Abstract
Chemotaxis of tumor cells in response to a gradient of extracellular ligand is an important step in cancer metastasis. The heterogeneity of chemotactic responses in cancer has not been widely addressed by experimental or mathematical modeling techniques. However, recent advancements in chemoattractant presentation, fluorescent-based signaling probes, and phenotypic analysis paradigms provide rich sources for building data-driven relational models that describe tumor cell chemotaxis in response to a wide variety of stimuli. Here we present gradient sensing, and the resulting chemotactic behavior, in a ‘cue-signal-response’ framework and suggest methods for utilizing recently reported experimental methods in data-driven modeling ventures.
Introduction
Although significant progress has been made in the cancer biology field, the mechanisms by which primary tumor cells metastasize to distant sites in the body are widely unknown. One important aspect of carcinoma invasion is the presence of multiple stimuli within the tumor microenvironment, including a wide variety of biochemical factors [1], biophysical effects of the extracellular matrix [2], and interstitial flow [3]. The spatiotemporal distributions of these cues are dynamic and demand rigorous analysis for interpretation. Tumor cells must sense a cue, most often through ligand binding, integrate signals, through activation of multiple intracellular networks, and respond through mobilization of cytoskeletal machinery and concomitant modification of surrounding matrix. Often it is only through simplification of experimental assays and implementation of mathematical and engineering principles can one begin to understand mechanisms of gradient sensing and response.
Directed cell migration, or chemotaxis, results from the ability of cells to process an extracellular cue through a complex intracellular signaling network to produce a coordinated and robust response. This inherent ‘cue-signal-response’ process represents a convenient paradigm for deconstructing the processes by which cells utilize gradients of chemical and mechanical stimuli within their microenvironment. We and others have taken advantage of this framework to build quantitative, data-driven, predictive models of cell response, analyzing cancer-relevant cell motility behaviors in two- and three-dimensional in vitro settings [4–7]; these types of relational models (see Box 1) have recently been extended to in vivo applications [8]. In this mini-review we concentrate on the quantitative cell biological and biochemical measurements that can serve as inputs (cues, signals, and responses) to modeling efforts with a focus on previously underutilized parameters that hold potential for improving predictive ability in application to chemotaxis in cancer. Recent advancements in experimental strategies for obtaining multi-dimensional and information-rich datasets will be emphasized. Particular mathematical techniques will not be discussed here; rather, readers are directed to reviews focused on the modeling approaches [9,10].
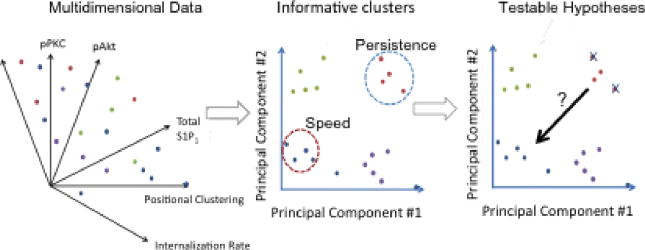
Box 1. Relational modeling for predicting chemotactic behavior.
Relational modeling can predict cell motility behaviors based upon measurement of intracellular signaling network state in vitro [4,5,7,74]. Modeling efforts begin through stimulation of motility through a set of ‘cues’, which can be addition of extracellular ligand, intracellular perturbation (siRNA or chemical inhibitor), and/or changes to the extracellular matrix. Highly multidimensional data is collected through biochemical or imaging techniques (panel 1). Representation of this type of data typically requires data-reduction techniques such as principal components analysis to reveal relationships between the measured ‘signals’ and the input ‘cues’ (panel 2). To gain insight to motility responses, signals and/or cues can be regressed against cellular ‘response’, often using techniques such as partial least squares or multilinear regression. These analyses correlate the clustered ‘signal’ and/or ‘cue’ data to a phenotypic output. For example, specific signals may be more representative of cell speed versus cell persistence (panel 2). Relational models are best utilized to generate hypotheses, such as specific ‘signals’ or ‘cues’ that may be inhibited to modulate cell motility (panel 3).
