Abstract
The ATPase subunits of the SWI/SNF chromatin remodeling enzymes, Brahma (BRM) and Brahma related gene 1 (BRG1), can induce cell cycle arrest in BRM and BRG1 deficient tumor cell lines, and mice heterozygous for Brg1 are predisposed to breast tumors, implicating loss of BRG1 as a mechanism for unregulated cell proliferation. To test the hypothesis that loss of BRG1 can contribute to breast cancer, we utilized RNA interference to reduce the amounts of BRM or BRG1 protein in the nonmalignant mammary epithelial cell line, MCF-10A. When grown in reconstituted basement membrane (rBM), these cells develop into acini that resemble the lobes of normal breast tissue. Contrary to expectations, knockdown of either BRM or BRG1 resulted in an inhibition of cell proliferation in monolayer cultures that was enhanced in three-dimensional rBM culture. This inhibition was strikingly enhanced in three-dimensional rBM culture, although some BRM depleted cells were later able to resume proliferation. Cells did not arrest in any specific stage of the cell cycle; instead, the cell cycle length increased by approximately 50%. Thus, SWI/SNF ATPases promote cell cycle progression in nonmalignant mammary epithelial cells.
INTRODUCTION
The mammalian SWI/SNF complexes are a family of chromatin remodeling enzymes that regulate gene expression by disrupting histone-DNA contacts in an ATP-dependent manner (Imbalzano et al., 1994; Kwon et al., 1994). The complexes are evolutionarily conserved in eukaryotes and contain either BRM (Brahma) or BRG1 (Brahma-related gene 1) as the central ATPase subunit (Khavari et al., 1993; Muchardt and Yaniv, 1993; Wang et al., 1996). SWI/SNF enzyme complexes include other proteins known as BRG1 and BRM-associated factors (BAFs) that can modulate the activity of the ATPase subunits and might provide gene-specific recruitment (Wang et al., 1996). The BRM and BRG1 proteins are highly similar, with a sequence identity of 74% in humans, and they display similar enzymatic properties (Chiba et al., 1994; Khavari et al., 1993; Muchardt and Yaniv, 1993; Phelan et al., 1999; Sif et al., 2001). Both are involved in developmental processes in plants, invertebrates, and vertebrates (reviewed in (de la Serna et al., 2006; Kwon and Wagner, 2007). Despite these similarities, the two alternative ATPase subunits can serve different functions in the regulation of differentiation, transcriptional control, and other important cell processes (Bultman et al., 2000; Kadam and Emerson, 2003; Reyes et al., 1998).
BRG1 and BRM are important for cell cycle arrest. Reintroduction of BRG1 or BRM into deficient tumor cell lines induces cell cycle arrest and a “flat cell” phenotype by a mechanism requiring RB family members (Dunaief et al., 1994; Strobeck et al., 2000b; Strober et al., 1996; Trouche et al., 1997; Zhang et al., 2000). RB and BRM (or BRG1) cooperate to repress E2F1 mediated activation (Trouche et al., 1997; Wang et al., 2002) and repress levels of CDK2, cyclin A, and cyclin E (Coisy et al., 2004; Roberts and Orkin, 2004; Strobeck et al., 2000a; Strobeck et al., 2000b). BRM can compensate for BRG1 loss in RB signaling pathways, suggesting a redundancy between the two factors in this mechanism of cell cycle control (Reisman et al., 2002; Strobeck et al., 2002).
Approximately 10% of mice heterozygous for Brg1 develop tumors, mostly mammary carcinomas (Bultman et al., 2008). This and earlier work (Bultman et al., 2000) firmly established BRG1 as a tumor suppressor in vivo. Although Brm deficient mice do not present with tumors, depending on the strain background, they can be physically larger, with an increased tissue and organ size due to increased proliferation (Reyes et al., 1998). In addition, immortalized fibroblasts derived from Brm deficient mouse embryos have a delayed and shorter S-phase, and a prolonged mitosis (Coisy-Quivy et al., 2006). Together, these previous studies indicate that BRG1 and BRM are negative regulators of cell cycle progression in culture and are likely to decrease proliferation in vivo. A logical prediction from this literature would be that the loss of BRG1 or BRM should lead to the loss of growth control, to hyperplasia, and to cancer progression.
The MCF-10A line immortilized spontaneously in culture from primary cells taken from a patient with fibrocystic disease (Soule et al., 1990). The MCF-10A line has a stable, near-diploid karyotype (Soule et al., 1990; Yoon et al., 2002), but has lost the p16 locus (Debnath et al., 2003; Yaswen and Stampfer, 2002). The cells express wild type p53 (Debnath et al., 2003; Merlo et al., 1995). MCF-10A cells cultured in three-dimensional reconstituted basement membrane culture (rBM) develop important features of normal breast tissue by a program of proliferation, cell cycle arrest, apical-basolateral polarization, and apoptosis to create a luminal space (Debnath et al., 2002; Debnath et al., 2003; Underwood et al., 2006). In addition, cell nuclei of mammary epithelial cells forming acini in three-dimensional rBM culture recapitulate the architecture of mammary epithelial cells in tissue (Lelievre et al., 1998).
To address the function of the SWI/SNF ATPases in normal mammary epithelial cells, we generated MCF-10A cells with inducible knockdowns of either BRG1 or BRM. The depletion of either ATPase subunit decreased the rate of cell proliferation without inducing a complete cell growth arrest in monolayer culture. The decrease in proliferation was amplified in three dimensional rBM culture. Further analysis demonstrated that the length of the cell cycle increased after depletion of either SWI/SNF ATPase, indicating a role for BRG1 and BRM as positive regulators of cell cycle progression.
MATERIALS AND METHODS
Cell culture
MCF-10A cells from the Karmanos Cancer Institute (Detroit, MI) were maintained in monolayer as described (Debnath et al., 2003).
Doxycycline-inducible BRG1 and BRM knockdowns
Generation of vectors
Lentiviral vectors were from the D. Trono lab (www.tronolab.unige.ch) and obtained from Addgene: pLV-tTRKRAB-Red encoding the TetR-KRAB regulator, pLVTHM, for cloning the shRNA, the packaging vector pCMV-dR8.91 and the Envelope vector pMD2.G. The shRNA sequences for BRG1 and BRM were from previously designed siRNAs (Rosson et al., 2005). Annealed oligonucleotides were cloned in the pSUPER.retro.puro vector then inserted between the EcoRI/ClaI sites of pLVTHM to express the shRNA under the control of the H1 promoter. The first forward sequence was 5’-GATCCCCGTGCGACATGTCTGCGCTGTTCAAGACACAGCGCAGACATGTCGCACTTTTTGGAAA-3’ where the underlined sequence is specific for the BRG1 ATPase domain. The second forward sequence was 5’-GATCCCCGTCTGAAGATCGTGCTGCTTTCAAGAGAAGCAGCACGATCTTCAGACTTTTTGGAAA-3’ where the underlined sequence is specific for the BRM ATPase domain. The control scrambled (SCRAM) forward sequence was 5’-ATCCCCCAGTTACTAGACGCGATCGTTCAAGAGACGATCGCGTCTAGTAACTGTTTTTA-3’.
For the double BRG1 and BRM knockdown, a new shBRM lentivector was engineered with Gateway® Technology (Invitrogen). The cassette tetO-H1-shBRM was removed from the pLVTHM and cloned in the Entry vector pENTR1A-no ccdB (a gift of Eric Campeau) used to transfer the shBRM expression cassette into a lentiviral destination vector. We used the promoter-less lentiviral destination vector pLenti 2X Puro DEST, clone #w16.1 (E. Campeau), which contains elements that allow packaging of the construct and a puromycin resistance marker for selection of stably transduced cells (Campeau et al., 2009). The LR Recombination Reaction was performed by using the Gateway LR clonase™ II enzyme Mix (Invitrogen).
Lentivirus production
5×106 293T cells were seeded in a 10 cm dish and transfected the following day with the Lipofectamine® 2000 reagent (Invitrogen). Viral supernatants were collected and 0.45 µm filtered at 48 and 72 hours post-transfection, then stored at −80°C.
Transduction of MCF-10A cells
MCF-10A cells at 75 % confluence were incubated for 16 hours with lentivirus diluted in growth media containing 8 µg/mL polybrene. The next day the viruses were removed, the cells were rinsed twice with PBS, and fresh media was added. Cells were passed twice before FACS sorting. To induce shRNA-GFP expression, Doxycycline was used at 0.01 to 0.5 µg/mL.
Cell sorting
To obtain a population of cells expressing the shRNA and GFP under doxycycline control, the cells were FACS sorted twice. The first sort was of uninduced cells and selected dsRED positive cells that constitutively expressed the tTR-KRAB protein coupled to the dsRed marker (constitutive expression). The second sort was performed two days after Doxycycline induction and selected both dsRED and GFP positive cells that expressed both shRNA and GFP after doxycycline induction. Cells were FACS selected prior to every experiment.
Western blot analysis
Whole cell extracts were prepared from MCF-10A cells in monolayer culture after trypsinization and two washes with 5 mL of cold 1X PBS. Then, cells were lysed in 100 µL Laëmmli lysis buffer (1 % SDS, 0.04 M Tris-HCl pH 6.8, 6 % glycerol, 0.003 % bromophenol blue, 0.015 M β-mercaptoethanol) for every106 cells, boiled for 5 minutes, and stored at −80°C. Proteins were separated on 7.5 % SDS-PAGE, transferred to a nitrocellulose membranes, and detected with primary antibodies and ECL detection (Amersham). The antibodies used were: BRG1 (dilution 1:1000, anti-serum de La Serna et al., 2000), BRM (dilution 1:1000, Abcam ab15597), PI3Kinase p85, H-SH2 domain (dilution 1:1000, Upstate cat. No. 06-496), GFP (dilution 1:1000, Roche cat. No. 1814460), p21 Waf1/Cip1(12D1) Rabbit mAb (dilution 1:1000, Cell Signaling 2947S), p53 (dilution 1:1000, Cell Signaling 9282), Phospho-p53 (Ser15) (dilution 1:1000, Cell Signaling 9284S), Phospho-p53 (Ser46) (dilution 1:1000, Cell Signaling 2521S), Phospho-p53 (Ser 20) (dilution 1:1000, Cell Signaling 9287S), Cyclin A (BF683) mouse mAb (dilution 1:1000, Cell Signaling 4656), mTOR (dilution 1:1000, Millipore 04-385), phospho-mTOR (Ser 2448) (dilution 1:1000, Millipore 09-213), p70 S6 kinase (S6K) (dilution 1:1000, Millipore 06-926),
Proliferation assays
Direct cell counting
Cells were grown 2 days in the presence (pre-induction) or absence of doxycycline (0.05 or 0.1 µg/mL) prior to seeding in a 12 well plate (4000 cells/well) with or without Doxycycline. Cells were counted daily after trypsin treatment using either a hemacytometer or a Z1 Coulter counter.
