Abstract
The processes involved in the development of complex multicellular communities, including the programmed elimination of individual cells during the formation of specialized structures, exhibits fundamental similarities between prokaryotic and eukaryotic organisms. Mechanistic similarities may also exist at the molecular level, as bacterial proteins hypothesized to be related to the apoptosis regulator Bax/Bcl-2 family, have been identified, fueling speculation about the existence of bacterial PCD. Here we review the regulatory networks controlling cell death and lysis in Staphylococcus aureus and examine the environmental parameters that might influence them during the development of a biofilm. We hypothesize that the heterogeneous environmental conditions found within a developing biofilm generate distinct physiological signals that coordinate the differential expression of cell death and lysis effectors.
Introduction
Bacterial programmed cell death (PCD) has been hypothesized to be involved in sporulation, genetic exchange, elimination of damaged and defective cells, control of viral infection, and limiting the rate of spontaneous mutations that have the potential to take over the population [1,2••,3]. PCD has also been proposed to function in many respects like it does in more complex organisms, in the development of multicellular structures, for example, in fruiting body formation by Myxococcus sp. [4–6]. Similarly, cell death and lysis within a biofilm population is well documented [7–12•], in many cases effecting biofilm architecture and leading to the release of cytoplasmic contents including genomic DNA [13,14]. This extracellular DNA (eDNA) can contribute to antibiotic resistance [15], promote horizontal gene transfer [16] and, by virtue of its adhesive properties, play an essential role in the biofilm matrix as an adherence molecule [14,17,18•,19].
The Staphylococcus aureus Cid/Lrg system
In Staphylococcus aureus, our understanding of molecular mechanisms controlling death and lysis during biofilm development is centered largely on the cidABC and lrgAB operons [12,14]. It was proposed that these operons may have an important role in the control of staphylococcal murein hydrolase activity and encode proteins analogous to the bacteriophage-encoded holins and antiholins [20,21]. Holins and antiholins are members of a large family of membrane proteins that are directly involved in regulation of bacteriophage-induced death and lysis. Specifically, they control the activity of bacteriophage-encoded murein hydrolases (i.e. endolysins) and the timing of lysis during bacteriophage infection by regulating the access of these enzymes to their substrate, peptidoglycan [22••]. This regulation can be achieved by one of two proposed mechanisms: through control of murein hydrolase transport across the cytoplasmic membrane or by mediating the release and activation of membrane-associated murein hydrolases [23–26]. Antiholins are homologous proteins (in fact, through a “dual start motif” they are commonly encoded in bacteriophage by the same gene encoding the holin) that have an inhibitory effect on holin activity [27]. Similarly opposing activities in controlling cell lysis and genomic DNA release during biofilm development have also been observed for the cid and lrg gene products, suggesting that cid encodes a holin and lrg encodes an antiholin [28] (see Figure 1). Additional support for holin-like properties of cid and lrg gene products was recently obtained by biochemical and molecular characterization of CidA and LrgA proteins that demonstrated the oligomerization and association of both CidA and LrgA with the cytoplasmic membrane [29]. Importantly, mutations in either the cid or lrg operon result in altered biofilm formation [12,14] suggesting that a careful balance between life and death is critical for biofilm development, not unlike development in more complex eukaryotic systems [30]. Given the similarities of holins and antiholins to the Bax/Bcl-2 family of proteins, it has been postulated that these seemingly unlinked proteins are also functionally related [30,31]. Support for this idea has recently come from the demonstration that Bax could functionally complement a holin deficiency in bacteriophage lambda [32]. Furthermore, the role of a plastid-associated “LrgB family” protein in plant senescence was recently described [33••] suggesting a broader role for these proteins than what was previously appreciated.