Cues
A major challenge of data-driven modeling is the need for generation of appropriate multivariate datasets, sufficiently broad in scope to comprehend a significant range of response behaviors driven by a spectrum of potentially germane cue conditions. More specifically, when studying a heterogeneous response, such as chemotaxis, how does one collect relevant data from enough cells that are exposed to a consistent set of cues? In contrast to macrogradients that persist over long distances, such as those that direct organogenesis [11], control lymphocyte egress [12], or are encountered in some wound healing models [13], most tumor cells encounter local gradients of signaling molecules arising from autocrine or paracrine secretion of chemokines and growth factors. This situation has been observed in vivo using intravital microscopy in murine mammary carcinoma, where tumor-associated macrophages induce tumor cell migration via secretion of Epidermal Growth Factor (EGF). In turn, tumor cells secrete Colony Stimulating Factor-1 (CSF-1), which is a potent chemoattractant for macrophages [14,15]. In vivo collection of stromal and carcinoma cells from ectopic and spontaneous tumors in response to growth factor diffusion from a Matrigel-based solution within a hollow-bore needle [16] has confirmed chemotaxis towards a variety of stimuli [14,17,18]. Several important points arise from these studies, including the demonstration of a saturable chemotactic response (thus setting boundary conditions on relevant in vitro studies), differential ligand requirement based upon receptor expression (lending insight into in vivo response across clinical subtypes), and demonstration of relative independence of biomechanical properties of the tumor (as the peak invasion across studies is roughly equivalent, but the position of collection is variable and does not account for tumor mechanics). These insights must be considered when designing experiments informative for model construction.
Close Encounters: Presentation of Chemoattractants in Tumors
Although it is clear that the mechanical properties of the tumor microenvironment play a role in tumor progression, we will focus here on cell-secreted ligands whose gradients could be influenced by their binding of extracellular matrix components. The most commonly studied in carcinoma are the EGF receptor family ligands (EGF, TGFα, HB-EGF, amphiregulin, NRG1), the angiogenic factor VEGF, the macrophage-motility factor CSF-1, the ‘scatter factor’ HGF, the antiproliferative factor TGFβ, and the cytokines CCL19, CCL21, and CXCL12 or SDF-1. Several methods of ligand presentation have been utilized to study chemotaxis of tumor cells in vitro, with Boyden or transwell chambers being the most common end-point assays [19]. Despite providing a convenient screening approach for evaluation of chemoattractant potency [20], there are several drawbacks to these assays, predominantly gradient instability [21,22]. To overcome this limitation several groups have developed microfluidic devices capable of generating stable gradients of chemoattractants that mimic the nonlinearity of early time points found with diffusion-based approaches [23–28]. These devices allow direct visualization of chemotaxis within well-defined gradients in both 2D- and 3D-culture formats and in some cases can be utilized for high-throughput data collection [29]. Alternatively, passive release of ligands from synthetic or natural hydrogels can also be used to create chemoattractant gradients [30,31].
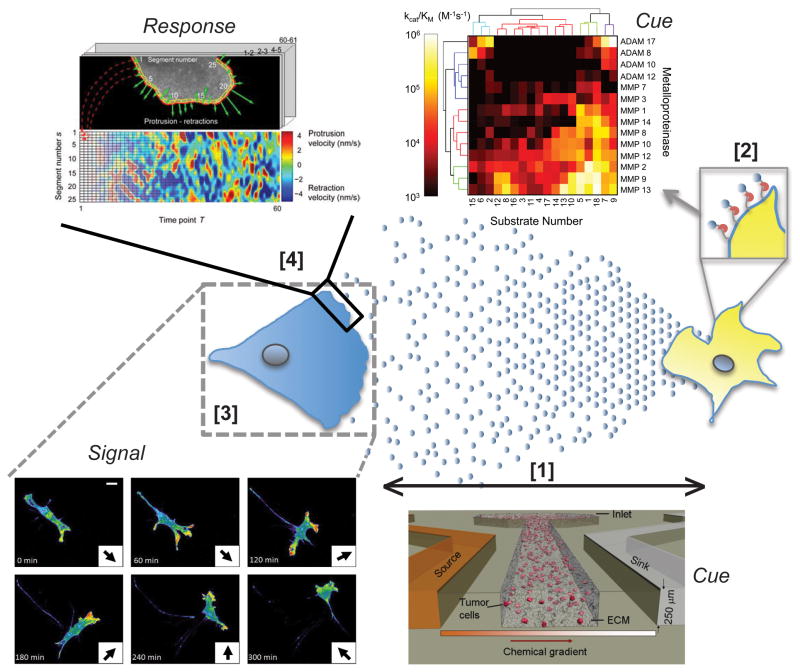
ECM-binding properties of cytokines and growth factors present an additional challenge for re-creating and characterizing chemoattractant gradients in vitro. For example, the heparin-associated matrix binding properties of CCL21, a chemokine that binds CCR-7, promotes formation of autologous autocrine gradients that are amplified in the presence of interstitial flow [3,32]. Creating appropriate CCL21 gradients in vitro is difficult due to the time required to reach steady-state gradient formation [26,27]. To address this issue, Haessler et al. created a microfluidic device incorporating a CCL21-containing agarose overlay. By allowing steady-state gradient formation to occur across the agarose, injection of a cell-containing ECM yields minimal system perturbation and leads to rapid gradient formation across the area of interest (see Figure 1) [26]. Such a device could be employed along with high-resolution imaging to study real-time dynamics of events during early stages of the chemotactic response in 3D culture.
