Abstract
Although mechanical stress is known to profoundly influence the composition and structure of the extracellular matrix (ECM), the mechanisms by which this regulation occurs remain poorly understood. We used a single-molecule magnetic tweezers assay to study the effect of force on collagen proteolysis by matrix metalloproteinase-1 (MMP-1). Here we show that the application of ~10 pN in extensional force causes a ~100-fold increase in proteolysis rates. Our results support a mechanistic model in which the collagen triple helix unwinds prior to proteolysis. The data and resulting model predict that biologically relevant forces may increase localized ECM proteolysis, suggesting a possible role for mechanical force in the regulation of ECM remodeling.
Mechanical stress is known to influence ECM remodeling during embryonic development1-4, aneurysm formation5, atherosclerosis6, and cancer metastasis7. However, the molecular pathways by which this regulation occurs remain poorly understood. ECM proteolytic degradation by matrix metalloproteinases (MMPs) is likewise important both during embryonic development8-10 and in the progression of a variety of diseases, notably cancer metastasis11. Prior crystallographic12, bulk enzymological13-17 and atomic force microscopy studies18 suggest that the collagen triple helix must be disrupted in order for MMP-catalyzed proteolysis to occur. These observations led us to investigate the possibility that mechanical load might directly modulate the rate at which MMPs cleave trimeric collagen.
The crystal structure of MMP-1 shows that its active site is too small to accommodate the collagen triple helix, implying that the collagen trimer must undergo a large conformational change during proteolysis12,13,19. The mechanism by which MMPs likely disrupt their substrates remains unclear. The “unwinding” description prevalent in the literature13 has recently been challenged by an alternative model in which MMPs capture spontaneously formed loops prior to proteolysis20. Experiments done on excised whole tissues21-27 or on reconstituted collagen gels28-31 yield conflicting results as to whether load speeds up21,22 or slows down23-27,29-31 proteolysis. A quantitative, single-molecule assay performed on a homogeneous substrate provides a logical means of reconciling these results.
We used a model collagen trimer (Fig 1a) and a high-throughput, single-molecule magnetic tweezers assay to study the effect of force proteolysis on single collagen model trimers (Fig 1b). We chose the cleavage of collagen I by MMP-1 (collagenase I) for our experiments because this is arguably the canonical combination of MMP and substrate. By sampling multiple fields of view, we achieve good experimental statistics (100s – 1000s of molecules per experiment). Matrix assisted laser desorption/ionization mass spectroscopy (MALDI-MS) and native polyacrylamide gel electrophoresis (PAGE) confirm the mass of collagen monomers (14,398 Da) and oligomerization (data not shown), respectively. MALDI-MS confirms that MMP-1 cleaves the model peptide at the recognition site (Supp Info). The concentrations of anti-myc surface attachment antibody, collagen, and magnetic beads were used such that a large majority of beads were attached to the coverslip via single attachments (Table S1).
Figure 1.
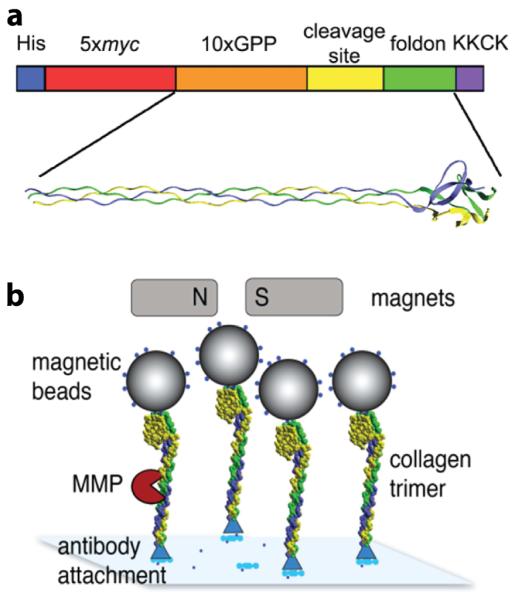
(a) Collagen model construct. The construct consists of a N-terminal 6xHis-tag for purification, followed by a 5x myc tag, (GPP)10 to enforce triple helix formation, the collagen α1 residues 772-786 (GPQGIAGQRGVVGL), which form the MMP-1 recognition site, the trimeric foldon sequence, and a C-terminal KKCK to facilitate biotinylation. (b) Single molecule force/proteolysis assay (not to scale). The magnetic tweezers generate load by pulling on the magnetic beads. MMP cuts collagen, causing bead detachment.