DNA quantification using the Cyquant Cell Proliferation kit (Invitrogen)
Cells were seeded in a 96-well plate at different densities in triplicate. One plate was prepared per day for the time course. Cell growth was stopped by removing the medium and freezing the plate immediately with storage at −80°C. The kit uses a proprietary green fluorescent dye, Cyquant® GR dye, which exhibits strong fluorescence enhancement when bound to DNA. Fluorescence was measured using a fluorescence microplate reader. A reference standard curve (with cell numbers from 50 to 50000 cells) was used to convert fluorescence values into cell numbers.
Immunofluorescent staining of monolayer cultures
Monolayer cultures were prepared following the methods of Wagner et al. (Wagner et al., 2003). Before mounting coverslips with Prolong Gold (Invitrogen), cells were stained with DAPI (2 µg/mL) and DRAQ5 (Alexis, 1:5000) DNA dyes in PBS for 5 minutes at room temperature then washed once with PBS.
Phospho-Histone H3 (serine 10) antibody was from Upstate Millipore. All the secondary antibodies (Alexa Fluor® 488 or 568 conjugated) were from Molecular Probes. For the observation of expressed fluorescent proteins, cells on coverslips were washed in PBS and directly fixed in 4% paraformaldehyde in CSK for 50 minutes, rinsed in TBS-0.05% Tween-20, and stained with DAPI and DRAQ5.
BrdU incorporation assay
Cell cycle length of non-synchronized MCF-10A cells was measured by labeling cells with Bromodeoxyuridine (BrdU). 2×105 cells were plated on a coverslip in each well of a 6-well dish. At time zero, BrdU was added to the culture medium to a final concentration of 20 µM. At each time point (1, 5, 10 and 15 hours of BrdU incorporation) cells were washed twice with cold DPBS, permeabilized with 0.5 % Triton X-100 in CSK buffer for 3 minutes, and fixed with 4% formaldehyde in CSK buffer for 20 minutes. All steps were performed on ice. Then cells were washed 3 times with PBS-0.5% Tween 20 for 10 minutes each at room temperature. DNA was denatured by 2N HCL for 30 minutes at 37°C, followed by 2 washes with PBS. All antibodies were diluted in TBS-1. Coverslips were incubated with anti-BrdU (clone BU-33, Sigma, 1:400) for 1 hour at 37°C or overnight at 4°C. The second antibody was goat anti-mouse IgG1 coupled to Alexa Fluor 568 (Invitrogen, 1:2000). After each antibody incubation, cells were rinsed 3 times in PBS containing 0.05 % Tween 20 for 10 minutes each at room temperature. Before mounting coverslips with Prolong Gold (Invitrogen), cells were stained with DAPI (2 µg/mL) and DRAQ5 (Alexis, 1:5000) diluted in PBS for 5 minutes at room temperature before a last wash with PBS. Images of 10 fields were taken at low magnification and BrdU positive cells were counted in each field with ImageJ. A linear regression was applied in order to extrapolate the time needed for 100% to incorporate BrdU. This time is the cell cycle length.
mRNA analysis
RNA was isolated from MCF-10A monolayer culture using Trizol (Invitrogen) and reverse transcribed. The cDNA was amplified using the Qiagen HotStarTaq Master Mix kit (Qiagen #203445) containing 0.1µg of specific primers and SYBR green. RT-PCR and real time PCR were performed using procedures previously described (Ohkawa et al., 2006). Primers for measuring GAS5 RNA levels were CAG TGT GGC TCT GGA TAG CA (forward) and TTA AGC TGG TCC AGG CAA GT (reverse).
Three-dimensional culture of MCF-10A cells on reconstituted basement membrane
MCF-10A cells were cultured in either Reduced Growth Factor Matrigel without phenol red (lot#11346, BD Biosciences) or Non-Reduced Growth Factor Matrigel with phenol red (lot#22704, BD Biosciences) following the procedures of Debnath et al. (Debnath et al., 2003). Briefly, for overlay cultures, cells were prepared for three dimensional rBM culture by growing to 20–30 % confluency in monolayer and seeding in a single cell suspension on 100ul of matrigel in a 35mm well at 7,000 to 15,000 cells/well or on 40ul matrigel in a 8-well chamber slide at 5000 cells/well. Cells in rBM were grown in assay media (Debnath et al., 2003) containing 2% horse serum, 5ng/ml EGF, and 2% Matrigel. All cultures were incubated at 37°C in a 5% CO2 humidified incubator for up to 20 days. Media was replaced every 2 to 4 days. Morphology was observed every 2 days via phase contrast microscopy. Acinar size was determined from phase contrast micrographs. Two diameters were measured per acinus with ImageJ software. The minimum number of measured acini per sample was 15.
Cell counting in rBM culture
Three dimensional cultures were incubated for 30–40 minutes in 37°C incubator with 0.25 % trypsin-EDTA. As soon as the Matrigel was dissolved, the acinar cell suspension was centrifuged 5 minutes at 1000 rpm, the supernatant was discarded and the pellet was resuspended with 0.05 % trypsin-EDTA then put back in the dish for incubation at 37°C. When a single cell suspension was observed, trypsin activity was stopped with Resuspension Media (DMEM/F12 containing 20 % horse serum and antibiotics) and cells were counted by trypan blue exclusion in a hemocytometer, or without trypan blue with an automatic cell counter.
RESULTS
Inducible knockdown of SWI/SNF ATPase subunits in MCF-10A human mammary epithelial cells
We engineered the conditional expression of a short hairpin (sh) sequence targeting BRG1 (Rosson et al., 2005) in the human breast epithelial cell line, MCF-10A (Soule et al., 1990). A doxycyclin-inducible lentivirus (LV) (Wiznerowicz and Trono, 2003) stably introduced the shRNA gene into MCF-10A cells. Two lentiviral constructs were used. The first expressed the tTR-KRAB transactivator and dsRed (LV-tetR-KRAB-dsRed), while the second expressed the shRNA and GFP under tTR-KRAB transcriptional repression (LV-shBRG1i-GFP). We used two different strategies of lentiviral infection without noting any significant difference in outcome. In some experiments, cells were modified in one step by a double infection with both lentiviruses. In other experiments, cells were engineered by sequential infection, first with LV-tetR-KRAB-dsRed and, after sorting for dsRed fluorescence, with LV-shBRG1i-GFP. Pools of cells were FACS sorted for dsRed fluorescence and doxycycline-induced GFP fluorescence, without a requirement for cloning or drug selection (Fig. S1).
This inducible knockdown system was used both in monolayer culture and in three-dimensional culture in reconstituted basement membrane. As expected, cultures maintained in the absence of doxycycline constitutively expressed dsRED which marks cells also expressing the TetR-KRAB repressor (Fig. 1). This repressor prevents expression of the GFP marker and the shRNA which are both encoded by the second virus. Addition of doxycycline induced expression of both GFP and the shRNA. Because of the molecular and functional similarity between BRG1 and the other SWI-SNF ATPase subunit, BRM, we also generated MCF-10A cells with an inducible BRM knockdown using the same lentiviral vector system. The control vector conditionally expressed a scrambled sequence shRNA.
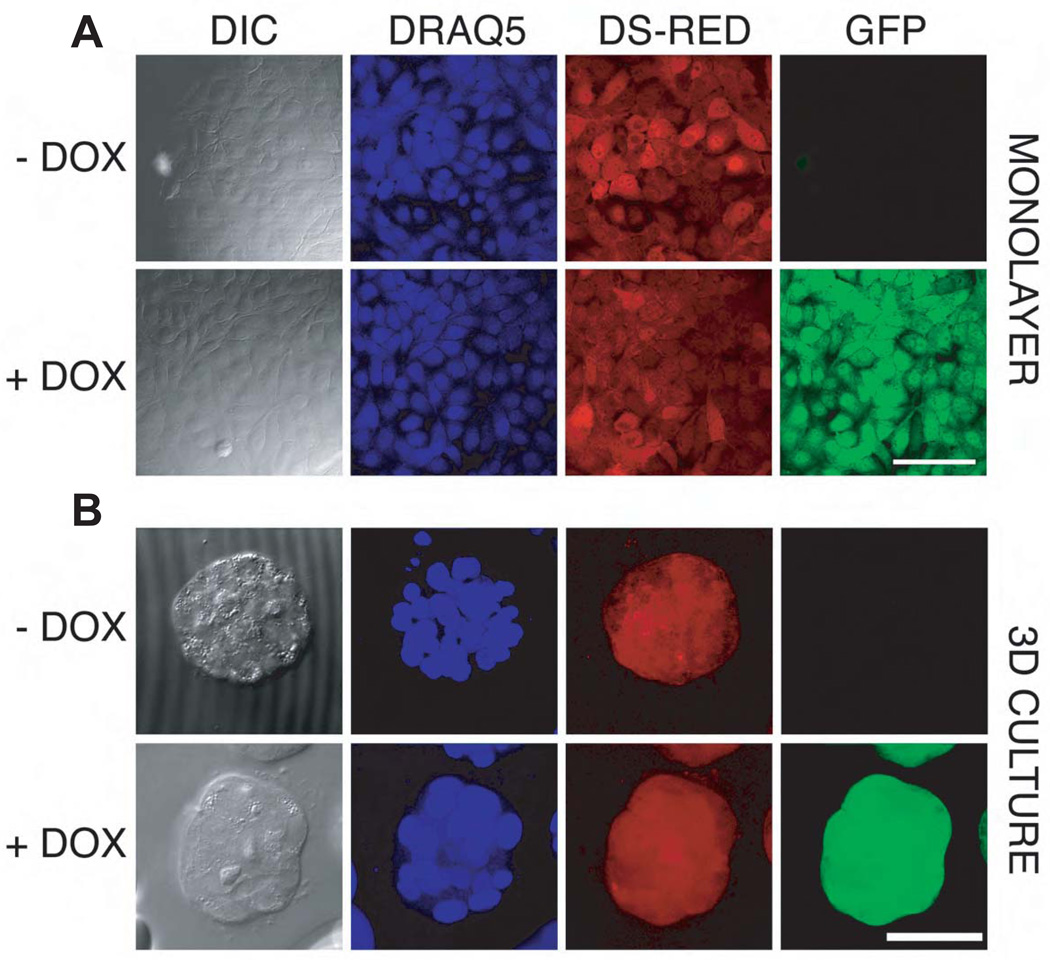
Figure 1. Doxycycline-Inducible shRNA expression in MCF-10A cells.