Figure 1. New experimental approaches for populating data-driven relational models of chemotaxis in cancer.
Growth factor gradients (circles) that arise from paracrine signaling between tumor-associated macrophages (yellow) and breast carcinoma cells (blue) represent one example of chemotaxis within tumors [14]. Relational models for probing the mechanisms underlying tumor cell chemotaxis can be conceptualized using a ‘cue-signal-response’ paradigm. [1] Utilization of recently developed, highly controllable 3D microfluidic platforms can provide physiologically relevant presentation of chemoattractant cues. [75]. Device diagram used with kind permission from Dr. Andrew Darling. [2] Quantification of protease activities utilizing a systems biology approach provides mechanism underlying gradient generation and cue presentation. A matrix of kinetic rate parameters measured with purified enzymes can be utilized to estimate specific protease activity in biological samples. Figure courtesy of M. Miller and D. Lauffenburger [41]. [3] New single cell analyses, including Signaling Vector Analysis [54,55], provide novel signaling metrics, such as the magnitude and direction of Akt-PH domain localization (pseudo-colored such that red is highest intensity). The resulting time-dependent vectors, inset arrows, can be correlated with input cues and phenotypic response. Figure used with kind permission from [55]. [4] Quantification of phenotypic response ultimately determines the applicability of any data-driven, relational model. Automated analyses, such as morphodynamic profiling [67], may provide unbiased quantitative data about acute cell response upon exposure to chemoattractant gradients. The metrics derived from analysis of edge dynamics can be correlated to cues, such as protease activity (chemoattractant release), or signals, such as growth factor receptor distribution (See Signals sub-section), to generate hypotheses through application of relational modeling. Figure reproduced with kind permission from [67].
Active Release: Accounting for Extracellular Proteases
Several families of extracellular proteases, including matrix metalloproteinases (MMPs), A Disintegrin and Metalloproteinases (ADAMs), serine proteases (such as matripase/MTSP-1), and cysteine proteases (such as cathepsins) are secreted by tumor cells (see [33] for a recent comprehensive review). Although their contributions to cancer progression and metastasis are well appreciated, therapeutic targeting of proteases in cancer has been unsuccessful [34]. The lack of systematic studies identifying and validating protease targets, which is a direct consequence of the complexity of the network and the paucity of specific, quantitative tools, likely underlies failure of protease-targeted drugs. In the context of gradient sensing, extracellular proteases -- often membrane-bound or secreted from tumor and stromal cells (Figure 1) -- can generate free ligand through cleavage of ECM-binding domains [35,36], enhance gradient formation through cleavage of ECM-binding ligands from the cell surface [37], and contribute to unmasking of cryptic ligand-like binding sites through direct cleavage of ECM proteins. Therefore, activities of extracellular proteases provide cues for gradient sensing, but have been largely neglected in data-driven relational models for cell migration.
Protease activity probes, based upon a decrease in fluorescence resonance energy transfer (FRET) upon substrate cleavage, have been employed to study protease activities in vitro [38,39]. However, identification of which protease is responsible for cleavage is problematic because of broad promiscuity for multiple substrates by many proteases [40] and substrate cross-recognition among several protease family members [38]. Miller et al. reported a multi-variate approach to deconvolving protease activity networks using combinatorial measurements coupled to a mathematical analysis permitting elucidation of specific protease activities [41]. The method involves construction of a matrix of kinetic rate parameters determined from purified enzymes with a library of FRET-based probes (see Figure 1). This parameter matrix is readily expandable to include additional proteases as desired depending on reagent availability. Although this method has to date been applied to two-dimensional culture systems, it can be extended to study of cells cultured in a range of ligand and matrix conditions -- thus permitting quantification of changes in protease activities during tumor progression.
Signals
Currently, most data-driven, relational models utilize population-level measurements to represent the signaling state of cells at various time-points after stimulation, often in the presence of perturbations such as RNAi or chemical inhibition [4–7,42]. Given the heterogeneity characteristic within tumors and tumor cell lines, this simplification may fail to capture signaling events occurring in the most dangerous cells, which may be rare within any given population. Development of more robust reporters and effective analysis tools [43] should facilitate building predictive models for cell gradient sensing that include quantitative measures of receptor distribution, protein localization, and spatiotemporal evolution of signaling states.
