellular toxins [42]. Here we have shown that loss of SCCmec also increased extracellular protease production in BH1CC, which in turn correlated with diminished Atl/FnBP-promoted biofilm forming capacity. It is also interesting to note that extracellular protease activity was similar in both 8325-4 pmecA HoR and BH1CC SCCmec pmecA, which both express a mecA-promoted biofilm phenotype and that as mentioned above BH1CC does not display the characteristic stationary phase induction of RNAIII typical of agr + strains [42]. Taken together these data indicate that the capacity of PBP2a to promote biofilm is dependent, at least in part, on the background levels of extracellular protease activity in individual strains.
The altered biofilm phenotype and repression of agr in 8325-4 pmecA HoR was accompanied by a significant reduction in virulence in a mouse model of device-related infection. This finding correlated with our previous data that the virulence of BH1CC was increased following excision of SCCmec in a mouse sepsis model [42]. Thus using two different strains and two different mouse infection models, our data indicate that expression of methicillin resistance reduces virulence potential in S. aureus. Furthermore although the 8325-4 pmecA HoR strain was equally capable of colonizing implanted biomaterials, it accumulated significantly more in peri-catheter tissue but was significantly less capable of causing invasive disease, resulting in a significantly reduced mortality rate than the wild type 8325-4. An intriguing backdrop to our findings are the recent reports that psm-mec locus, which is located adjacent to the mecA locus on type II SCCmec elements, also represses virulence and promotes the S. aureus biofilm phenotype [26], [27], [28]. Thus both mecA-encoded PBP2a and the psm-mec encoded phenol-soluble modulin-mec (PSM-mec) can independently repress virulence and promote biofilm in MRSA. The psm-mec-encoded RNA is able to repress expression of the chromosomally encoded PSMα, while both the psm-mec RNA and the PSM-mec protein repress colony spreading ability and promote biofilm [26], [27], [28].
Given that healthcare-associated MRSA strains are typically responsible for infections in immunocompromised patients in which numerous implanted medical devices are used for organ and life support and in which the use of antimicrobial drugs is high, these findings reveal a sophisticated level of adaptation by MRSA to the hospital environment. The reduced metabolic costs associated with PBP2a-induced repression of Agr and exoprotein production coupled with PBP2a-mediated biofilm production, may confer advantages on MRSA strains. Agr defective strains are known to be less virulent [48], [49] and a number of studies [50], [51] have suggested that, in immunocompromised patients, Agr defective strains may have advantages including reduced metabolic costs of exoprotein production, increased biofilm production [52] and increased FnBP expression [53] with resulting effects on host cell invasion/immune evasion [54]. Indeed biofilm-forming S. epidermidis strains, which express fewer exoproteins and exotoxins (and are consequently less virulent), are also significant pathogens in this patient group. Thus sacrificing virulence for antibiotic resistance is not necessarily a disadvantage for MRSA and may in fact benefit the pathogen in this clinical setting. Furthermore MRSA strains have retained the capacity for biofilm production, albeit using surface protein adhesins rather than PNAG, which is important given that the majority of bloodstream infections in hospital patients are associated with implanted medical devices.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All mouse protocols were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Centre. All surgery was performed under anesthesia and all efforts were made to minimize suffering.
Bacterial strains, plasmids and growth conditions
The S. aureus strains and the plasmids used in the manipulation of these strains are described in Table 1. Escherichia coli strains were grown at 37°C on LB medium supplemented, when required, with ampicillin (100 µg/ml) or kanamycin (50 µg/ml). S. aureus strains were grown at 30°C or 37°C on Brain-Heart Infusion (BHI) (Oxoid) medium supplemented when required with chloramphenicol (10 µg/ml), tetracycline (10 µg/ml), kanamycin (10 µg/ml) and oxacillin (0–100 µg/ml). BHI broth was supplemented where indicated with 1% glucose or 4% NaCl.
Table 1. Strains and plasmids used in this study.