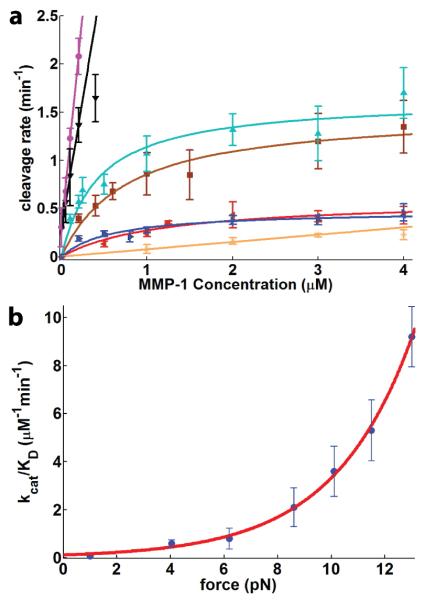
Proteolysis of a tethering collagen trimer results in bead detachment from the coverslip. We measured bead detachment as a function of time and MMP-1 concentration (Fig 2). The observed bead detachment kinetics are well-fit by a single exponential plus a constant: f(t) = ae−kt +c, where f(t) is the fraction of beads still attached at time t, k is the detachment rate, and c likely reflects non-specifically attached beads. The observation of a single detachment rate k is consistent with a single, rate-limiting step in trimer proteolysis. Bead detachment kinetics at a constant force and varying MMP-1 concentration are well-described by a hyperbolic fit: (Fig 3a)
| (Eqn. 1) |
Figure 2.
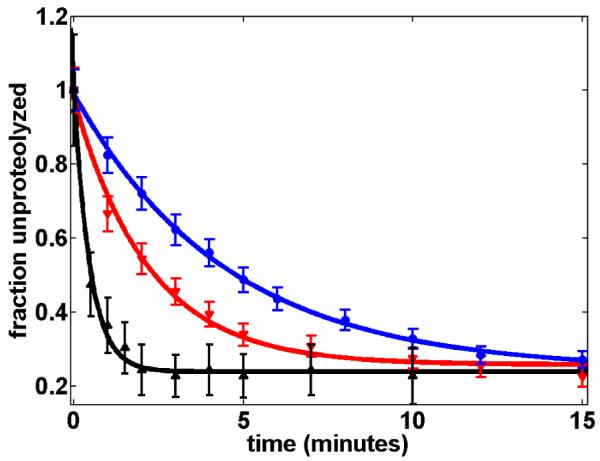
Fraction of beads attached to cover slips at 1.0 pN (3 μM MMP-1; blue), 6.2 pN (3 μM MMP-1; red) and 13 pN (0.2 μM MMP-1; black). Detachment rates are 0.22 ± 0.02 min−1 (1 pN) and 0.46 ± 0.09 min−1 (6.2 pN) and 2.08 ±0.18 min−1 (13 pN).
Figure 3.
(a) Kinetics of collagen cleavage by MMP-1 (purple = 13.0 pN, black = 11.5 pN, cyan = 10.1 pN, brown = 8.6 pN, red = 6.2 pN, blue = 4.0 pN, orange = 1.0 pN). Data recorded at 10.1, 8.6, 6.2 and 4.0 pN were fit to Eqn 1. The slope of the linear regime was used to calculate kcat/KD for data recorded at 13 pN, 11.5 pN and 1 pN. The error bars are one standard deviation, calculated using bootstrap analysis32. (b) The apparent bimolecular rate constant kcat/KD for collagen proteolysis by MMP-1 increases exponentially with force (see text).