Initiation of Signal Propagation: Distribution of Cell Surface Receptors
Regardless of cue presentation, cells begin information processing at the cell membrane. In the case of most biochemical ligands (e.g., growth factors or cytokines), cell surface receptors initiate signaling flux upon ligand binding, so that the original spatial distribution of unligated receptor could influence initial gradient sensing. This may not be the case for cytokine sensing in mammalian cells as G-protein coupled receptors are uniformly distributed along the plasma membrane regardless of chemokine gradient [44]. Similarly, receptor tyrosine kinases (RTKs), such as the EGF receptor, are typically distributed evenly along the cell perimeter in vitro in the absence of signaling gradients [45]. However, despite a uniform total receptor distribution as visualized using epiflourescence microscopy techniques, bias towards increased receptor clustering at the cell periphery has been inferred using fluorescently-tagged receptor or single-particle tracking and TIRF microscopy [46,47]. Therefore, relevant receptor distribution studies might be those describing redistribution of receptors in the first minutes after exposure to a graded cue. The spatiotemporal parameters of receptor distribution, trafficking, and ligand binding have not yet been incorporated into relational models of cell migration. Nor have these parameters been combined with large proteomic or genomic datasets to obtain a systems view of how the biophysical presentation of cytokine and growth factor receptors directly impact specific cell responses. Increased accessibility of live-cell, high resolution imaging modalities should enable correlation of near-single molecule dynamic data to downstream signaling pathways.
Choosing a direction: Receptor proximal signaling in single cells
In the simplest sense, chemotaxis is driven through coordinated cycles of actin polymerization at sites of ligand-bound receptor and substrate adhesion [48]. Our knowledge of the molecules involved in the chemotactic response has increased significantly in the last decade, and a review of many of the known mechanisms underlying chemotaxis in cancer was recently published [1]. Here, we will concentrate on novel experimental and quantitative methods for studying the underlying signaling cascades.
An early step in the chemotactic response of mammary carcinoma cells is the activation of PLCγ, which facilitates initial actin polymerization through a local increase in filament severing activity by PI(4,5)P2-associated proteins [49]. This activity of PLCγ is required for directional migration in response to a chemotactic cue [50,51]. Generally, phosphorylation of PLCγ is used as a proxy for hydrolysis of PI(4,5)P2, but van Rheenen et al. showed through FRET-based assays that local hydrolysis of PI(4,5)P2 leading to spatially-restricted cofilin-actin interaction sets the directional compass of carcinoma cells responding to gradients [52]. The amplitude of this response likely varies within heterotypic cultures, especially considering that cells from the same tumor can exhibit dramatically different responses to growth factor stimulation [53].
In lower organisms as well as tumor cells, a second wave of polymerization-competent actin barbed end generation occurs and is dependent on signaling through the PI3K pathway [49]. The activation of PI3K, and the generation of PI(3,4,5)P3, is recognized as a primal step in cell polarization. A recent series of reports from Haugh and colleagues demonstrates a technique to monitor motility parameters as a function of PI3K activity on an individual-cell basis [54–56]. The evolution of spatiotemporal PI3K-mediated PI(3,4,5)P3 generation was tracked using a fluorescently tagged Akt-PH domain in randomly migrating and chemotaxing cells. A custom algorithm revealed significant positive correlation between a motility parameter (the chemotactic index) and a PI3K-specific parameter (the signaling index) that is a measure of the directionality of signal strength based upon localization and fluorescence intensity (see Figure 1). This correlation was independent of growth factor gradient, suggesting that the location of PI3K signaling is indicative of directional migration regardless of a spatiotemporal cue context [54]. These results agree with the polarized role of PI3K in previous models of motility [57], but extend the insight to include mechanistic coupling between stochastic generation of signal and the subsequent migration directionality in mammalian cells [55].
Assessing signaling activities using ectopic expression of biosensors remains compromised by potential artifactual effects. Another, perhaps more generally reliable and accessible, approach to obtain information-rich, single-cell, spatially relevant data for relational models, is the employment of high-content imaging [58]. Two examples illustrate the usefulness of this approach for studying directed migration. First, the localization of active (phosphorylated) cofilin is inversely related with its ability to sever actin filaments and promote early membrane protrusion, rendering cell-averaged measures of phospho-cofilin uninformative about fast cell migration responses [59,60]. Second, chemotaxis is dependent on detachment of the cell rear from the underlying substrate [48], and cleavage of adhesion components by m-calpain is integral to this process [61,62]. Localization of m-calpain to PI(4,5)P2 at the cell membrane that regulates this activity, instead of phosphorylation level of either the inhibitory or activating phospho-serine sites [63]. Therefore, localization of at least certain signaling proteins can produce increased insights concerning chemotaxis.