| Strain/Plasmid | Characteristic(s) | Source/reference |
| 8325-4 | NCTC8325 cured of prophages. 11-bp deletion in rsbU. | [33] |
| RN4220 | Restriction deficient mutant of 8325-4. | [64] |
| SH1000 | 8325-4 with repaired defect in rsbU. | [33] |
| HG003 | NCTC8325 with repaired tcaR and rsbU genes. | [36] |
| 8325-4 pmecA HeR | 8325-4 carrying pmecA expressing heterogeneous resistance to oxacillin. | [42] |
| 8325-4 pmecA HoR | 8325-4 carrying pmecA expressing homogeneous resistance to oxacillin. | This study |
| ICA1 | ΔicaADBC::Tcr; isogenic mutant of 8325-4. | [3] |
| DU5883 | fnbA::Tcr,fnbB::Emr; isogenic mutant of 8325–4. | [65] |
| SKM1 | ΔsrtA::Emr; isogenic mutant of RN4220. | [66] |
| JT1392 | Δatl::Cmr, isogenic mutant of RN4220. | [67] |
| spa::Kmr | spa::Kmr, isogenic mutant of 8325-4. | [68] |
| BH1CC | MRSA clinical isolate. Biofilm positive. SCCmec type II, MLST type 8, clonal complex 8. | [4] |
| BH1CC ΔSCCmec | Isogenic mutant of BH1CC lacking SCCmec. | [61] |
| BH10(03) | Biofilm positive, homogenoeously methicillin resistant MRSA. SCCmec type IV, clonal complex 22. | [4] |
| DAR13 | Biofilm positive, homogenoeously methicillin resistant (HoR) MRSA. SCCmec type IV, clonal complex 8. | [4] |
| DAR168 | Biofilm positive, HoR MRSA. SCCmec type I, clonal complex 8. | [4] |
| DAR34 | Biofilm positive, HoR MRSA. SCCmec type II, clonal complex 8. | [4] |
| DAR199 | Biofilm positive, HoR MRSA. SCCmec type II, clonal complex 30. | [4] |
| DAR112 | Biofilm positive, HoR MRSA. SCCmec type III, clonal complex 239. | [4] |
| DAR9 | Biofilm positive, heterogeneously methicillin resistant (HeR) MRSA. SCCmec type I, clonal complex 5. | [4] |
| DAR26 | Biofilm positive, HeR MRSA. SCCmec type IV, clonal complex 5. | [4] |
| DAR173 | Biofilm positive, HeR MRSA. SCCmec type II, clonal complex 5. | [4] |
| MSSA476 | Community associated-MSSA. Novel SCC element lacking mecA. | [69] |
| LAC | Community associated, HeR MRSA USA300. | [35] |
| 15981 | Clinical MSSA isolate. Biofilm positive. rsbU +. | [70] |
| E. coli TOPO | recA1 endA1 lac [F′ proAB lac1q Tn10 (tetr)] | Invitrogen |
| Plasmids | ||
| pSR | Shuttle vector carrying the SCCmec ccrA and ccrB recombinase genes | [15] |
| pRB474 | Low copy E. coli-Staphylococcus shuttle vector. Apr (E. coli), Cmr (Staphylococcus). | [71] |
| pSAmecA5 | 2,850 bp PCR fragment containing mecA gene amplified using primers SAmecA3 and SAmecA4 and cloned into pCR-Blunt II-TOPO (Invitrogen). | [42] |
| pmecA | 2,867 bp EcoRI fragment containing mecA from pSAmecA5 subcloned into the EcoRI site of pRB474. | [42] |
| pmecAS403A | pmecA expressing a PBP2a S403A mutant | This study |
δ-haemolytic activity was visualized on sheep blood BHI agar as described by Traber et al. [41]. Briefly clinical S. aureus isolates were inoculated onto a lawn of S. aureus RN4220, which produces only β-haemolysin. The RN4220-expressed β-haemolysin enhances lysis of red blood cells by δ-haemolysin, while inhibiting α-haemolysin, thus enabling detection of δ-haemolytic activity by clinical S. aureus isolates.
Genetic techniques
Genomic and plasmid DNA was prepared using Wizard Genomic DNA and plasmid purification kits (Promega). Prior to DNA extraction cells were pre-treated with 5–10 µl of a 1 mg/ml concentration of lysostaphin (Ambi products, New York) in 100 µl 50 mM EDTA to facilitate lysis. Restriction and DNA modifying enzymes (Roche, UK and New England Biolabs, MA) were used according to the manufacturer's instructions. The serine residue at amino acid 403 in the active site of PBP2a was mutated to alanine using Phusion polymerase (NEB) and the primers mecAS403A_For and mecAS403A_Rev (Table 2). Plasmid pmecA was used as the template. Successful mutagenesis of the DNA sequence encoding the S403 residue resulted in the introduction of a new DraIII restriction enzyme site and candidate plasmids harbouring the mutation were digested with this enzyme before being confirmed by sequencing. The mutated plasmid designated pmecAS403A.