In this system dsRED and the TET-KRAB regulator are constitutively expressed from the same vector while GFP fluorescence is expressed only after doxycycline induction of the shRNA targeting BRG1.
Cells were sorted after 2 days of doxycycline induction (0.1 µg/mL). Then, after 5 days of culture without doxycycline, cells were seeded on coverslips and induced or not with 0.05 µg/mL doxycycline for 3 days before fixing with formaldehyde and staining nuclei with DRAQ5. Confocal image stacks, shown here for the MCF-10A-SCRAM control cell line), were collected for both monolayer (Panel 1A, scale bar = 100 µm.) and 3D culture (Panel 1B, scale bar= 50 µm.). The micrographs of the acinus (Panel B) were maximum intensity projections. Confocal settings were held constant so that linear quantitative comparisons could be made between samples with and without doxycycline induction.
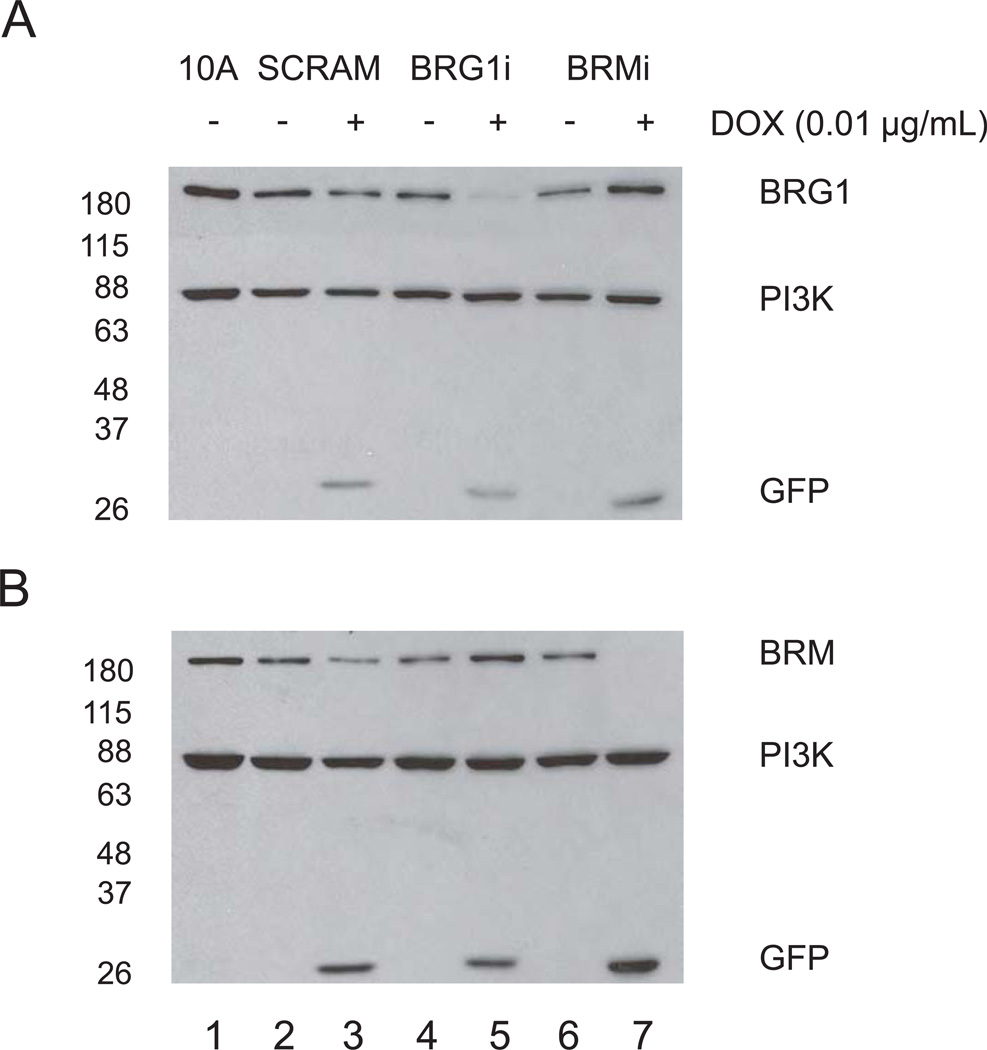
We evaluated the efficiency of the BRG1 and BRM knockdown by Western blotting (Fig. 2). Protein lysates were obtained from cells expressing the BRG1 shRNA, (BRG1i), the BRM shRNA (BRMi), or the scrambled sequence control shRNA (SCRAM), with or without induction with 0.01 µg/mL doxycycline in monolayer culture for 3 days. As shown in Fig. 2A, the protein level of BRG1 was efficiently knocked down in BRG1i cells (compare lanes 4 and 5), but was not decreased in the BRMi cells (compare lanes 6 and 7). Similarly, BRM levels (Fig. 2B) were not decreased in BRG1i cells but were greatly reduced in BRMi cells, demonstrating the specificity of each shRNA. GFP expression was monitored as an additional marker of doxycycline induction. Protein levels were measured by quantification of Western blot signals. The knockdown of BRG1 protein in BRG1i cells was determined to be 75%, while the knockdown of BRM was 90% in BRMi cells, with minor variations between experiments. Optimizing this inducible system, we determined that 48 hours were needed to get maximal protein decrease (Fig. S2) at an optimal concentration of doxycycline of between 0.01 and 0.05 µg/mL (Fig. S3). We also noticed a small but consistent increase in the amount of BRM protein in the BRG1i cell line (Fig. 2B, compare lanes 4 and 5) and a similar small but reproducible increase in the amount of BRG1 in BRMi cells (Fig. 2A, compare lanes 6 and 7). These observations suggest a compensation effect in protein levels of the two SWI/SNF ATPase subunits BRG1 and BRM.
Figure 2. Doxycycline-inducible knockdown of BRG1 and BRM in MCF-10A cells.
Total protein was extracted from cells treated or not for 3 days with 0.01 µg/ml doxycycline. BRG1 (A), BRM (B) and GFP protein expression were examined by Western blotting. The protein of 0.5 × 105 cells per lane was separated by SDS-PAGE (7.5 %) and probed with the indicated antibodies. PI3 kinase was the loading control.
One concern with siRNA technology is the capacity of dsRNA to trigger a non-specific interferon response in some cellular systems (Bridge et al., 2003; Diebold et al., 2003; Sledz et al., 2003). The inclusion of the scrambled sequence SCRAM controls in every experiment controlled for these effects, but in order to further validate the system we directly measured the level of mRNAs coding for the interferon response genes IFITM1, MX1, and OAS1. The results showed that the interferon response was not activated by any of the shRNAs in this experimental system (data not shown).
BRG1 or BRM knockdown impedes the early proliferation stage of MCF-10A acinus formation
Normal MCF-10A mammary epithelial cells, when cultured in three dimensional reconstituted basement membrane (rBM) culture, reproduce important features of normal breast tissue in a well characterized temporal and spatial program (Debnath et al., 2002; Debnath et al., 2003; Petersen et al., 1992; Weaver et al., 1995; Weaver et al., 2002; Weaver et al., 1997). An initial stage of proliferation produces loosely connected groups of cells and is followed by cell cycle arrest. Acini form from these groups of cells by basal deposition of a basement membrane and luminal clearance of cells not apposing the basement membrane. Malignant changes in cells alter this program of development in rBM culture with the formation of structures having an altered morphology, loss of cell cycle arrest, and basement membrane-independent cell survival (Imbalzano et al., 2009).