Responses
The final input variable for cue-signal-response relational models, and the most important for connection to physiological applications, is the cell phenotypic response(s) elicited by imposed signaling gradients. One consequence of building relational models that result in identification of important signals (or cues) is the issue of context dependency. When measuring motility phenotypes it is important to consider the relevance of experimental conditions to the physiological question that is being interrogated. Classic two-dimensional chemotaxis assays exhibit release profiles and kinetics that are predictable based upon diffusive transport phenomenon [64,65], which usually are inappropriate for tumor cell chemotaxis due to fast dissipation of the imposed gradient (min) in comparison to the relatively slow rate of tumor cell migration in vitro (hours). Thus we raise here recently developed methods for quantifying directed motility parameters arising from acute cell response to chemokine and long-term chemotaxis.
Measuring the output: New methods to quantify gradient sensing phenotype
Among the first quantifiable cell responses to a chemotactic cue is formation of a lamellipodial protrusion. Membrane protrusion response to paracrine factors occurs in vivo (see Supplemental Movie 3 from [15]) and has been examined in three-dimensional culture following perturbation in adhesion-related proteins [66]. Unbiased characterization of lamellipodial dynamics in response to growth factors and cytokines has proven challenging, especially in unmodified cells (those not expressing a fluorescently tagged protein), due to the heterogeneous shape of the extending cell protrusion in most cell types. The manual nature of these analyses has limited their applicability in correlative studies due to small sample size. Recently, morphodynamic profiling [67] (see Figure 1) approaches for quantification of leading edge dynamics in resting cells and in response to various perturbations have provided significant insight to the roles of several proteins important in the chemotactic response [59]. Thus far only the localized dynamics of signaling proteins occurring near the leading edge have been analyzed with respect to the detailed membrane activity maps [43,59,67–70]. These data could also be analyzed in combination with network-wide signaling measurements and measurements of protein secretion and proteolytic activity (on a population-level as a first pass) to construct network maps informative of changes in lamellipodial dynamics. We have observed that the initial membrane protrusion response across a panel of triple negative breast cancer cell lines is highly predictive of subsequent 3D migration in extracellular matrix (Meyer, A.S., Hughes-Alford, S.K., Lauffenburger, D.A., unpublished work). Therefore, insight gained from using morphodynamic profiling techniques applied to cells in a gradient of chemoattractant might provide predictive power in regards to paracrine and autocrine sensing in 3D culture.
It is attractive to hypothesize that the intracellular signaling pathways underlying random and directed migration are similar [54,71], and that migration mode is governed by signal initiation that could result from cue gradients, receptor presentation [46] or stochastic variability [55,57,71]. Indeed, there is substantial evidence for a set of common pathway participants, although the utilization of individual components may differ among cell types. Gruver et al. recently proposed a simple analysis technique based upon distinct changes in cellular morphology that characterize cell motility, random and directed, using a single scaling law derived from bimodal analysis [72]. Migration can be parsed into the time spent moving in a persistent manner versus that spent reorienting the cell polarity. When compared, these characteristic times are inversely correlated and fit an exponentially decreasing curve. The authors show that the model is robust to abrupt changes in cell direction, which offers an advance over the commonly used ‘persistence time’ parameter found by fitting migration paths to a persistent random-walk model [73].
Conclusions and Future Perspectives
Data that can be generated with single-cell experimental methods are optimal for estimation of molecular-level mechanistic process parameters. However, the realization of fully mechanistic network models encompassing multiple processes and their regulatory pathways remains a future hope. Accordingly, our notion of integrating these kinds of data into relational multi-variate network models can begin to bridge the spatiotemporal knowledge gap that currently exists in the era of high-throughput genomic and proteomic analysis. How does one incorporate single cell measurements with population level data to make predictions about cell phenotype? One advantage of single-cell measurements is the realization of a distribution of response. It is then possible to use measures of error, or distribution width, to characterize the heterogeneity within a specific signaling or response measurement. These data may allow identification of ‘rare cell’ populations, such as those that respond in the bottom or top 5% of the distribution, and might represent the most aggressive tumor cells.
Although the gradient sensing mechanisms in chemotaxis are productively conceptualized in a ‘cue-signal-response’ framework, in reality the process for building data-driven, predictive models is iterative. For example, challenges in measuring the appropriate ‘responses’ that arise from well-planned ‘cues’ mandate preliminary experiments to determine what experimental conditions should be used when measuring the signaling dataset. The most useful signaling data may not come from experiments that are exactly the same as those used to measure the phenotypic response. For instance, the similarity between the leading edge of a cell in chemotaxis versus chemokinetic stimuli suggests that measurement of the signaling network under the latter conditions is likely be to equally as informative and more tractable on a large scale. Certainly, careful consideration of trade-offs between experimental consistency and physiological relevance will aid in most effective construction of data-driven multi-variate relational models for tumor cell chemotaxis.