Table 2. Oligonucleotide primers used in this study.
| Target gene | Primer names | Primers sequence (5′-3′) |
| mecA | SAmecA1 | AGTTGTAGTTGTCGGGTTTGG |
| SAmecA2 | GCATTGTAGCTAGCCATTCCTT | |
| mecA | SAmecA3 | TGACGATTCCAATGACGAAC |
| SAmecA4 | GCATCTGGCATGCATACACT | |
| mecA | SAmecAFOR | TTGGAACGATGCCTATCTCA |
| SAmecAREV | TCCAGGAATGCAGAAAGACC | |
| 16S rRNA | SA_16S_for | ATGCAAGTCGAGCGAAC |
| SA_16S_rev | TGTCTCAGTTCCAGTGTGGC | |
| RNAIII | RNAIII_2_for | GGAGTGATTTCAATGGCACA |
| RNAIII_2_rev | CATGGTTATTAAGTTGGGATGG | |
| icaA | SAicaEON1 | TACCTTGCCTAACCCGTAC |
| SAicaEON2 | GTTGGCTCAATGGGGTCTAA | |
| icaR | SAicaR1 | TTCTCAATATCGATTTGTATTGTCAAC |
| SAicaR2 | TGTCAGGCTTCTTGTTCAATG | |
| mecA | mecAS403_For | ACAACTTCACCAGGTTGCACTCAAAAAATATTA |
| mecAS403R_Rev | TAATATTTTTTGAGTGCAACCTGGTGAAGTTGT | |
| gdpP | gdpP_For | CGCATGGTTCAGACGATAAA |
| gdpP_Rev | GCTTCGGCAATTTGTTTTGT |
Transformations of plasmid DNA into E. coli and Staphylococcus strains were performed as described previously [55]. MWG Biotech, Germany or Sigma-Aldrich, Ireland supplied oligonucleotide primers used for PCR and RT-PCR (Table 2).
Biofilm and protease assays
Semi-quantitative measurements of biofilm formation were determined using Nunclon tissue culture treated (Δ surface) 96-well polystyrene plates (Nunc, Denmark), based on the methods of Christensen et al. [56] and Ziebuhr et al. [57] with the following modification. Bacteria were grown in individual wells of 96-well plates at 37°C in BHI medium or BHI supplemented with 4% NaCl or 1% glucose. The serine protease inhibitor dichloroisocoumarin was added to BHI glucose media at concentrations of 0.004–0.5 mM, where indicated. After 24 h of growth, the plates were washed vigorously three times with distilled H2O to remove unattached bacteria and dried for 1 hour at 60°C, as recommended by Gelosia et al. [58] as described previously [4], [55]. The absorbance of the adhered, stained biofilms was measured at A 492 using a microtitre plate reader. Each strain was tested at least three times and average results are presented. Mouse monoclonal anti-PBP2a antibody (CalBioreagents, CA), DNase I (Sigma) or polyanethole sodium sulfonate (PAS) (Sigma) were added to biofilm cultures at the start of the assay as indicated. Biofilm stability against proteinase K (Sigma), sodium-meta-periodate (Sigma) was tested as described previously [10], [59], [60].
Bacteria were grown on Congo red agar (CRA) plates, which are composed of BHI agar supplemented with 5% sucrose (Sigma) and 0.8 mg of Congo red/ml (Sigma) to distinguish between PNAG-producing (black, dry colony morphology) and non-PNAG-producing (red, smooth colony morphology) phenotypes as described previously [4], [55].
Protease activity in culture supernatants was measured using a protease assay kit (Calbiochem, Germany) according to the manufacturer's instructions.
To analyze biofilm formation under flow conditions, we utilized the BioFlux 1000 microfluidic system (Fluxion Biosciences Inc., South San Francisco, CA) which allows automated image acquisition within specialized multi-well plates. To grow biofilms, the microfluidic channels were primed with 50% BHI supplemented with 4% NaCl or 1% glucose at 10.0 dyn/cm2. Channels were seeded at 2 dyn/cm2 with 107 CFU from overnight cultures of 8325-4 pmecA HeR and 8325-4 pmecA HoR. The plate was then incubated at 37°C for 1 hour to allow cells to adhere. Excess inoculums were removed and 2 ml of 50% BHI supplemented with 4% NaCl or 1% glucose was added to the input wells. Biofilms were grown at 37°C with a flow of fresh media at a constant shear of 0.7 dyn/cm2. Images were taken every 5 minutes for 18 hours at 200× magnification under brightfield.
Isolation and analysis of a homogeneous oxacillin resistant derivative of S. aureus 8325-4
The pmecA plasmid [61] was introduced by electroporation into 8325-4. The 8325-4 pmecA strain exhibited heterogeneous oxacillin resistance (HeR) characterized by a minority of cells with an MIC>100 µg/ml and the majority of cells with an MIC<1 µg/ml. To obtain high-level oxacillin resistant derivatives, dilutions of 8325-4 pmecA cultures were grown on BHI agar containing increasing concentrations of oxacillin (0–100 µg/ml) and the number of colony forming units counted after 24 h growth at 37°C. High-level, homogeneous oxacillin resistant derivatives (HoR) were subcultured from oxacillin 100 µg/ml plates. Curing the pmecA plasmid from 8325-4 pmecA HoR was achieved by 48 h growth in antibiotic free media at 45°C and isolation of chloramphenicol susceptible colonies followed by plasmid profile analysis.