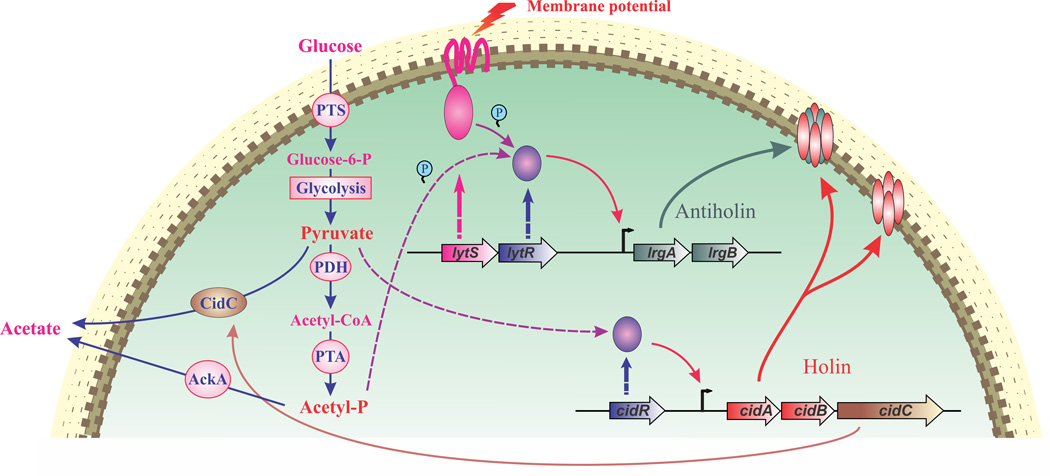
Figure 1. The Cid/Lrg regulatory network.
The products of cidABC and lrgAB operons involved in the control of cell death and lysis in Staphylococcus aureus. The cidA and lrgA genes encode homologous hydrophobic proteins that are hypothesized to function as holins and antiholins, respectively. The cidB and lrgB genes encode homologous hydrophobic proteins with unknown functions. The cidC gene encodes pyruvate oxidase that decarboxylates pyruvate to acetate. The LytSR two-component regulatory system senses decreases in membrane potential and responds by inducing lrgAB transcription. The intermediate product of overflow metabolism (shown on the left side) acetyl phosphate (Acetyl-P) is proposed to activate LytR independent of LytS. The LysR-type transcription regulator, CidR, induces cidABC transcription during overflow metabolism in response to the accumulation of intracellular pyruvate and/or acetate. Enzymes presented in this scheme that are involved in overflow metabolism in S. auereus: PTS, phosphoenolpyruvate-dependent phosphotransferase system; PDH, pyruvate dehydrogenase complex; PTA, phosphotransacetylase; AckA, acetate kinase.
Regulation of the Staphylococcus aureus Cid/Lrg system
The presence of a system controlling bacterial cell fate is almost certainly going to be associated with a complex regulatory mechanism. Indeed, there are two regulatory networks controlling cell death and lysis mediated by the action of Cid/Lrg system including the LytSR two-component regulatory system and the LysR-type transcriptional regulator known as CidR.
The LytSR two-component regulatory system
Our studies of the S. aureus cid and lrg operons were initiated over a decade ago, with the identification of the LytSR two-component regulatory system that affects murein hydrolase activity and autolysis [34]. It has been shown that disruption of the lytSR genes causes increased autolysis and altered levels of murein hydrolase activity, suggesting that the LytS and LytR proteins are involved in controlling autolysis in S. aureus by affecting the intrinsic murein hydrolase activity associated with the cell [34]. The LytSR system consists of a sensor histidine protein kinase (LytS) and a response regulator (LytR) and has been shown to function, in part, by inducing lrgAB expression [35]. Current evidence suggests that LytS senses some aspect of membrane potential, since agents that dissipate this component of the proton motive force induce lrgAB expression in a lytSR-dependent manner [36]. It has also been shown that lrgAB expression is strongly induced in a lytSR-dependent manner during overflow metabolism (i.e. under conditions of excess of glucose and oxygen) and coincides with accumulation of acetate in the media [37]. Interestingly, recent studies demonstrate that under conditions of overflow metabolism, LytR alone can complement lytSR inactivation and induce lrgAB transcription (Sharma-Kuinkel and Bayles; unpublished results). These observations could be explained by a model in which a small phosphodonor molecule, such as acetyl phosphate, directly phosphorylates LytR independent of LytS (Figure 1). This is in agreement with the findings that many other well-characterized response regulators can be activated by small phosphodonor molecules such as acetyl phosphate or carbamoyl phosphate [38–41]. Additional support for this model comes from the fact that during aerobic growth of S. aureus, glucose is predominantly catabolized through the Pta-AckA pathway, of which acetyl phosphate is an intermediate [42•]. Thus, the ability of LytR alone to induce lrgAB transcription during overflow metabolism may be a consequence of increased acetyl phosphate pools and the direct phosphorylation of LytR by this molecule.