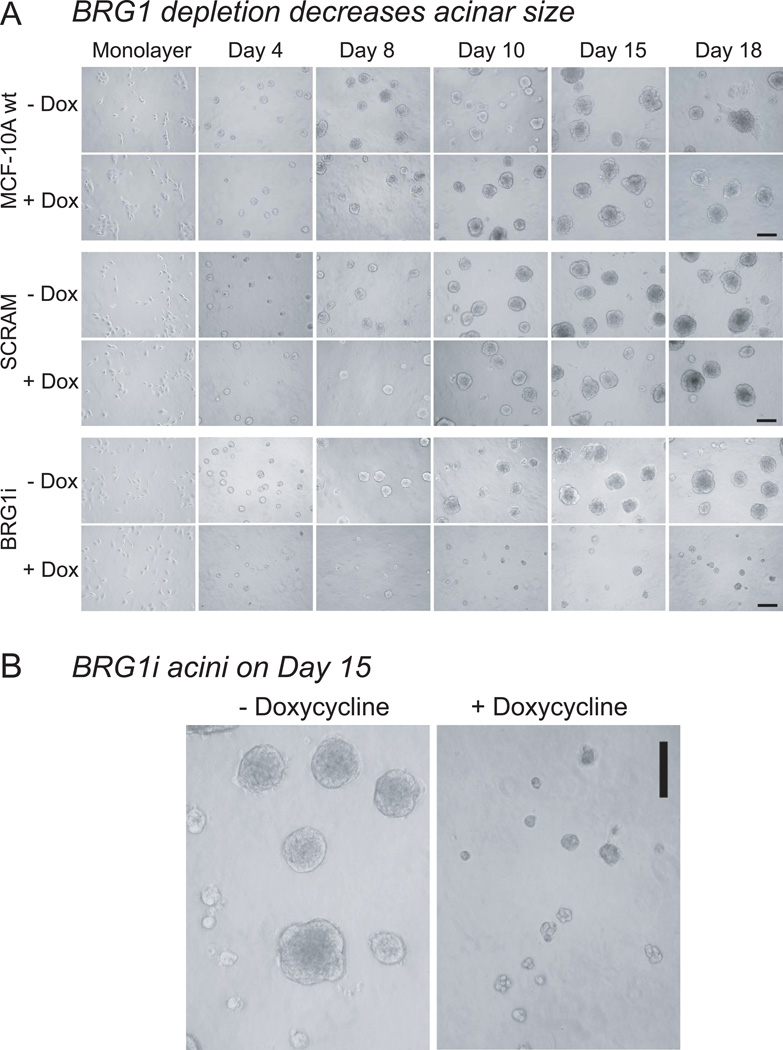
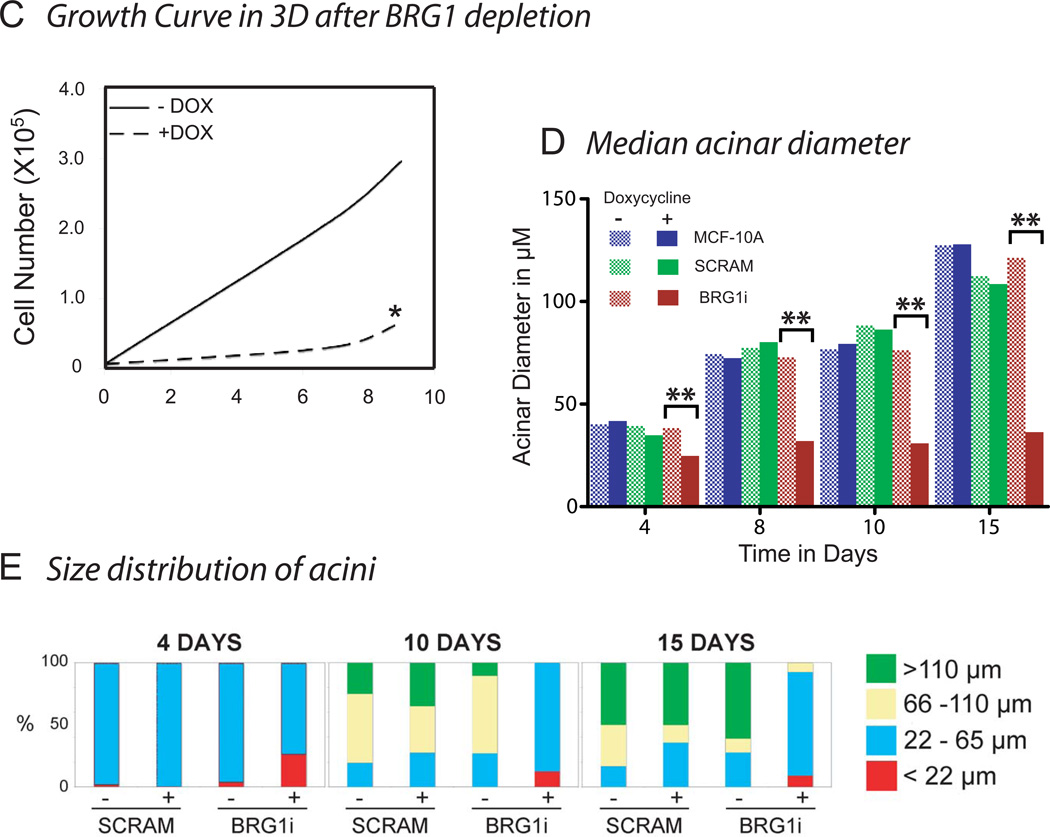
To determine whether the depletion of BRG1 caused these malignant alterations in acinar development, we cultured wild type MCF-10A human mammary epithelial cells, BRG1i cells expressing the doxycycline-inducible shRNA targeting BRG1, and control SCRAM cells in three-dimensional reconstituted basement membrane (rBM) culture. These cells were preinduced with doxycycline two days before being plated on a layer of rBM with an overlay of 2% rBM in culture medium and were maintained in culture for 18 days (Debnath et al., 2003). Twenty-four hours after establishing these overlay cultures, the cells from all three cell lines (wild type MCF-10A, BRG1i and SCRAM) had attached to the rBM. After two days, all the cell lines formed spherical masses of cells. The diameter of these structures was measured. As shown in Figs. 3A, B, and C, from 4 days of culture, the size of the multicellular structures of BRG1i cells expressing the shBRG1 after doxycycline induction were smaller than the structures formed from non-induced BRG1i cells or from controls (SCRAM and wild type MCF-10A). This size difference increased with time in culture (Fig. 3D, E). Dead cells were rare as determined by dye-exclusion. Cell counting, performed after digestion of the extracellular matrix and dissociation of the cell masses, revealed a dramatic decrease in proliferation (Fig. 3C).
Figure 3. Decreased Proliferation after BRG1 knockdown in rBM culture.
(A) MCF-10A cells were seeded with or without doxycycline in Matrigel. The micrograph for day 0 shows the initial monolayer culture. Every two days, media with or without doxycycline were replaced and phase contrast micrographs were taken. Decreased proliferation was observed from day 4 in MCF-10A cells expressing the shRNA targeting BRG1 (+doxycycline). Size bar : 150 mm
(B) Representative micrographs of MCF-10A BRG1i acini grown in the presence or absence of doxycycline for 15 days. These pictures were among those used to make the measurements of acinar diameter presented in panel D. Size bar : 150 µm.
(C) The three dimensional Matrigel cultures were trypisinized to obtain a single cell suspension at different times after seeding (day 4 and day 9). Then, cells were counted both using a hemacytometer with trypan blue to detect dead cells, and with an automatic cell counter. The star * indicates that no increase of trypan blue positive dead cells was detected in the BRG1i cells grown with doxycycline.
(D) Median acinar diameter during differentiation. For each cell sample the median acinus size was calculated (n ≥10 acini with 2 measurements of diameter for each acinus). Statistical analysis was by a Student’s t-test. The comparisons marked ** had a high degree of statistical significance (p ≤ 0.01).
(E) Size distribution of acini. The percentage of acini with a diameter smaller than 22 µm, contained in the interval 22 to 65 µm, 65 to 110 µm, or greater than 110 µm were calculated.
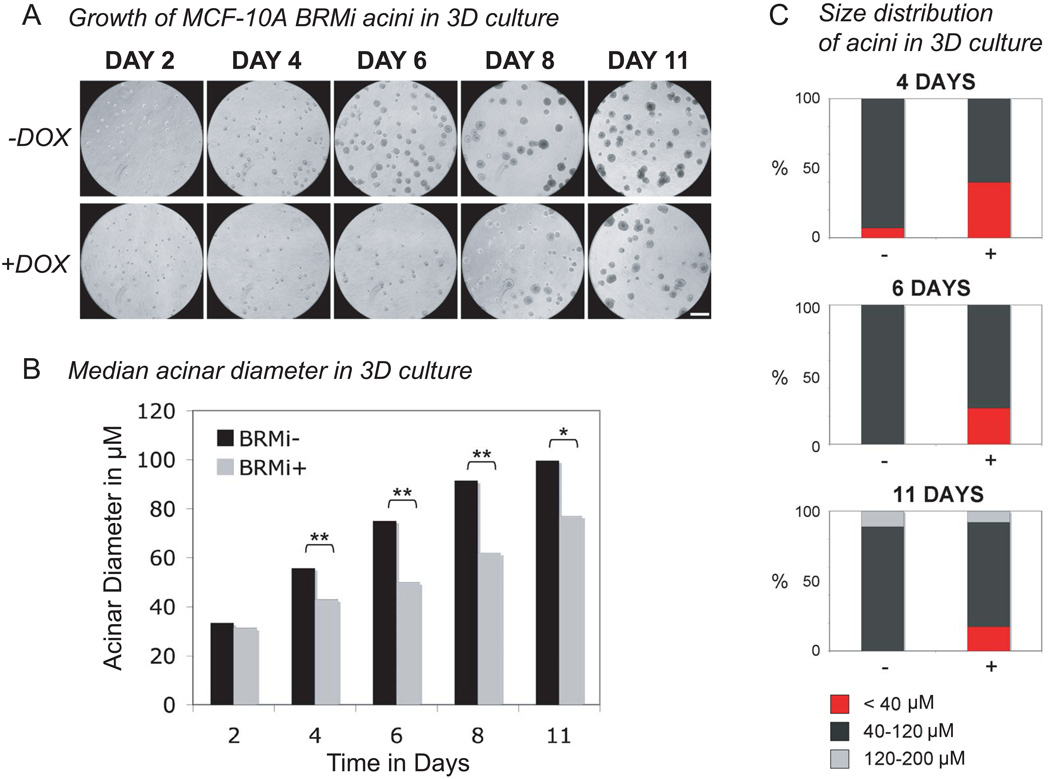
BRMi cells with a doxycycline-inducible knockdown of BRM and control SCRAM cells were grown in three-dimensional rBM culture, with conditions matched to the BRG1 knockdown experiments. As illustrated in Fig. 4A, the BRMi cells expressing the shBRM with doxycycline gave smaller multicellular masses than the uninduced BRMi without doxycycline and the SCRAM control cells. These changes were clear from day 4 in culture. The diameter (Fig. 4B) and the size distribution (Fig. 4C) of the multicellular structures were determined. In the early proliferation stage of differentiation (day 0 to day 8), the MCF-10A cells expressing the shBRM (+ doxycycline) recapitulated the striking decrease in proliferation observed previously with the BRG1 knockdown MCF-10A cells. An analysis of size distributions (Fig. 4C, compare fraction of acini with a diameter < 40 µm) showed a larger fraction of small multicellular structures after BRM knockdown than was observed in the controls from day 4 to day 11. However, unlike BRG1i cells, the difference in size between BRMi cells induced to deplete BRM and uninduced cells decreased with time in culture (see Fig. 4A after day 8). A subset of cells escaped from the growth defect and formed multicellular structures that were larger than normal acini. Subsequent work has established that these escaping structures have developed a tumor like phenotype via altered integrin expression (V. M. Weaver, manuscript in preparation).