The genomes of the 8325-4 pmecA HeR and 8325-4 pmecA HoR were sequenced using an Illumina Genome Analyzer as per manufacturer's instructions (Illumina, San Diego, CA, USA) generating 5.7 million and 7.5 million reads for 8325-4 pmecA HoR and 8325-4 pmecA HeR, respectively. The genome sequences were then mapped back to the S. aureus NCTC8325 (CP000253) genome sequence using the short read aligner Bowtie (http://bowtie-bio.sourceforge.net/index.shtml & PMID:19261174) allowing up to 2 mismatches per uniquely mapped read. Single nucleotide polymorphisms (SNPs) were identified using samtools programs [PMID:19505943] in regions with a read depth of greater than 4. Identified SNPs were confirmed by PCR amplification followed by capillary electrophoresis sequencing of all candidate polymorphic regions from 8325-4 pmecA HeR and 8325-4 pmecA HoR.
RNA purification and real time RT-PCR
Cultures were grown in BHI glucose after which cells were collected and immediately stored at −20°C in RNAlater (Ambion) to ensure maintenance of RNA integrity prior to purification. Cells were pelleted and resuspended in 100 µl of 200 mM Tris HCl (pH 7.8) supplemented with 800 µg/ml lysostaphin for 2 min to weaken the cell wall prior to lysis. Total RNA was subsequently isolated using the Qiagen RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Residual DNA present in the RNA preparations was removed using Ambion recombinant Turbo DNase. Purified RNA was eluted and stored in RNAsecure resuspension solution (Ambion) and the integrity of the rRNA confirmed by agarose gel electrophoresis. RNA concentration was determined using a Nanodrop spectrophotometer.
Real-time reverse transcription-PCR (RT-PCR) was performed on a LightCycler instrument using the RNA amplification kit Sybr Green I (Roche Biochemicals, Switzerland) following the manufacturer's recommended protocol. RT was performed at 61°C for 30 min, followed by a denaturation step at 95°C for 30 sec and 35 amplification cycles of 95°C for 20 sec, 50°C for 20 sec and 72°C for 20 sec. Melting curve analysis was performed at 45°C to 95°C (temperature transition, 0.1°C per sec) with stepwise fluorescence detection. For LightCycler RT-PCR, RelQuant software (Roche Biochemicals) was used to measure relative expression of target genes. 16S rRNA was used as an internal standard in real-time RT-PCR experiments. Each experiment was performed at least three times and average data with standard deviations are presented.
PNAG assays
PNAG assays were performed as described elsewhere [62]. Briefly 5 ml overnight cultures (approximately 5×109 bacteria) were collected by centrifugation, resuspended in 500 µl of 0.5 M EDTA and boiled for 5 min. The cell debris was again centrifuged and the supernatant treated with 20 µg proteinase K at 65°C for 1 h. The proteinase K was inactivated by boiling for 5 min and the samples diluted as appropriate before application onto nitrocellulose (pre-wetted in TBS) using a vacuum blotter. The blots were dried, re-wet in TBS, and blocked for 1 h in 5% skimmed milk. The primary antibody (1∶5,000 dilution of rabbit anti-PNAG (a kind gift from Tomas Maira Litran) in TBST+1% skimmed milk) was then applied to the membrane for 1 h. Horseradish peroxidase linked anti-rabbit IgG secondary antibody (1∶5,000 dilution in TBST+1% skimmed milk) was then incubated with the membrane for 1 h. A chemiluminescence kit (Amersham) was used to generate light via the HRP-catalyzed breakdown of luminal and detected using a BioRad Fluor-S Max CCD camera system.
Detection of PBP2a by Western blot
Western blot analysis of PBP2a was performed as described elsewhere [63] using overnight bacterial cultures (20 ml) grown at 37°C in BHI glucose. The A 600 of the overnight cultures was measured to ensure that similar cell densities were present prior to protein extraction. The cells were harvested by centrifugation, washed with 50 mM Tris (Sigma), 150 mM NaCl and 5 mM MgCl (pH 7.5) and resuspended in the same buffer. Lysostaphin (200 µg/µl), RNase (10 µg/µl) and DNase (20 µg/µl) were added to the cell suspension and incubated at 37°C for 30 min. The cells were disrupted by sonication on ice and the insoluble cell fraction was pelleted by ultracentrifugation at 80000× g for 40 min before being resuspended in 50 mM sodium phosphate (pH 7.0) containing 6 M urea.