The CidR regulator
The analysis of the S. aureus genome sequence revealed an open reading frame located immediately upstream of cidABC locus, designated cidR, that encodes a putative LysR-type transcriptional regulator (LTTR) [43]. Similar to other members of the LTTR family, the CidR amino acid sequence contains a conserved N-terminal helix-turn-helix motif responsible for DNA binding [44] and a putative C-terminal regulatory domain that is involved in binding of a small co-inducer molecule [44–46]. Inactivation of the cidR gene has a negative effect on cidABC transcription and results in reduced murein hydrolase activity and survival during stationary phase [43]. Northern blot analysis of cid transcription revealed the presence of two overlapping transcripts designated cidABC and cidBC. Expression of the smaller transcript is dependent on sigma factor B [47], while the full-length cidABC transcript is induced by growth in the presence of excess glucose and/or acetic acid and is CidR dependent [37,43]. Furthermore, comparison of the transcriptional profiles of the cidR mutant with the parental strain revealed that only two operons, alsSD and cidABC, were upregulated by CidR [48]. The products of the alsS and alsD genes are acetolactate synthase and acetolactate decarboxylase, respectively, and like the cidC-encoded pyruvate oxidase, these proteins are involved in catabolism of pyruvate formed under excess glucose conditions. However unlike pyruvate oxidase that decarboxylates pyruvate to acetate [49], these enzymes belong to the 2,3-butanediol pathway, converting pyruvate to the neutral byproduct, acetoin [48,50]. Since members of the LTTR family typically require a small co-inducer molecule for activation of their target genes [44], we speculate that pyruvate or an intermediate of pyruvate metabolism might serve as this molecule (Figure 1).
Control of Cid/Lrg expression during biofilm development
Given the nature of the signals known to induce expression of the Cid/Lrg system, we propose that control of cid and lrg expression depends upon the varied environmental conditions that exist within different micro-niches of a biofilm [51•,51,53•,54,55••,56]. But how might the biofilm micro-environment affect cid and lrg expression? With respect to cid expression, induction in planktonic cultures was observed under excess glucose [37,43], where the high rate of glycolysis inhibits aerobic respiration and induces carbon flow through fermentation pathways, a phenomenon known as the Crabtree effect [57••]. Thus, we considered the possibility that the signal sensed by CidR may be related to fermentative metabolism, which would occur most commonly under hypoxic conditions. Indeed, cell death and lysis are commonly observed in the interior of large biofilm structures, where oxygen levels would presumably be limiting [13,14]. In support of this notion, growth of S. aureus under hypoxic conditions resulted in the CidR-dependent induction of cid expression (manuscript in preparation). Furthermore, real-time microsensor measurements of local metabolic activities in ex vivo dental biofilms show strong acidification of the anoxic layer of biofilm in response to the addition of sucrose [54]. The organic acids responsible for this acidification may also provide signals relevant to LytSR-mediated control of lrgAB transcription, as it is well known that weak acids can serve as effective protonophores capable of transporting hydrogen ions through the cytoplasmic membrane barrier and releasing them into the cytoplasm, thus, dissipating the membrane potential [58••,59,60].
Intriguingly, as was mentioned above, CidR function is not limited to the regulation of the cidA and cidB genes. It also includes regulation of genes involved in diverse pathways of pyruvate catabolism, leading to the generation of acetate and acetoin [48–50]. Hence it is plausible that disruption of the delicate balance in expression of these metabolic genes might also be involved in the control of cell death and lysis [31]. Additionally, DNA microarray analyses revealed that LytSR TCS system in both S. aureus and S. epidermidis regulates expression of a wide variety of genes involved in major cellular processes like energy, carbohydrate, and nucleotide metabolism [35,61]. These results suggest that LytSR may also affect cell viability and adaptation to an altered environment, not only by preventing cell lysis through up regulation of the synthesis of antiholin molecules, but also by regulating expression of genes controlling the metabolic state of the bacteria.