Figure 4. MCF-10A cells induced to knock down BRM formed acini with a more variable median size in rBM culture.
(A) MCF-10A cells with an inducible BRM knockdown or control scrambled shRNA were seeded in Matrigel overlay culture. Cultures were fed every 2 days with medium containing 0.05µM doxycycline or medium without doxycycline. Every 2 days phase contrast micrographs were taken of the live cultures. Smaller acini were observed from day 4 in the MCF-10A cells expressing the shRNA targeting BRMi (+doxycycline). No differences in size were observed in MCF-10A cells expressing scrambled shRNA in the presence or absence of doxycycline (data not shown). Size bar : 250 µm Most acini were smaller after the induced knockdown of BRM, but a few acini were able to escape this decrease in proliferation.
(B) For each cell line and condition, the median acinar diameter was calculated from micrographs including the one in panel A. Two measurements of diameter were averaged for each acinus and more than 24 acini were measured for each sample and time point. Statistical analysis was by a Student’s t-test. The comparisons marked by one star (*) were significant (p < 0.05) and those marked by two stars (**) were highly significant (p ≤ 0.01).
(C) The size distribution of acini in 3D culture is shown for the data of panels A and B. The percent of acini with a diameter inferior to 40 µm, contained in the interval 40 to 120 µm, and contained in the interval 120 to 200 µm were calculated.
BRG1 and BRM knockdown decreases proliferation in monolayer culture
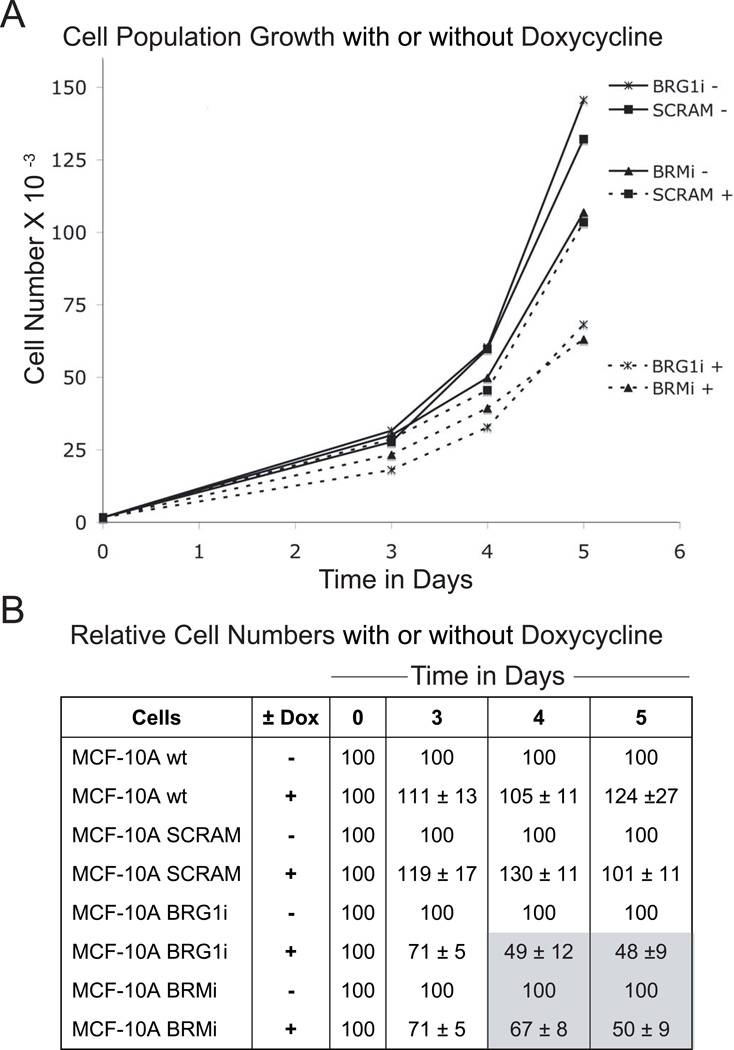
To ascertain whether growth in three dimensional rBM culture was required for this unexpected decrease in proliferation after BRM or BRG1 depletion, we grew the same cell lines with or without doxycycline induction in monolayer culture. After 2 days of pre-induction, we seeded the different cell lines at the same density in 12-well dishes. Each day from day 2 to day 6, cells were trypsinized and counted. A 50% decrease in cell number was observed four or five days after seeding (Fig. 5A), roughly matching the 4 day delay observed in three dimensional culture (see Figs. 3 and 4).
Figure 5. The knockdown of BRG1 slowed proliferation in MCF-10A cells grown in monolayer culture.
(A) Growth curves for MCF-10A cells after BRG1 or BRM shRNA knockdown, or expressing a scrambled sequence shRNA (SCRAM). Cells were pre-incubated for 2 days with 0.05 µg/ml doxycycline (marked +) before being seeded in a 12-well dish at 1500 cells per well at day 0. Control cells (marked −) were not treated with doxycycline and did not express the shRNA. Cells were trypsinized and then counted each day from day 2 to day 5.
(B) Cell proliferation was measured every day by quantifying DNA(Cyquant kit) in parallel with the cell counting of panel A. The decrease of proliferation was quantified by calculating the ratio (in percent) of the cell number with doxycycline to the cell number of the matched doxycycline-free control.
In order to more precisely quantify the proliferation decrease after BRG1 or BRM knockdown, we used a second method that was more robust and sensitive than simple cell counting, eliminated the need for trypsinization, and allowed larger numbers of replicates of each sample. This method measured the density of adherent cells using a dye that fluoresces strongly when bound to cellular nucleic acids, and confirmed the previous results obtained from cell counting. There was a 50% proliferation decrease by day 5 after BRG1 or BRM knockdown (Fig. 5B).
To eliminate the possibility that BRG1 knockdown was only causing the detachment of cells, we manually counted the unattached cells suspended in the culture medium during the 5 days of the proliferation assay. Viability was measured by trypan blue exclusion. There was no increase in the number of detached live or dead cells after knockdown. We also examined whether cellular senescence might explain the reduced proliferation. β-galactosidase staining marks senescent cells (Dimri et al., 1995) and we observed no increase in β -galactosidase positive cells after knockdown. We also stained the cell cultures with toluidine blue to observe cell morphology and detect structural changes characteristic of senescent or apoptotic cells. No changes were observed that were characteristic of senescence or programmed cell death.
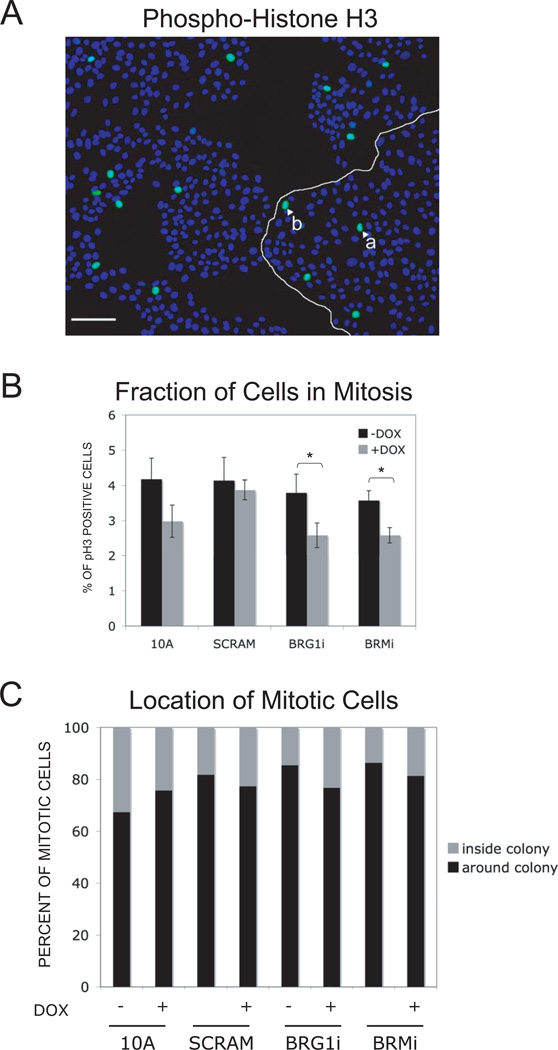
A decrease of proliferation without complete arrest might be due to an abnormal stimulation of cell contact inhibition. A delay of four days for a proliferation decrease to become significant might be explained by this hypothesis. Induction of contact inhibition depends on cell density and might need some time after cell seeding to develop (Gray et al., 2008; Liu et al., 2006; Nelson and Chen, 2003). To determine whether contact inhibition was involved in the decrease of cell proliferation after BRG1 or BRM knockdown, we plated the cell lines on glass coverslips in 6-well plates. After 5 days of doxycycline induction, we immunostained the cells with an antibody that specifically recognizes the mitosis-specific serine 10 phosphorylation of histone H3 (Fig. 6A) (Ajiro and Nishimoto, 1985; Ajiro et al., 1983; Eberlin et al., 2008; Goto et al., 1999; Li et al., 2005; Shibata et al., 1990; Wei et al., 1999). The fraction of cells in mitosis and the location of those mitotic cells within the epithelial colonies was measured (Fig. 6). The overall percentage of cells in mitosis was low, as would be expected after 5 days in culture, but the number of mitotic cells after BRG1 or BRM knock down was reduced (p<0.05) relative to the uninduced control (Fig. 6B). This confirmed the growth inhibition caused by BRG1 or BRM depletion using a third method, and showed that the decrease in proliferation was not caused by a mitotic arrest.
Figure 6. The knockdown of BRG1 or of BRM reduced the percentage of cells MCF-10A cells in monolayer culture that were in mitosis.