Here k is the proteolysis rate, kcat is the maximal turnover rate (min−1), [MMP] is the MMP-1 concentration, and KD is an effective dissociation constant for MMP-1. Although the mechanism of collagen trimer cleavage is likely more complex (Supp Info), a simple kinetic framework is consistent with our data:
where M is MMP, C is collagen, MC is the uncut collagen-MMP complex, P is the cleaved collagen product, KD = k−1/k1, and the cleavage rate kcat is a function of force (F).
A plot of kcat/KD vs. force is well-fit by a single exponential, suggesting that a single force-sensitive step dominates the observed kinetics (Fig 3b). Although we do not rule out force dependence for KD, our present data are adequately described with a single force-dependent kcat (Supp Info):
| (Eqn. 2) |
Here F is the applied load, D is the change in length of the collagen upon stretching (Supp Info) and kBT is the thermal energy (4.2 pN nm). The ratio kcat/KD gives an apparent bimolecular rate constant at limiting MMP-1 concentration.
We observe an 81±3-fold increase (error calculated using the error in D) in kcat/KD at 13 pN of load. A fit to the above equation yields an extrapolated kcat(F=0)/KD of 0.11±0.10 μM−1min−1, similar to the value reported in bulk measurements (0.3 μM−1 min−1)33, and D = 1.42 ± 0.25 nm. We note that the rise per amino acid is 0.29 nm in trimeric collagen model peptides34, the contour length per amino acid is 0.4 nm in unfolded proteins35, and the MMP-1 recognition site is 14 residues long. Together, these observations predict an increase in length of 1.5 nm if the MMP-1 recognition site unwinds and stretches to its full extent, a figure that is in excellent agreement with our measured D.
We propose a model in which a 1.4 nm increase in length, or “stretch,” precedes proteolysis (Fig 4). Our present data are also consistent with comparable MMP-1 affinities for the relaxed and stretched trimer conformations. Finally, a model in which the collagen trimer is cleaved in a single processive encounter most easily explains the single-exponential bead detachment kinetics that we observe under all the conditions assayed. Together, these observations support the model shown in Fig 4, in which MMP-1 cleaves a transient, stretched collagen conformation during one processive encounter. Mechanical force stabilizes the stretched intermediate, accounting for the exponential increase in proteolysis rates with applied load. Our model is consistent bulk enzymological studies that were also interpreted to support the idea that a structural transition within the trimer is the rate-limiting step in proteolysis15,16,33.
Figure 4.
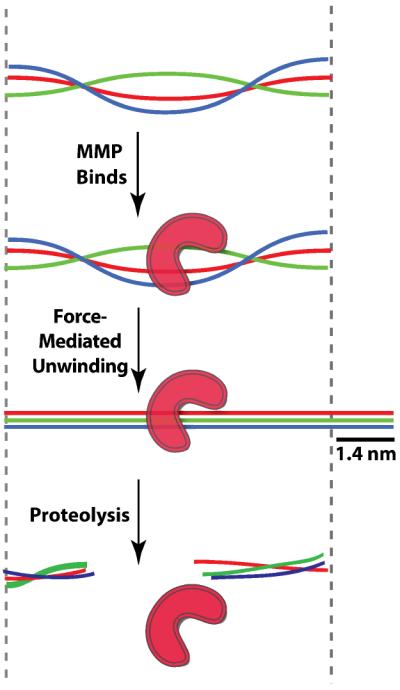
Proposed mechanism of collagen proteolysis. Applied load stabilizes a stretched, proteolytically accessible collagen conformation.
Several bulk studies show modest, ~two-fold decreases in proteolysis rates with mechanical load23-27,29-31. These studies are arguably more difficult to interpret owing to the greater structural and molecular complexity of the samples. Despite this proviso, apparent differences with our results plausibly stem from the structural differences between isolated collagen trimers and collagen fibrils, which contain hundreds of trimers36. For example, triple helix unwinding is likely facile in our experimental geometry, but may well be more constrained within the intact fibrils present in most bulk measurements. Tensile load on the fibrils may further constrain helix unwinding, thus leading to decreased proteolysis rates. This picture is consistent with a report in which slower proteolysis in excised corneal tissue under applied load was argued to correlate with a transition in mechanical properties from entropic to energetic elasticity24. It is interesting to speculate that mechanical stress may thus protect load-bearing fibrils from digestion while simultaneously hastening the degradation of isolated trimers. Such a mechanism would facilitate ECM remodeling without compromising tissue mechanical integrity.