The membrane proteins (25 µg) were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred onto Immobilon-P membranes (Millipore) at 12 V for 30 mins. The primary antibody (1∶5000 dilution of a mouse anti-PBP2a monoclonal antibody (Denka Seiken) in TBS+0.1% Tween) was applied to the membranes overnight. A horseradish peroxidase conjugated anti-mouse IgG secondary antibody (1∶1000 in TBST+1% skim milk) was incubated with the membranes for 1 hour. A chemiluminescence kit (Amersham) was used as described above.
Mouse infection experiments
A mouse model of device-related infection [39] was used to compare virulence of S. aureus 8325-4 and 8325-4 pmecA HoR. Briefly, the flanks of anesthetized 6-week old male C57BL/6 mice were shaved, and the skin cleansed with povidone-iodine. Using aseptic technique, a 1-cm segment of 14-gauge polyethylene intravenous catheter was implanted into the subcutaneous space (two per mouse and the incision closed with Vetbond (3M, Minneapolis, MN). Next, 1×107 or 1×108 S. aureus were injected into the catheter lumen. Eight mice were used to test each inoculum and strain. Survival over 7 days was measured. Animals were euthanized before the end of the experiment using the following indices for evaluating whether a moribund state had been achieved: extreme lethargy, failure to demonstrate typical avoidance behaviour when handled, ulceration of the infection site through the skin, excessive loss of body weight (i.e. >20%), and/or labored breathing.
The high level of mortality associated with infection of 8325-4 prompted us to repeat the above experiment over 18 hours using an inoculum of 1×107 bacteria before sacrificing all animals and enumerating the numbers of bacteria associated with the catheter, surrounding tissue, blood, liver, spleen and kidneys. To do this, the catheters were aseptically removed, placed in sterile microcentrifuge tubes with 1 ml of phosphate buffered saline (PBS), vortexed for one minute, and quantitatively cultured on tryptone soya agar (TSA). In addition, peri-catheter tissue, liver, kidneys and spleen were dissected, weighed, homogenized and quantitatively cultured on TSA. Finally, bacteria present in blood were also quantitatively cultured on TSA.
TNF-α and IL-6 levels were measured in the peri-catheter tissue using TNF-α (OptiEIA, BD Bioscience) or IL-6 (Duoset; R&D Systems) ELISA kits according to the manufacturer's instructions. Results were normalized to the total amount of tissue recovered.
Statistical analysis
Two-tailed, two-sample equal variance Student's t-tests (Microsoft Excel 2007) were used to determine statistically significant differences in biofilm forming capacity and relative gene expression.
For the animal experiments descriptive statistics (including mean, standard deviation, median minimum and maximum) values were calculated for each strain at each location i.e. catheter, peri-catheter tissue, blood, liver, spleen and kidneys. Log10 transformation of CFU data from catheter and tissue was used to ensure normal distribution. A two-sample t-test was conducted to compare the log-transformed CFU values for 8325-4 and 8325-4 pmecA HoR from either catheter or peri-catheter tissue. Nonparametric Wilcoxon rank sum tests were used to compare 8325-4 and 8325-4 pmecA HoR CFU data from blood, liver, spleen and kidneys.
Supporting Information
Biofilm phenotypes of DAR13 (CC8, SCCmec type IV), DAR168 (CC8, SCCmec type I), DAR34 (CC8, SCCmec type II), BH10(03) (CC22, SCCmec type IV), DAR199 (CC30, SCCmec type II) and DAR112 (CC239, SCCmec type III), together with their respective ΔSCCmec mutants and the ΔSCCmec mutants complemented with pmecA. Biofilms were grown for 24 h at 37°C in BHI glucose in hydrophilic 96-well polystyrene plates. The data presented are the average of three independent experiments and standard deviations are shown.
(TIF)
Immunoblot analysis of PNAG production in whole cell extracts of 8325-4, HG003 and 8325-4 ΔicaADBC grown overnight at 37°C in BHI media.
(TIF)
Biofilm formation by 8325-4 pmecA HeR and 8325-4 pmecA HoR under flow conditions after 15 h growth in BHI glucose or BHI NaCl. Biofilms were grown in a BioFlux 1000 microfluidics system. Images were taken at 200× magnification under brightfield illumination.