(A) In this micrograph, nuclei are stained blue with DAPI and mitotic cells are identified by immunostaining for serine 10 phosphorylated Histone H3 (green) Size bar : 100 mm. The number of mitotic cells was determined, along with their position in the colonies. For example, on this picture, the white arrows show one peripheral (a) and one internal mitotic (b) cell. (B) This histogram compares the percentage of mitotic cells at all positions within colonies after 7 days in the presence or absence of doxycycline. The star (*) indicates a difference that had statistical significance (p ≤ 0.05). (C) The position of mitotic cells in MCF-10A monolayer colonies was scored from micrographs including that of panel A. The compiled data quantifies the percentage of mitotic cells at the periphery of the colony and the percentage at interior positions. For each cell sample n ≥2000 cells.
The localization of mitotic cells was scored according to whether they were inside the colony, crowded by other cells, or whether they were at the edge of the colony. As seen in Fig. 6A–C, mitotic cells were preferentially located at the edges of colonies. Quantification (Fig. 6C) was consistent with previous studies reporting contact inhibition in MCF-10A cells (Liu et al., 2006). The percent of mitotic cells inside the colonies was about 20% and this was not significantly changed by knockdown of either BRG1 or BRM. Taken together these data are inconsistent with the hypothesis that a decrease in cell proliferation after BRG1 or BRM knockdown was caused by the hyper-activation of cell contact inhibition.
Reduction of BRG1 or BRM levels lengthens the cell cycle
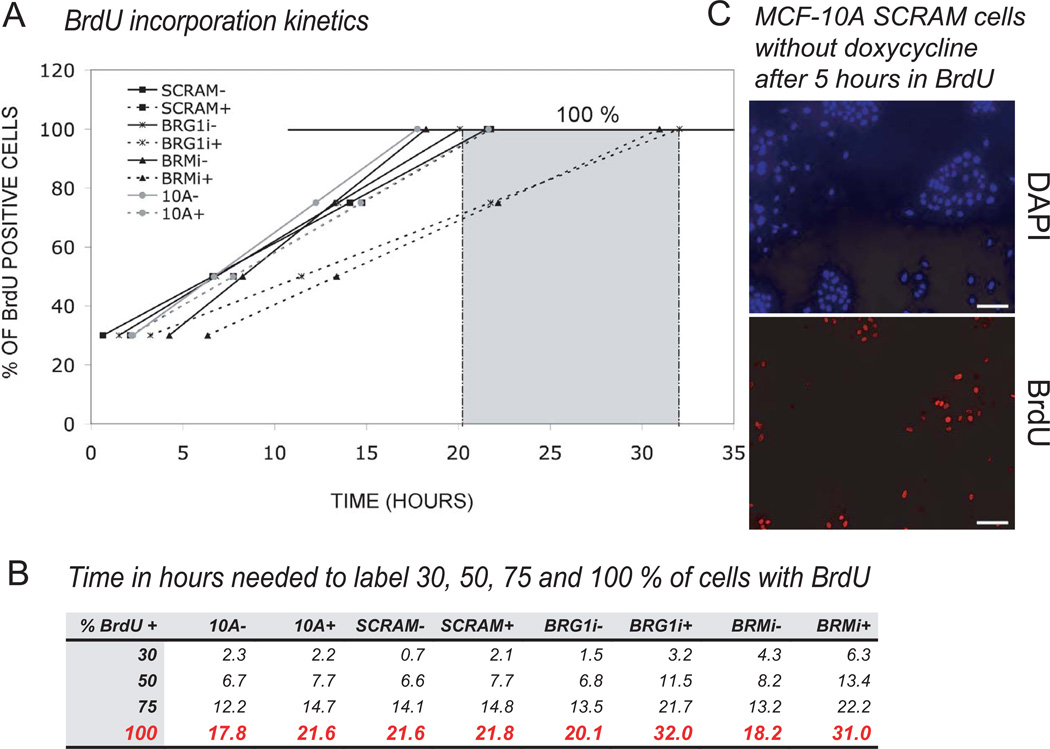
FACS sorting of propidium iodide- and DAPI-stained cells showed no significant changes in the fraction of cells in each phase of the cell cycle after reduction of BRG1 or BRM levels. In order to measure the length of the cell cycle, we performed a time course experiment after pulse labeling cells with BrdU (Fig. 7). BrdU is only incorporated into DNA during S phase, so we were able to measure the cell cycle length of each cell line by counting the fraction of BrdU positive cells at different times after pulse labeling. Using a linear regression method, we extrapolated the results to 100% incorporation, which corresponded to the time needed for one complete cell cycle (Fig. 7A). The normal cell cycle length of wild type MCF-10A cells, control SCRAM cells, and uninduced BRG1 and BRM knockdown cells (no doxycycline) was 18 to 22 hours (Fig. 7A, B). The induced BRG1 and BRM knockdown cells exhibited a longer cell cycle of 31–32 hours. We concluded that an extra 10–14 hours were needed to complete one cell cycle for MCF-10A cells depleted for either BRG1 or BRM and, consequently, that BRG1 and BRM regulate cell cycle length.
Figure 7. The knockdown of BRG1 or of BRM increased the length of the cell cycle as determined by pulse labeling with BrdU.
(A) Cell cycle length was measured from a time course of BrdU incorporation. The graph plots the percent of BrdU positive cells as a function of time in hours. A linear extrapolation to the time when 100 % of cells were BrdU positive cells calculated the mean length of a single cell cycle.
(B) Calculated according to the linear regression method, the time in hours needed for 30, 50, 75 and 100 % of cells to become BrdU positive.
(C) A representative micrograph from which the measurements of panels A and B were made shows MCF-10A cells with the SCRAM control shRNA in absence of doxycycline after 5 hours in BrdU. The number of BrdU positive cells (lower) and the number of total cells (upper, DAPI fluorescence) were counted. Seven fields were analyzed for each group in each of two independents experiments.
A BRG1-BRM double knockdown is lethal
The reciprocal increase of BRM protein level in BRG1 knock- down MCF-10A cells and an increase in BRG1 levels in BRM knockdown cells (Fig. 2) implicates a compensatory mechanism for these two ATPase subunits. This suggests that a more severe phenotype might result from knocking down both BRM and BRG1. To evaluate this, we created a double knockdown in MCF-10A cells. MCF-10A cells with a doxycycline-inducible knockdown of BRG1 were infected with a lentivirus expressing both shRNA targeting BRM and a puromycin resistance gene. After adding doxycycline and puromycin to the media, most of these cells died and the remaining drug-resistant cells did not show the expected decrease in BRM protein. Presumably, the drug treatment conditions selected for cells that escaped BRM knockdown. As a control, MCF-10A cells expressing only the TetR-KRAB regulator were infected under the same conditions. After selection and doxycycline induction, there was minimal cell death and these cells showed the expected BRM knockdown (data not shown).
Upregulation of GAS5 in cells depleted for BRM
To address the mechanisms reducing proliferation rates in MCF10-A cells with reduced BRG1 or BRM, a number of cell cycle regulators were examined. Most of these were not altered after BRG1 or BRM knockdown.
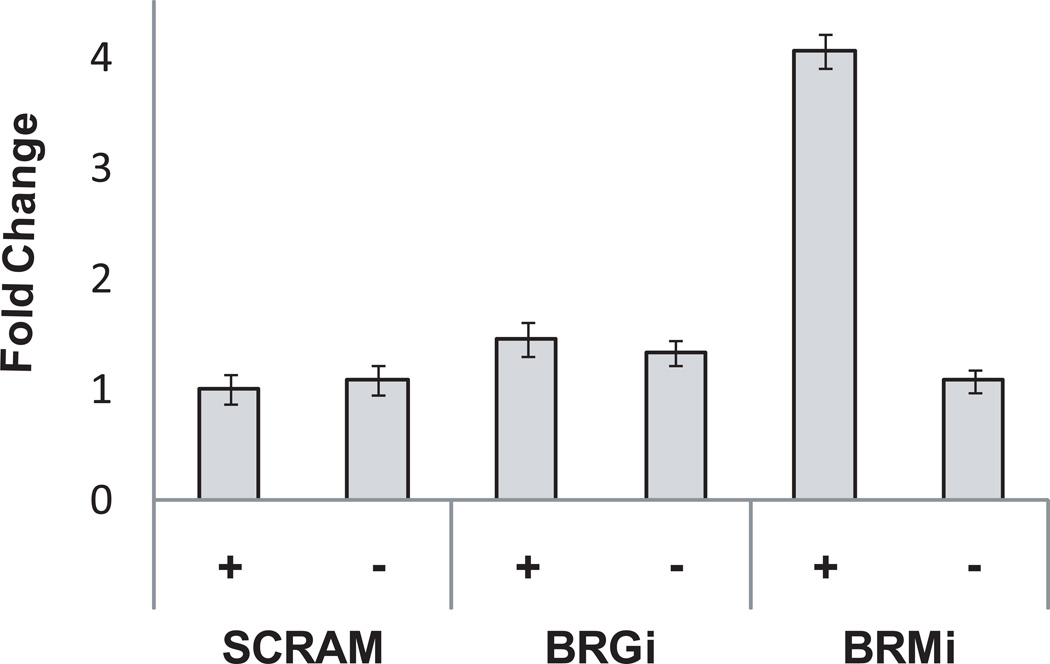
One difference observed was in the levels of GAS5 in BRM, but not BRG1, deficient cells. GAS5 is an alternatively spliced, long non-coding RNA with several snoRNAs in its introns (Coccia et al., 1992; Muller et al., 1998; Raho et al., 2000; Smith and Steitz, 1998). Overexpression of specific GAS5 transcripts can induce cell cycle arrest in some cell lines and sensitize cells to apoptotic signals (Mourtada-Maarabouni et al., 2008) (Mourtada-Maarabouni et al., 2009). GAS5 was reported to be down-regulated in breast cancer derived cell lines (Mourtada-Maarabouni et al., 2009). Q-PCR using primers for regions of GAS5 common to all splice variants showed that GAS5 levels were up-regulated in BRM depleted MCF-10A cells, but not BRG1 depleted cells (Fig. 8). Pathways converging on GAS5 are not yet known, but our data establish a correlation between the overexpression of a noncoding RNA that can negatively regulate cell cycle progression and the decreased proliferation of MCF-10A cells after BRM reduction.
Figure 8. Overexpression of GAS5 RNA in BRM, but not BRG1, deficient cells.