The rapid increase proteolysis rates with load that we observe may have direct biological relevance. Individual ECM proteins likely experience loads comparable or greater to the 13 pN used in the present study: cells exert forces up to 10 nN per focal adhesion37, individual integrin-ECM protein interactions range from 20 to 100 pN in strength38, and fibronectin partially unfolds in response to cellular traction forces39. Both cellular force production and MMPs appear to be essential for tumor cell motility in three dimensions40,41, and cell motility and MMP activity are coordinated at the transcriptional level42. Recent studies likewise show that the proteolysis of von Willebrand factor is also force sensitive over biologically relevant force ranges43. A direct linkage between micro-scale mechanical forces and local ECM remodeling could thus have important consequences in cell, developmental and cancer biology.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Burroughs Wellcome Career Award at the Scientific Interface (ARD), the National Institutes of Health through the NIH Director’s New Innovator Award Program 1-DP2-OD007078 (ARD), the William Bowes Jr. Stanford Graduate Fellowship (ASA), and the Stanford Cardiovascular Institute Younger Predoctoral Fellowship (JC). The authors thank Diego Ramallo, Zev Bryant, K. C. Huang, and Miriam Goodman for their insightful advice during manuscript preparation and James Spudich for the loan of microscopy equipment.
Footnotes
SUPPORTING INFORMATION Detailed information on materials and methods used. Single molecule force proteolysis, magnetic tweezers calibration and detailed kinetic models.
REFERENCES
- (1).Wozniak MA, Chen CS. Nat Rev Mol Cell Biol. 2009;10:34. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Nat Cell Biol. 2008;10:429. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- (3).Latimer A, Jessen JR. Matrix Biol. 2010;29:89. doi: 10.1016/j.matbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- (4).Sherwood DR. Trends Cell Biol. 2006;16:250. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- (5).Xiong W, Knispel R, MacTaggart J, Greiner TC, Weiss SJ, Baxter BT. J Biol Chem. 2009;284:1765. doi: 10.1074/jbc.M806239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hahn C, Schwartz MA. Nat Rev Mol Cell Biol. 2009;10:53. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ingber DE. Semin Cancer Biol. 2008;18:356. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Glasheen BM, Kabra AT, Page-McCaw A. Proc Natl Acad Sci U S A. 2009;106:2659. doi: 10.1073/pnas.0804171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Guha A, Lin L, Kornberg TB. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tomlinson ML, Guan P, Morris RJ, Fidock MD, Rejzek M, Garcia-Morales C, Field RA, Wheeler GN. Chem Biol. 2009;16:93. doi: 10.1016/j.chembiol.2008.12.005. [DOI] [PubMed] [Google Scholar]
- (11).Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lloyd LF, Skarzynski T, Wonacott AJ, Cawston TE, Clark IM, Mannix CJ, Harper GP. J Mol Biol. 1989;210:237. doi: 10.1016/0022-2836(89)90304-5. [DOI] [PubMed] [Google Scholar]
- (13).Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. EMBO J. 2004;23:3020. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tam EM, Moore TR, Butler GS, Overall CM. J Biol Chem. 2004;279:43336. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- (15).Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. J Biol Chem. 2006;281:38302. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