(TIF)
Impact of homogeneous oxacillin resistance on the biofilm phenotypes and δ-hemolytic activity of the MSSA strains 15981 and MSSA476. (A) Biofilm phenotypes of 15981, 15981 pmecA HeR, 15981 pmecA HoR and 15981 pmecA HoR (cured). (B) δ hemolytic activity of 15981 pmecA HeR and 15981 pmecA HoR on sheep blood agar (C) Dispersal of 5981, 15981 pmecA HeR, 15981 pmecA HoR and 15981 pmecA HoR (cured) biofilms by sodium metaperiodate and proteinase K. (D) Biofilm phenotypes of MSSA476, MSSA476 pmecA HeR, MSSA476 pmecA HoR and MSSA476 pmecA HoR (cured). (E) δ hemolytic activity of MSSA476 pmecA HeR and MSSA476 pmecA HoR on sheep blood agar. (F) Dispersal of MSSA476, MSSA476 pmecA HeR, MSSA476 pmecA HoR and MSSA476 pmecA HoR (cured) biofilms by sodium metaperiodate and proteinase K. All biofilms were grown for 24 h in BHI, BHI NaCl and BHI glucose on hydrophilic polystyrene. Experiments were repeated three times and average data are shown.
(TIF)
Impact of homogeneous oxacillin resistance on the biofilm phenotypes and δ-hemolytic activity of HeR MRSA strains. (A) Dispersal of USA300 strain LAC and USA300 strain LAC HoR biofilms by sodium metaperiodate and proteinase K. (B) δ hemolytic activity of USA300 HeR and USA300 HoR on sheep blood agar. (C and D) Dispersal of DAR26 wild type (HeR, CC5, SCCmec type IV) and DAR26 HoR biofilms by sodium metaperiodate and proteinase K. (E) δ hemolytic activity of DAR26 HeR and DAR26 HoR on sheep blood agar. (F and G) Dispersal of DAR173 wild type (HeR, CC5, SCCmec type II) and DAR173 HoR biofilms by sodium metaperiodate and proteinase K. (H) δ hemolytic activity of DAR173 HeR and DAR173 HoR on sheep blood agar. (I and J) Dispersal of DAR9 wild type (HeR, CC5, SCCmec type and DAR9 HoR biofilms by sodium metaperiodate and proteinase K. (K) δ hemolytic activity of DAR9 HeR and DAR9 HoR on sheep blood agar. HoR strains were isolated and cultured in oxacillin 100 µg/ml-supplemented media. USA300 strain LAC biofilms were grown in BHI glucose only. DAR26, DAR173 and DAR9 biofilms were grown in BHI, BHI NaCl and BHI glucose. All biofilms were grown for 24 h at 37°C on hydrophilic polystyrene. Experiments were repeated three times and average data are shown.
(TIF)
Acknowledgments
We thank Dr. O. Schneewind for S. aureus SKM1, Dr. M. Sugai for S. aureus JT1392, Prof T. Foster for S. aureus DU5883, LAC, MSSA476 and spa::Kmr and T. Maira Litran for rabbit anti-PNAG serum. We also thank T. Kielian, M. Hanke, L. Kinkead, J. Lindgren, A. Nuxoll, K. Wilcox for assistance with the animal experiments and F. Yu for statistical analysis of the animal data. We acknowledge the helpful comments and experimental advice of L. Holland, P. Houston, E. O'Neill, S. Rowe, B. Conlon, H. McCarthy, S. Hogan, J. Geoghegan, T. J. Foster and J. Mitchell during this study.
Footnotes
The authors have declared that no competing interests exist.