Real-time PCR analysis of GAS5 RNA levels in each cell line. The data represent the mean plus or minus a standard deviation for three independent experiments.
Cyclin A has been implicated in cell cycle control by BRG1 during RB-mediated cell cycle arrest (Strobeck et al., 2000b; Zhang et al., 2000) and is a direct target of BRM in some cells (Coisy et al., 2004). We observed no differences in the expression level of cyclin A in either BRG1 or BRM deficient MCF10-A cells (Fig. S4A). The mTOR pathway can also affect cell cycle progression via control of translation (Ma and Blenis, 2009). Western blot analysis of mTOR, phopho-mTOR, and the mTOR downstream target, p70 S6K, showed no changes due to BRG1 or BRM depletion (Fig. S4B).
A recent report indicated that BRG1 depletion activated p53 in several tumor cell lines (Naidu et al., 2009). We examined total p53 and serine-15 phosphorylated p53 protein levels in BRG1 and BRM depleted MCF10-A cells by Western blot, but observed no increase (Fig. S4C) as might be expected if the p53 pathway were activated.
DISCUSSION
BRG1 and BRM depletion reduces proliferation in MCF-10A mammary epithelial cells
We knocked down BRG1 and BRM in MCF-10A cells, an immortalized but largely normal mammary epithelial cell line (Soule et al., 1990; Yoon et al., 2002). Grown in three-dimensional reconstituted basement membrane culture, MCF-10A cells form acini that resemble structures in normal breast tissue (Debnath et al., 2002). Contrary to expectations, we did not observe an increased rate of proliferation in either monolayer or three dimensional rBM culture. rBM cultures of breast tumor derived cells (Weaver et al., 1997) or of malignant cells engineered from MCF-10A cells (Dawson et al., 1996; Miller et al., 1993; Santner et al., 2001; Strickland et al., 2000) form larger, disorganized structures without proliferation arrest or lumen formation (Imbalzano et al., 2009). BRG1 depleted MCF-10A cells did not have a tumor-like phenotype. Instead, the cells grew more slowly in monolayer culture and, in rBM culture, they failed to expand or form acini. MCF-10A cells with reduced BRM levels also proliferated more slowly on average in both monolayer and for the first 6 days in three dimensional rBM culture.
Multiple methods confirmed the decrease in proliferation of both BRG1 and BRM knockdown MCF-10A cells. In monolayer culture, cells having reduced levels of BRG1 or BRM did not arrest in any specific phase of the cell cycle. BrdU incorporation kinetics definitively showed that cells having reduced levels of BRG1 or BRM simply took longer to traverse the cell cycle. The conclusions that can be reached from these studies are that BRG1 and BRM act as positive regulators of cell cycle progression and BRG1 is required for acinus formation in three dimensional rBM culture.
MCF-10A cells expressing shRNA targeting either BRG1 or BRM showed nearly identical decreases in proliferation and increases in cell cycle length in monolayer culture. However, the cells remained proliferative, which suggests that either protein is sufficient to support some proliferation. The proteins are similar structurally and, while some functions are unique to one or the other ATPase (Kadam and Emerson, 2003), there are also circumstances where the one ATPase can compensate for the absence of the other (Reyes et al., 1998; Strobeck et al., 2002). Given that cells died when we attempted to knock down both BRG1 and BRM, and that in the absence of one protein the levels of the other increased (Fig. 2), we propose that either BRG1 or BRM is necessary to promote cell proliferation and that the cells undergo some form of compensation to increase the levels of the remaining protein when the other is knocked down.
Recently, two reports have established a BRG1 requirement for cell proliferation. These reports differ from the present study in the cell types examined, which suggests that the function of BRG1, and likely BRM, is cell context dependent. In several tumor cell lines with wild type p53, the depletion of BRG1, but not the depletion of BRM, led to activation of p53 and cell senescence (Naidu et al., 2009). In contrast, our results are in a non-tumorigenic cell and do not implicate the p53 pathway. A second study using adult fibroblasts from mice that are deficient for Brm and/or Brg1 showed that the absence of Brg1, but not the absence of Brm, decreased genome integrity, leading to aberrant mitoses and decreased proliferative capacity (Bourgo et al., 2009). In contrast, our results implicate both BRG1 and BRM in promoting cell proliferation.
Efforts to address the mechanisms responsible for decreased proliferation showed that neither the p53 nor the mTOR pathways were altered in BRG1 or BRM knockdown MCF-10A cells. We did find that BRM, but not BRG1, deficient cells contained elevated levels of the large non-coding RNA, GAS5. GAS5 is an inhibitor of cell cycle progression, but is also reported to sensitizes cells to apoptotic signaling (Mourtada-Maarabouni et al., 2009). We observed no apoptosis after BRM knockdown. The correlation between cell cycle length and elevated GAS5 RNA was observed only for BRM deficient MCF-10A cells, despite the similar decrease in cell proliferation rate observed after BRG1 knockdown. The mechanisms by which BRG1 and BRM alter cell cycle progression may be different.
BRG1 in breast cancer progression
There are numerous links between BRG1, BRM and cancer. While Brg1 null mice are embryonic lethal, heterozygous Brg1 mice have an increased susceptibility to epithelial tumors of the breast (Bultman et al., 2000; Bultman et al., 2008). Tissue specific knockout of one Brg1 allele in the lung potentiates tumor formation in an induced carcinogenesis model (Glaros et al., 2008). In contrast, Brm null mice are viable and do not have increased tumor rates, a result attributed to compensation for Brm loss by elevated levels of Brg1 (Reyes et al., 1998). In addition, the knockout of Brm does not cause additional tumors in Brg1+/− mice (Bultman et al., 2008). Despite the lack of tumor formation in Brm null mice, the levels and localization of BRM have prognostic value in staging human lung tumors (Fukuoka et al., 2004; Reisman et al., 2003).
Our results strongly suggest that, at least in normal mammary epithelial cells, reduction of BRG1 or BRM protein levels does not cause a loss of proliferation control leading to accelerated growth. These are different results than those obtained in prior studies using tumor cells, where loss of BRG1/BRM accelerates proliferation. This transformation-specific difference might be due to additional genetic lesions accumulating in cancer cells prior to loss of BRG1 or BRM function.
In conclusion, both BRG1 and BRM are positive regulators of normal mammary cell cycle progression. Despite previous studies indicating that loss of one Brg1 allele predisposes mice to breast tumors, significant reduction of BRG1 levels in normal but immortalized human mammary epithelial cells does not promote properties associated with tumor or transformed cells. This suggests that reduction of BRG1 levels is not sufficient for mammary epithelial cell transformation. BRG1 is required for acinus formation in three-dimensional rBM culture, a model system that recapitulates many aspects of normal breast tissue development. These results are consistent with previous observations that functional SWI/SNF ATPases are necessary for the development and differentiation of many tissue types (reviewed in de la Serna et al., 2006). We propose that SWI/SNF ATPases, and BRG1 in particular, contribute to normal cell growth and differentiation, whereas the contribution of BRG1 deficiency to oncogenesis requires additional genetic changes.
Supplementary Material
MCF-10A cells were infected a lentivirus expressing the TET-KRAB repressor coupled to a dsRed selection marker and with a second lentivirus expressing, under TET-KRAB regulation, an shRNA sequence coupled with a GFP selection marker under TET-KRAB regulation. The cells which integrated both lentiviruses constitutively expressed the TET-KRAB protein with dsRed fluorescence, but also expressed the shRNA and GFP fluorescence after three days of induction with doxycycline (right pattern).
shRNA expression, measured by bicistronic GFP production, and a decrease of BRG1 levels were observed by 48 hours. The shRNA MCF-10A-BRG1i cells were grown in 6-well plate in the presence or absence of 0.5 µg/mL doxycycline, At the indicated time in days, cells were lysed and proteins were separated on a 7.5% SDS-PAGE gel for Western blotting. Proteins were detected with the following antibodies : αBRG1 rabbit polyclonal-immunoserum (de La Serna et al., 2000), αPI3K rabbit polyclonal antibody (Upstate Biosciences), α-tubulin mouse monoclonal (Sigma), and αGFP mouse monoclonal (Abcam).
The MCF-10A-BRG1i cell line was treated with different doses of doxycycline (0.01, 0.05, 0.1, 0.25 and 0.5 µg/mL) and protein extracts were prepared after 3 days. Control cells (marked −) were never treated with doxycycline. Protein lysates were separated on a 7.5% SDS-PAGE gel, after Western blotting the proteins were revealed with the following antibodies : αBRG1 rabbit polyclonal-immunoserum (de La Serna et al., 2000), αPI3K rabbit polyclonal (Upstate Biosciences), and a αGFP mouse monoclonal (Roche).
Total protein was extracted from cells treated or not for 3 days with 0.01 µg/ml doxycycline. Equal numbers of cells were loaded in each lane before electrophoresis and blotting. (A) Western blot analysis of Cyclin A1. (B) Western blot analysis of m-TOR, phosphorylated m-TOR, and p70 S6 kinase. (C) Western blot analysis of total p53 and Serine-10 phosphorylated p53. For all panels, PI 3 kinase probed on the same Western blot was the loading control.
ACKNOWLEDGMENTS
This work was funded by the National Cancer Institute (PO1 CA82834). We thank Eric Campeau (UMass Medical School) for contributing lentivirus vectors, including the pLenti 2X Puro DEST vector of his design, and for his advice and support. Timothy Pasek provided valuable technical assistance. Samisubbu Naidu and Elliott Androphy provided helpful discussion and antibodies, and Jean Underwood generously provided microscopy support. Core facilities were subsidized by Diabetes Endocrinology Research Center grant P30 DK32520.