- (16).Minond D, Lauer-Fields JL, Nagase H, Fields GB. Biochemistry. 2004;43:11474. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- (17).Bhaskaran R, Palmier MO, Lauer-Fields JL, Fields GB, Van Doren SR. Journal of Biological Chemistry. 2008;283:21779. doi: 10.1074/jbc.M709966200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Rosenblum G, Van den Steen PE, Cohen SR, Bitler A, Brand DD, Opdenakker G, Sagi I. PLoS One. 2010;5:e11043. doi: 10.1371/journal.pone.0011043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Iyer S, Visse R, Nagase H, Acharya KR. J Mol Biol. 2006;362:78. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nerenberg PS, Salsas-Escat R, Stultz CM. Proteins. 2008;70:1154. doi: 10.1002/prot.21687. [DOI] [PubMed] [Google Scholar]
- (21).Ellsmere JC, Khanna RA, Lee JM. Biomaterials. 1999;20:1143. doi: 10.1016/s0142-9612(99)00013-7. [DOI] [PubMed] [Google Scholar]
- (22).Jesudason R, Sato S, Parameswaran H, Araujo AD, Majumdar A, Allen PG, Bartolak-Suki E, Suki B. Biophys J. 2010;99:3076. doi: 10.1016/j.bpj.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Willett TL, Labow RS, Avery NC, Lee JM. Ann Biomed Eng. 2007;35:1961. doi: 10.1007/s10439-007-9375-x. [DOI] [PubMed] [Google Scholar]
- (24).Zareian R, Church KP, Saeidi N, Flynn BP, Beale JW, Ruberti JW. Langmuir. 2010;26:9917. doi: 10.1021/la100384e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nabeshima Y, Grood ES, Sakurai A, Herman JH. J Orthop Res. 1996;14:123. doi: 10.1002/jor.1100140120. [DOI] [PubMed] [Google Scholar]
- (26).Lotz JC, Hadi T, Bratton C, Reiser KM, Hsieh AH. Eur Spine J. 2008;17:1149. doi: 10.1007/s00586-008-0721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wyatt KE, Bourne JW, Torzilli PA. J Biomech Eng. 2009;131:051004. doi: 10.1115/1.3078177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Huang C, Yannas IV. J Biomed Mater Res. 1977;11:137. doi: 10.1002/jbm.820110113. [DOI] [PubMed] [Google Scholar]
- (29).Ruberti JW, Hallab NJ. Biochem Biophys Res Commun. 2005;336:483. doi: 10.1016/j.bbrc.2005.08.128. [DOI] [PubMed] [Google Scholar]
- (30).Flynn BP, Bhole AP, Saeidi N, Liles M, Dimarzio CA, Ruberti JW. PLoS One. 2010;5(8):e12337. doi: 10.1371/journal.pone.0012337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bhole AP, Flynn BP, Liles M, Saeidi N, Dimarzio CA, Ruberti JW. Philos Transact A Math Phys Eng Sci. 2009;367:3339. doi: 10.1098/rsta.2009.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Efron B, Tibshirani R. An introduction to the bootstrap. Chapman & Hall; New York: 1993. [Google Scholar]
- (33).Han S, Makareeva E, Kuznetsova NV, Deridder AM, Sutter MB, Losert W, Phillips CL, Visse R, Nagase H, Leikin S. J Biol Chem. 2010;285:22276. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Stetefeld J, Frank S, Jenny M, Schulthess T, Kammerer RA, Boudko S, Landwehr R, Okuyama K, Engel J. Structure. 2003;11:339. doi: 10.1016/s0969-2126(03)00025-x. [DOI] [PubMed] [Google Scholar]
- (35).Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, Fernandez JM. Biophys J. 2007;92:225. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bozec L, van der Heijden G, Horton M. Biophys J. 2007;92:70. doi: 10.1529/biophysj.106.085704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Stricker J, Sabass B, Schwarz US, Gardel ML. J Phys Condens Matter. 2010;22:194104. doi: 10.1088/0953-8984/22/19/194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li F, Redick SD, Erickson HP, Moy VT. Biophys J. 2003;84:1252. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. PLoS. Biol. 2007;5:2243. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Biophys J. 2008;95:4077. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, Allen E, Chung D, Weiss SJ. J Cell Biol. 2004;167:769. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ota I, Li XY, Hu Y, Weiss SJ. Proc Natl Acad Sci U S A. 2009;106:20318. doi: 10.1073/pnas.0910962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Science. 2009;324:1330. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.