This study was funded by grants RP/2005/197 and HRA_POR/2010/90 from the Irish Health Research Board (www.hrb.ie) and a major grant award (2004) from the Healthcare Infection Society (www.his.org.uk) to J. P. O'Gara. E. M. Waters was the recipient of a Scholarship from the Irish Research Council for Science Engineering and Technology (www.ircset.ie). We are grateful to the Society for General Microbiology for a travel grant to support a research visit by E. M. Waters to the laboratory of P. D. Fey. Work in the laboratory of P. D. Fey was supported in part by funds from NIH/NIAID P01A1083211. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 2.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzpatrick F, Humphreys H, O'Gara JP. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol. 2005;43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297:117–122. doi: 10.1016/j.ijmm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 6.O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 10.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 11.Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O'Gara JP, et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, et al. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, et al. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75:187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- 15.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.IWG-SCC. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers HF, Hackbarth CJ. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1987;31:1982–1988. doi: 10.1128/aac.31.12.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers HF. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J Infect Dis. 1999;179(Suppl 2.):S353–359. doi: 10.1086/513854. [DOI] [PubMed] [Google Scholar]
- 20.Sabath LD, Wallace SJ. The problems of drug-resistant pathogenic bacteria. Factors influencing methicillin resistance in staphylococci. Ann N Y Acad Sci. 1971;182:258–266. doi: 10.1111/j.1749-6632.1971.tb30662.x. [DOI] [PubMed] [Google Scholar]
- 21.Berger-Bachi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983;154:479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami K, Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989;171:874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maki H, Yamaguchi T, Murakami K. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol. 1994;176:4993–5000. doi: 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuirolo A, Plata K, Rosato AE. Development of homogeneous expression of resistance in methicillin-resistant Staphylococcus aureus clinical strains is functionally associated with a beta-lactam-mediated SOS response. J Antimicrob Chemother. 2009;64:37–45. doi: 10.1093/jac/dkp164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plata KB, Rosato RR, Rosato AE. Fate of mutation rate depends on agr locus expression during oxacillin-mediated heterogeneous-homogeneous selection in methicillin-resistant Staphylococcus aureus clinical strains. Antimicrob Agents Chemother. 2011;55:3176–3186. doi: 10.1128/AAC.01119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5:e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaito C, Omae Y, Matsumoto Y, Nagata M, Yamaguchi H, et al. A novel gene, fudoh, in the SCCmec region suppresses the colony spreading ability and virulence of Staphylococcus aureus. PLoS One. 2008;3:e3921. doi: 10.1371/journal.pone.0003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, et al. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 2011;7:e1001267. doi: 10.1371/journal.ppat.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galdbart JO, Allignet J, Tung HS, Ryden C, El Solh N. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J Infect Dis. 2000;182:351–355. doi: 10.1086/315660. [DOI] [PubMed] [Google Scholar]
- 30.Mempel M, Feucht H, Ziebuhr W, Endres M, Laufs R, et al. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob Agents Chemother. 1994;38:1251–1255. doi: 10.1128/aac.38.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mempel M, Muller E, Hoffmann R, Feucht H, Laufs R, et al. Variable degree of slime production is linked to different levels of beta-lactam susceptibility in Staphylococcus epidermidis phase variants. Med Microbiol Immunol (Berl) 1995;184:109–113. doi: 10.1007/BF00224346. [DOI] [PubMed] [Google Scholar]
- 32.Christensen GD, Baddour LM, Madison BM, Parisi JT, Abraham SN, et al. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J Infect Dis. 1990;161:1153–1169. doi: 10.1093/infdis/161.6.1153. [DOI] [PubMed] [Google Scholar]
- 33.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, et al. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths JM, O'Neill AJ. Loss of function of the GdpP protein leads to joint {beta}-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:579–81. doi: 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67:2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, et al. Methicillin resistance reduces the virulence of HA-MRSA by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadykov MR, Olson ME, Halouska S, Zhu Y, Fey PD, et al. Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J Bacteriol. 2008;190:7621–7632. doi: 10.1128/JB.00806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Xiong YQ, Sadykov MR, Fey PD, Lei MG, et al. Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infect Immun. 2009;77:4256–4264. doi: 10.1128/IAI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, et al. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun. 2008;76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, et al. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother. 2006;50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marti M, Trotonda MP, Tormo-Mas MA, Vergara-Irigaray M, Cheung AL, et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 49.Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci U S A. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, et al. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis. 2008;198:1171–1174. doi: 10.1086/592051. [DOI] [PubMed] [Google Scholar]
- 51.Sakoulas G, Moise PA, Rybak MJ. Accessory gene regulator dysfunction: an advantage for Staphylococcus aureus in health-care settings? J Infect Dis. 2009;199:1558–1559. doi: 10.1086/598607. [DOI] [PubMed] [Google Scholar]
- 52.Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 53.Saravia-Otten P, Muller HP, Arvidson S. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol. 1997;179:5259–5263. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards AM, Potts JR, Josefsson E, Massey RC. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. PLoS Pathog. 2010;6:e1000964. doi: 10.1371/journal.ppat.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conlon KM, Humphreys H, O'Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002a;184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziebuhr W, Heilmann C, Gotz F, Meyer P, Wilms K, et al. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gelosia A, Baldassarri L, Deighton M, Van Nguyen T. Phenotypic and genotypic markers of Staphylococcus epidermidis virulence. Clin Microbiol Infect. 2001;7:193–199. doi: 10.1046/j.1469-0691.2001.00214.x. [DOI] [PubMed] [Google Scholar]
- 59.Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60:2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holland LM, Conlon B, O'Gara JP. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology. 2011;157:408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 61.Collins J, Rudkin J, Recker M, Pozzi C, O'Gara JP, et al. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 2010;4:577–584. doi: 10.1038/ismej.2009.151. [DOI] [PubMed] [Google Scholar]
- 62.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186:2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyle-Vavra S, Yin S, Daum RS. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2006;262:163–171. doi: 10.1111/j.1574-6968.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- 64.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 65.Greene C, McDevitt D, Francois P, Vaudaux PE, Lew DP, et al. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 66.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi J, Komatsuzawa H, Yamada S, Nishida T, Labischinski H, et al. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol Immunol. 2002;46:601–612. doi: 10.1111/j.1348-0421.2002.tb02741.x. [DOI] [PubMed] [Google Scholar]
- 68.Higgins J, Loughman A, van Kessel KP, van Strijp JA, Foster TJ. Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol Lett. 2006;258:290–296. doi: 10.1111/j.1574-6968.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- 69.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, et al. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 71.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biofilm phenotypes of DAR13 (CC8, SCCmec type IV), DAR168 (CC8, SCCmec type I), DAR34 (CC8, SCCmec type II), BH10(03) (CC22, SCCmec type IV), DAR199 (CC30, SCCmec type II) and DAR112 (CC239, SCCmec type III), together with their respective ΔSCCmec mutants and the ΔSCCmec mutants complemented with pmecA. Biofilms were grown for 24 h at 37°C in BHI glucose in hydrophilic 96-well polystyrene plates. The data presented are the average of three independent experiments and standard deviations are shown.