LITERATURE CITED
- Ajiro K, Nishimoto T. Specific site of histone H3 phosphorylation related to the maintenance of premature chromosome condensation. Evidence for catalytically induced interchange of the subunits. J Biol Chem. 1985;260(29):15379–15381. [PubMed] [Google Scholar]
- Ajiro K, Nishimoto T, Takahashi T. Histone H1 and H3 phosphorylation during premature chromosome condensation in a temperature-sensitive mutant (tsBN2) of baby hamster kidney cells. J Biol Chem. 1983;258(7):4534–4538. [PubMed] [Google Scholar]
- Beale LS. Examination of sputum from a case of cancer of the pharynx and the adjacent parts. Archives of Medicine, London. 1860;2:44. [Google Scholar]
- Bourgo RJ, Siddiqui H, Fox S, Solomon D, Sansam CG, Yaniv M, Muchardt C, Metzger D, Chambon P, Roberts CW, Knudsen ES. SWI/SNF-Deficiency Results in Aberrant Chromatin Organization, Mitotic Failure, and Diminished Proliferative Capacity. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-12-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27(4):460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22(10):1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L, Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12(8):3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coisy M, Roure V, Ribot M, Philips A, Muchardt C, Blanchard JM, Dantonel JC. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2alpha. Mol Cell. 2004;15(1):43–56. doi: 10.1016/j.molcel.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Coisy-Quivy M, Disson O, Roure V, Muchardt C, Blanchard JM, Dantonel JC. Role for Brm in cell growth control. Cancer Res. 2006;66(10):5069–5076. doi: 10.1158/0008-5472.CAN-05-0596. [DOI] [PubMed] [Google Scholar]
- Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148(1):313–319. [PMC free article] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20(8):2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111(1):29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424(6946):324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79(1):119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Eberlin A, Grauffel C, Oulad-Abdelghani M, Robert F, Torres-Padilla ME, Lambrot R, Spehner D, Ponce-Perez L, Wurtz JM, Stote RH, Kimmins S, Schultz P, Dejaegere A, Tora L. Histone H3 tails containing dimethylated lysine and adjacent phosphorylated serine modifications adopt a specific conformation during mitosis and meiosis. Mol Cell Biol. 2008;28(5):1739–1754. doi: 10.1128/MCB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrants AK. Chromatin remodelling and actin organisation. FEBS Lett. 2008;582(14):2041–2050. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- Feoli F, Paesmans M, Van Eeckhout P. Fine needle aspiration cytology of the breast: impact of experience on accuracy, using standardized cytologic criteria. Acta Cytol. 2008;52(2):145–151. doi: 10.1159/000325472. [DOI] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, Hewitt S, Travis WD, Jen J. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10(13):4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Giardina C, Renzulli G, Serio G, Caniglia DM, Lettini T, Ferri C, D'Eredita G, Ricco R, Delfino VP. Nuclear morphometry in node-negative breast carcinoma. Anal Quant Cytol Histol. 1996;18(5):374–382. [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68(10):3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, Inagaki M. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274(36):25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- Gray DS, Liu WF, Shen CJ, Bhadriraju K, Nelson CM, Chen CS. Engineering amount of cell-cell contact demonstrates biphasic proliferative regulation through RhoA and the actin cytoskeleton. Exp Cell Res. 2008;314(15):2846–2854. doi: 10.1016/j.yexcr.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroske G, Dimmer V, Friedrich K, Meyer W, Thieme B, Theissig F, Kunze KD. Nuclear image analysis of immunohistochemically stained cells in breast carcinomas. Histochem Cell Biol. 1996;105(6):479–485. doi: 10.1007/BF01457662. [DOI] [PubMed] [Google Scholar]
- Hill DA, Chiosea S, Jamaluddin S, Roy K, Fischer AH, Boyd DD, Nickerson JA, Imbalzano AN. Inducible changes in cell size and attachment area due to expression of a mutant SWI/SNF chromatin remodeling enzyme. J Cell Sci. 2004;(117)(Pt 24):5847–5854. doi: 10.1242/jcs.01502. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370(6489):481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Imbalzano KM, Tatarkova I, Imbalzano AN, Nickerson JA. Increasingly transformed MCF-10A cells have a progressively tumor-like phenotype in three-dimensional basement membrane culture. Cancer Cell Int. 2009;9(1):7. doi: 10.1186/1475-2867-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23(8):403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lelievre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci U S A. 1998;95(25):14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DW, Yang Q, Chen JT, Zhou H, Liu RM, Huang XT. Dynamic distribution of Ser-10 phosphorylated histone H3 in cytoplasm of MCF-7 and CHO cells during mitosis. Cell Res. 2005;15(2):120–126. doi: 10.1038/sj.cr.7290276. [DOI] [PubMed] [Google Scholar]
- Liu WF, Nelson CM, Pirone DM, Chen CS. E-cadherin engagement stimulates proliferation via Rac1. J Cell Biol. 2006;173(3):431–441. doi: 10.1083/jcb.200510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Marshall PN. Papanicolaou staining--a review. Microsc Acta. 1983;87(3):233–243. [PubMed] [Google Scholar]
- Merlo GR, Basolo F, Fiore L, Duboc L, Hynes NE. p53-dependent and p53-independent activation of apoptosis in mammary epithelial cells reveals a survival function of EGF and insulin. J Cell Biol. 1995;128(6):1185–1196. doi: 10.1083/jcb.128.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FR, Soule HD, Tait L, Pauley RJ, Wolman SR, Dawson PJ, Heppner GH. Xenograft model of progressive human proliferative breast disease. J Natl Cancer Inst. 1993;85(21):1725–1732. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121(Pt 7):939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AJ, Chatterjee S, Teresky A, Levine AJ. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine chromosome 1. Mamm Genome. 1998;9(9):773–774. doi: 10.1007/s003359900862. [DOI] [PubMed] [Google Scholar]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009;28(27):2492–2501. doi: 10.1038/onc.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Chen CS. VE-cadherin simultaneously stimulates and inhibits cell proliferation by altering cytoskeletal structure and tension. J Cell Sci. 2003;116(Pt 17):3571–3581. doi: 10.1242/jcs.00680. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. Embo J. 2006;25(3):490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89(19):9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3(2):247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Raho G, Barone V, Rossi D, Philipson L, Sorrentino V. The gas 5 gene shows four alternative splicing patterns without coding for a protein. Gene. 2000;256(1–2):13–17. doi: 10.1016/s0378-1119(00)00363-2. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21(8):1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4(2):133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. J Cell Physiol. 2005;205(2):286–294. doi: 10.1002/jcp.20397. [DOI] [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65(2):101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Shibata K, Inagaki M, Ajiro K. Mitosis-specific histone H3 phosphorylation in vitro in nucleosome structures. Eur J Biochem. 1990;192(1):87–93. doi: 10.1111/j.1432-1033.1990.tb19199.x. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15(5):603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086. [PubMed] [Google Scholar]
- Strickland LB, Dawson PJ, Santner SJ, Miller FR. Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers. Breast Cancer Res Treat. 2000;64(3):235–240. doi: 10.1023/a:1026562720218. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Fribourg AF, Puga A, Knudsen ES. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene. 2000a;19(15):1857–1867. doi: 10.1038/sj.onc.1203510. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, Knudsen ES. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000b;97(14):7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, Kowalik TF, Weissman BE, Knudsen ES. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277(7):4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- Strober BE, Dunaief JL, Guha, Goff SP. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16(4):1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A. 1997;94(21):11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JM, Imbalzano KM, Weaver VM, Fischer AH, Imbalzano AN, Nickerson JA. The ultrastructure of MCF-10A acini. J Cell Physiol. 2006;208(1):141–148. doi: 10.1002/jcp.20639. [DOI] [PubMed] [Google Scholar]
- Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci U S A. 2003;100(6):3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21(55):8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10(17):2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Howlett AR, Langton-Webster B, Petersen OW, Bissell MJ. The development of a functionally relevant cell culture model of progressive human breast cancer. Semin Cancer Biol. 1995;6(3):175–184. doi: 10.1006/scbi.1995.0021. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2(3):205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97(1):99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77(16):8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaswen P, Stampfer MR. Molecular changes accompanying senescence and immortalization of cultured human mammary epithelial cells. Int J Biochem Cell Biol. 2002;34(11):1382–1394. doi: 10.1016/s1357-2725(02)00047-x. [DOI] [PubMed] [Google Scholar]
- Yoon DS, Wersto RP, Zhou W, Chrest FJ, Garrett ES, Kwon TK, Gabrielson E. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer. Am J Pathol. 2002;161(2):391–397. doi: 10.1016/S0002-9440(10)64194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101(1):79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MCF-10A cells were infected a lentivirus expressing the TET-KRAB repressor coupled to a dsRed selection marker and with a second lentivirus expressing, under TET-KRAB regulation, an shRNA sequence coupled with a GFP selection marker under TET-KRAB regulation. The cells which integrated both lentiviruses constitutively expressed the TET-KRAB protein with dsRed fluorescence, but also expressed the shRNA and GFP fluorescence after three days of induction with doxycycline (right pattern).
shRNA expression, measured by bicistronic GFP production, and a decrease of BRG1 levels were observed by 48 hours. The shRNA MCF-10A-BRG1i cells were grown in 6-well plate in the presence or absence of 0.5 µg/mL doxycycline, At the indicated time in days, cells were lysed and proteins were separated on a 7.5% SDS-PAGE gel for Western blotting. Proteins were detected with the following antibodies : αBRG1 rabbit polyclonal-immunoserum (de La Serna et al., 2000), αPI3K rabbit polyclonal antibody (Upstate Biosciences), α-tubulin mouse monoclonal (Sigma), and αGFP mouse monoclonal (Abcam).
The MCF-10A-BRG1i cell line was treated with different doses of doxycycline (0.01, 0.05, 0.1, 0.25 and 0.5 µg/mL) and protein extracts were prepared after 3 days. Control cells (marked −) were never treated with doxycycline. Protein lysates were separated on a 7.5% SDS-PAGE gel, after Western blotting the proteins were revealed with the following antibodies : αBRG1 rabbit polyclonal-immunoserum (de La Serna et al., 2000), αPI3K rabbit polyclonal (Upstate Biosciences), and a αGFP mouse monoclonal (Roche).
Total protein was extracted from cells treated or not for 3 days with 0.01 µg/ml doxycycline. Equal numbers of cells were loaded in each lane before electrophoresis and blotting. (A) Western blot analysis of Cyclin A1. (B) Western blot analysis of m-TOR, phosphorylated m-TOR, and p70 S6 kinase. (C) Western blot analysis of total p53 and Serine-10 phosphorylated p53. For all panels, PI 3 kinase probed on the same Western blot was the loading control.