Acknowledgments
We thank Aaron Meyer for critical reading and suggestions. Special thanks to Dr. Andrew Darling and Professor Mingming Wu for figure contribution. Funding was provided by DOD BCRP fellowship BC087781 to SKA, and by National Institutes of Health grants U54-CA112967 and R01-GM081336 to DAL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez JI, Kang I, You WK, McDonald DM, Weaver VM. In situ force mapping of mammary gland transformation. Integr Biol (Camb) 2011;3:910–921. doi: 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shields JD, Fleury ME, Yong C, Tomei AA, Randolph GJ, Swartz MA. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11:526–538. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Kim H-D, Meyer AS, Wagner JP, Alford SK, Wells A, Gertler FB, Lauffenburger DA. Signaling network state predicts twist-mediated effects on breast cell migration across diverse growth factor contexts. Mol Cell Proteomics. 2011;10:M111.008433. doi: 10.1074/mcp.M111.008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. Modeling HER2 effects on cell behavior from mass spectrometry phosphotyrosine data. PLoS Comput Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim H-D, Grantcharova V, Lauffenburger DA, White FM. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alford SK, Wang Y, Feng Y, Longmore GD, Elbert DL. Prediction of sphingosine 1-phosphate-stimulated endothelial cell migration rates using biochemical measurements. Ann Biomed Eng. 2009;38:2775–2790. doi: 10.1007/s10439-010-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. This report is the first to demonstrate the applicability of data-driven relational modeling techniques to ‘cue-signal-response’ mechanisms in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 10.Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput Biol. 2011;7:e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- 12.Schwab SR, Pereira JP, Matloubian M, Xu Y, Huang Y, Cyster JG. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 13.McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CC, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 14.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 15.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 16.Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–5404. [PubMed] [Google Scholar]
- 17.Hernandez L, Smirnova T, Kedrin D, Wyckoff J, Zhu L, Stanley ER, Cox D, Muller WJ, Pollard JW, Van Rooijen N, et al. The EGF/CSF-1 paracrine invasion loop can be triggered by heregulin beta1 and CXCL12. Cancer Res. 2009;69:3221–3227. doi: 10.1158/0008-5472.CAN-08-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–9506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zigmond SH, Foxman EF, Segall JE. Chemotaxis assays for eukaryotic cells. Curr Protoc Cell Biol. 2001;Chapter 12(Unit 12):11. doi: 10.1002/0471143030.cb1201s00. [DOI] [PubMed] [Google Scholar]
- 20.Ott TR, Pahuja A, Lio FM, Mistry MS, Gross M, Hudson SC, Wade WS, Simpson PB, Struthers RS, Alleva DG. A high-throughput chemotaxis assay for pharmacological characterization of chemokine receptors: Utilization of U937 monocytic cells. J Pharmacol Toxicol Methods. 2005;51:105–114. doi: 10.1016/j.vascn.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Buettner HM, Lauffenburger DA, Zigmond SH. Measurement of leukocyte motility and chemotaxis parameters with the Millipore filter assay. J Immunol Methods. 1989;123:25–37. doi: 10.1016/0022-1759(89)90026-4. [DOI] [PubMed] [Google Scholar]
- 22.Keenan TM, Folch A. Biomolecular gradients in cell culture systems. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SJ, Saadi W, Lin F, Minh-Canh Nguyen C, Li Jeon N. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300:180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 24.Mosadegh B, Saadi W, Wang S-J, Jeon NL. Epidermal growth factor promotes breast cancer cell chemotaxis in CXCL12 gradients. Biotechnol Bioeng. 2008;100:1205–1213. doi: 10.1002/bit.21851. [DOI] [PubMed] [Google Scholar]
- 25.Huang CP, Lu J, Seon H, Lee AP, Flanagan LA, Kim HY, Putnam AJ, Jeon NL. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip. 2009;9:1740–1748. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Haessler U, Pisano M, Wu M, Swartz MA. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci U S A. 2011;108:5614–5619. doi: 10.1073/pnas.1014920108. By carefully considering the properties of the cytokine presented, Haessler et al. modify and existing 3D microfluidic device (reference 75) to differentiate responses to two CCR-7 ligands relevant in immune cell and tumor cell chemotaxis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polacheck WJ, Charest JL, Kamm RD. Interstitial flow influences direction of tumor cell migration through competing mechanisms. Proc Natl Acad Sci U S A. 2011;108:11115–11120. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. A platform for assessing chemotactic migration within a spatiotemporally defined 3D microenvironment. Lab Chip. 2008;8:1507–1515. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyvantsson I, Vu E, Lamers C, Echeverria D, Worzella T, Echeverria V, Skoien A, Hayes S. Image-based analysis of primary human neutrophil chemotaxis in an automated direct-viewing assay. J Immunol Methods. 2011;374:70–77. doi: 10.1016/j.jim.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raja WK, Gligorijevic B, Wyckoff J, Condeelis JS, Castracane J. A new chemotaxis device for cell migration studies. Integr Biol (Camb) 2010;2:696–706. doi: 10.1039/c0ib00044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Irvine DJ. Engineering chemoattractant gradients using chemokine-releasing polysaccharide microspheres. Biomaterials. 2011;32:4903–4913. doi: 10.1016/j.biomaterials.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel DD, Koopmann W, Imai T, Whichard LP, Yoshie O, Krangel MS. Chemokines have diverse abilities to form solid phase gradients. Clin Immunol. 2001;99:43–52. doi: 10.1006/clim.2000.4997. [DOI] [PubMed] [Google Scholar]
- 33.Affara NI, Andreu P, Coussens LM. Delineating protease functions during cancer development. Methods Mol Biol. 2009;539:1–32. doi: 10.1007/978-1-60327-003-8_1. [DOI] [PubMed] [Google Scholar]
- 34.Dorman G, Cseh S, Hajdu I, Barna L, Konya D, Kupai K, Kovacs L, Ferdinandy P. Matrix metalloproteinase inhibitors: a critical appraisal of design principles and proposed therapeutic utility. Drugs. 2010;70:949–964. doi: 10.2165/11318390-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 35.Koshikawa N, Mizushima H, Minegishi T, Iwamoto R, Mekada E, Seiki M. Membrane type 1-matrix metalloproteinase cleaves off the NH2-terminal portion of heparin-binding epidermal growth factor and converts it into a heparin-independent growth factor. Cancer Res. 2010;70:6093–6103. doi: 10.1158/0008-5472.CAN-10-0346. [DOI] [PubMed] [Google Scholar]
- 36.Szabo R, Rasmussen AL, Moyer AB, Kosa P, Schafer JM, Molinolo AA, Gutkind JS, Bugge TH. c-Met-induced epithelial carcinogenesis is initiated by the serine protease matriptase. Oncogene. 2011;30:2003–2016. doi: 10.1038/onc.2010.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37:551–566. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moss ML, Rasmussen FH. Fluorescent substrates for the proteinases ADAM17, ADAM10, ADAM8, and ADAM12 useful for high-throughput inhibitor screening. Anal Biochem. 2007;366:144–148. doi: 10.1016/j.ab.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 39.Moss ML, Powell G, Miller MA, Bin Q, Sang QX, De Strooper B, Tesseur I, Lichtenthaler SF, Taverna M, Zhong JL, et al. ADAM9 inhibition increases membrane activity of ADAM10 and controls {alpha}secretase processing of amyloid precursor protein. J Biol Chem. 2011 doi: 10.1074/jbc.M111.280495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- 41•.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb) 2010;3:422–438. doi: 10.1039/c0ib00083c. This study presents the first systems approach to accurately deconvolve and quantify protease activity within mixtures of MMPs and ADAMs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauken CM, Caplan MR. Temporal differences in Erk1/2 activity distinguish among combinations of extracellular matrix components. Acta Biomater. 2011;7:3973–3980. doi: 10.1016/j.actbio.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 43.Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol. 2011;12:749–756. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Servant G, Weiner OD, Neptune ER, Sedat JW, Bourne HR. Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis. Mol Biol Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailly M, Wyckoff J, Bouzahzah B, Hammerman R, Sylvestre V, Cammer M, Pestell R, Segall JE. Epidermal growth factor receptor distribution during chemotactic responses. Mol Biol Cell. 2000;11:3873–3883. doi: 10.1091/mbc.11.11.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. A key paper demonstrating localization dependent EGFR pre-dimerization can modulate growth factor binding and cell response. [DOI] [PubMed] [Google Scholar]
- 47.Rappoport JZ, Simon SM. Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J Cell Sci. 2009;122:1301–1305. doi: 10.1242/jcs.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 49.Mouneimne G, Soon L, DesMarais V, Sidani M, Song X, Yip SC, Ghosh M, Eddy R, Backer JM, Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouneimne G, DesMarais V, Sidani M, Scemes E, Wang W, Song X, Eddy R, Condeelis J. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 51.Chen P, Xie H, Sekar MC, Gupta K, Wells A. Epidermal growth factor receptor-mediated cell motility: phospholipase C activity is required, but mitogen-activated protein kinase activity is not sufficient for induced cell movement. J Cell Biol. 1994;127:847–857. doi: 10.1083/jcb.127.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, Desmarais V, Yip SC, Backer JM, Eddy RJ, Condeelis JS. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell. 2008;15:813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Melvin AT, Welf ES, Wang Y, Irvine DJ, Haugh JM. In chemotaxing fibroblasts, both high-fidelity and weakly biased cell movements track the localization of PI3K signaling. Biophys J. 2011;100:1893–1901. doi: 10.1016/j.bpj.2011.02.047. References 54, 55, and 56 track the development of Signaling Vector Analysis, a tool that combines TIRF-based imaging of PIP3 binding probes and long-term chemotaxis or random motility measurements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Weiger MC, Ahmed S, Welf ES, Haugh JM. Directional persistence of cell migration coincides with stability of asymmetric intracellular signaling. Biophys J. 2010;98:67–75. doi: 10.1016/j.bpj.2009.09.051. References 54, 55, and 56 track the development of Signaling Vector Analysis, a tool that combines TIRF-based imaging of PIP3 binding probes and long-term chemotaxis or random motility measurements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56••.Weiger MC, Wang CC, Krajcovic M, Melvin AT, Rhoden JJ, Haugh JM. Spontaneous phosphoinositide 3-kinase signaling dynamics drive spreading and random migration of fibroblasts. J Cell Sci. 2009;122:313–323. doi: 10.1242/jcs.037564. References 54, 55, and 56 track the development of Signaling Vector Analysis, a tool that combines TIRF-based imaging of PIP3 binding probes and long-term chemotaxis or random motility measurements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Echeverria V, Meyvantsson I, Skoien A, Worzella T, Lamers C, Hayes S. An automated high-content assay for tumor cell migration through 3-dimensional matrices. J Biomol Screen. 2010;15:1144–1151. doi: 10.1177/1087057110378890. [DOI] [PubMed] [Google Scholar]
- 59.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13:646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 61.Wells A, Huttenlocher A, Lauffenburger DA. Calpain proteases in cell adhesion and motility. Int Rev Cytol. 2005;245:1–16. doi: 10.1016/S0074-7696(05)45001-9. [DOI] [PubMed] [Google Scholar]
- 62.Huttenlocher A, Palecek SP, Lu Q, Zhang W, Mellgren RL, Lauffenburger DA, Ginsberg MH, Horwitz AF. Regulation of cell migration by the calcium-dependent protease calpain. J Biol Chem. 1997;272:32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- 63•.Leloup L, Shao H, Bae YH, Deasy B, Stolz D, Roy P, Wells A. m-Calpain activation is regulated by its membrane localization and by its binding to phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2010;285:33549–33566. doi: 10.1074/jbc.M110.123604. This elegant study provides a mechanism for tail retraction in migrating cells and provides another example of the importance of polarized phosphoinositide signaling within a chemotaxing cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Postma M, van Haastert PJ. Mathematics of experimentally generated chemoattractant gradients. Methods Mol Biol. 2009;571:473–488. doi: 10.1007/978-1-60761-198-1_31. [DOI] [PubMed] [Google Scholar]
- 65.Lauffenburger DA, Tranquillo RT, Zigmond SH. Concentration gradients of chemotactic factors in chemotaxis assays. Methods Enzymol. 1988;162:85–101. doi: 10.1016/0076-6879(88)62067-2. [DOI] [PubMed] [Google Scholar]
- 66.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tkachenko E, Sabouri-Ghomi M, Pertz O, Kim C, Gutierrez E, Machacek M, Groisman A, Danuser G, Ginsberg MH. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol. 2011;13:660–667. doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, Blenis J. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell. 2011;41:661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70••.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. This elegant study combines morphodynamic analysis with activity probes for Rho GTPases to map localized activity underlying the differential roles of Rac, RhoA, and Cdc42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tranquillo RT, Lauffenburger DA, Zigmond SH. A stochastic model for leukocyte random motility and chemotaxis based on receptor binding fluctuations. J Cell Biol. 1988;106:303–309. doi: 10.1083/jcb.106.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Gruver JS, Potdar AA, Jeon J, Sai J, Anderson B, Webb D, Richmond A, Quaranta V, Cummings PT, Chung CY. Bimodal analysis reveals a general scaling law governing nondirected and chemotactic cell motility. Biophys J. 2010;99:367–376. doi: 10.1016/j.bpj.2010.03.073. The motility metric derived in this study overcomes a significant limitation in the classically used persistent random walk model by providing a way to describe abrupt direction changes during migration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunn GA. Characterising a kinesis response: time averaged measures of cell speed and directional persistence. Agents Actions Suppl. 1983;12:14–33. doi: 10.1007/978-3-0348-9352-7_1. [DOI] [PubMed] [Google Scholar]
- 74.Kumar N, Zaman MH, Kim H-D, Lauffenburger DA. A high-throughput migration assay reveals HER2-mediated cell migration arising from increased directional persistence. Biophys J. 2006;91:L32–34. doi: 10.1529/biophysj.106.088898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haessler U, Kalinin Y, Swartz MA, Wu M. An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies. Biomed Microdevices. 2009;11:827–835. doi: 10.1007/s10544-009-9299-3. [DOI] [PubMed] [Google Scholar]