(TIF)
Immunoblot analysis of PNAG production in whole cell extracts of 8325-4, HG003 and 8325-4 ΔicaADBC grown overnight at 37°C in BHI media.
(TIF)
Biofilm formation by 8325-4 pmecA HeR and 8325-4 pmecA HoR under flow conditions after 15 h growth in BHI glucose or BHI NaCl. Biofilms were grown in a BioFlux 1000 microfluidics system. Images were taken at 200× magnification under brightfield illumination.
(TIF)
Impact of homogeneous oxacillin resistance on the biofilm phenotypes and δ-hemolytic activity of the MSSA strains 15981 and MSSA476. (A) Biofilm phenotypes of 15981, 15981 pmecA HeR, 15981 pmecA HoR and 15981 pmecA HoR (cured). (B) δ hemolytic activity of 15981 pmecA HeR and 15981 pmecA HoR on sheep blood agar (C) Dispersal of 5981, 15981 pmecA HeR, 15981 pmecA HoR and 15981 pmecA HoR (cured) biofilms by sodium metaperiodate and proteinase K. (D) Biofilm phenotypes of MSSA476, MSSA476 pmecA HeR, MSSA476 pmecA HoR and MSSA476 pmecA HoR (cured). (E) δ hemolytic activity of MSSA476 pmecA HeR and MSSA476 pmecA HoR on sheep blood agar. (F) Dispersal of MSSA476, MSSA476 pmecA HeR, MSSA476 pmecA HoR and MSSA476 pmecA HoR (cured) biofilms by sodium metaperiodate and proteinase K. All biofilms were grown for 24 h in BHI, BHI NaCl and BHI glucose on hydrophilic polystyrene. Experiments were repeated three times and average data are shown.
(TIF)
Impact of homogeneous oxacillin resistance on the biofilm phenotypes and δ-hemolytic activity of HeR MRSA strains. (A) Dispersal of USA300 strain LAC and USA300 strain LAC HoR biofilms by sodium metaperiodate and proteinase K. (B) δ hemolytic activity of USA300 HeR and USA300 HoR on sheep blood agar. (C and D) Dispersal of DAR26 wild type (HeR, CC5, SCCmec type IV) and DAR26 HoR biofilms by sodium metaperiodate and proteinase K. (E) δ hemolytic activity of DAR26 HeR and DAR26 HoR on sheep blood agar. (F and G) Dispersal of DAR173 wild type (HeR, CC5, SCCmec type II) and DAR173 HoR biofilms by sodium metaperiodate and proteinase K. (H) δ hemolytic activity of DAR173 HeR and DAR173 HoR on sheep blood agar. (I and J) Dispersal of DAR9 wild type (HeR, CC5, SCCmec type and DAR9 HoR biofilms by sodium metaperiodate and proteinase K. (K) δ hemolytic activity of DAR9 HeR and DAR9 HoR on sheep blood agar. HoR strains were isolated and cultured in oxacillin 100 µg/ml-supplemented media. USA300 strain LAC biofilms were grown in BHI glucose only. DAR26, DAR173 and DAR9 biofilms were grown in BHI, BHI NaCl and BHI glucose. All biofilms were grown for 24 h at 37°C on hydrophilic polystyrene. Experiments were repeated three times and average data are shown.
(TIF)