Concluding remarks
Although we are making progress in understanding the molecular signals involved in the regulation of the genes encoding the Cid/Lrg regulatory system, the mechanisms dictating the differential control of cell death and lysis within a developing biofilm remain poorly understood. Clearly, by virtue of the functional nature of these genes, this cannot be an “all or nothing” phenomenon. Therefore, what processes determine which cells will live and replicate within a biofilm, and which cells will die and lyse, remains an important unanswered question. Although studies of the function and regulation of the S. aureus Cid and Lrg proteins have provided a unique perspective on cell death and lysis during biofilm development, we are only at the early stages of appreciating the impact of the biofilm environment on these processes. A more complete understanding of the consequences of these physiological changes and how they contribute to the coordination of metabolic signals leading to cell death and lysis will undoubtedly provide new perspectives on biofilm development.
Highlights.
> We discuss the roles of the Staphylococcus aureus cid and lrg operons in death and lysis during biofilm development. > We describe the function of the S. aureus LytSR and CidR regulatory proteins in the control of cid and lrg expression. > We hypothesize that the metabolic heterogeneity observed within the biofilm environment determines the differential expression of cid and lrg, which in turn, dictates which cells die and lyse.
Acknowledgments
We thank Vinai Chittezham Thomas and Kari Nelson for their useful comments and suggestions. This research was supported by grants P01-AI083211 and R01-A1038901 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Doyle RJ, Koch AL. The functions of autolysins in the growth and division of Bacillus subtilis. Crit Rev Microbiol. 1987;15:169–222. doi: 10.3109/10408418709104457. [DOI] [PubMed] [Google Scholar]
- 2. Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. This review is one of the first to introduce the concept of bacterial PCD and its potential role in developmental processes and biofilm formation.
- 3.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbluh A, Rosenberg E. Role of autocide AMI in development of Myxococcus xanthus. J Bacteriol. 1990;172:4307–4314. doi: 10.1128/jb.172.8.4307-4314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller C, Dworkin M. Effects of glucosamine on lysis, glycerol formation, and sporulation in Myxococcus xanthus. J Bacteriol. 1991;173:7164–7175. doi: 10.1128/jb.173.22.7164-7175.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Hall-Stoodley L, Stoodley P. Developmental regulation of microbial biofilms. Curr Opin Biotechnol. 2002;13:228–233. doi: 10.1016/s0958-1669(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 9.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry JA, Cvitkovitch DG, Levesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mai-Prochnow A, Evans F, Dalisay-Saludes D, Stelzer S, Egan S, James S, Webb JS, Kjelleberg S. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol. 2004;70:3232–3238. doi: 10.1128/AEM.70.6.3232-3238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. First description of the role of cell lysis and DNA release in S. aureus biofilm development
- 13.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother. 2009;53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 17.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 18. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. First experimental evidence for the importance of extracellular DNA in Pseudomonas aeruginosa biofilm development: inhibition of biofilm formation and destabilization of an established biofilm by DNase I treatment.
- 19.Vilain S, Pretorius JM, Theron J, Brozel VS. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol. 2009;75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice KC, Firek BA, Nelson JB, Yang SJ, Patton TG, Bayles KW. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. This review summarizes advances in the understanding of the physical and biochemical properties, function, and regulation of holins.
- 23.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci U S A. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu M, Struck DK, Deaton J, Wang IN, Young R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci U S A. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 27.Blasi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 28.Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranjit DK, Endres JL, Bayles KW. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol. 2011;193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 32.Pang X, Moussa SH, Targy NM, Bose JL, George NM, Gries C, Lopez H, Zhang L, Bayles KW, Young R, Luo X. Active Bax and Bak are functional holins. Genes Dev. 2011;25:2278–2290. doi: 10.1101/gad.171645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang Y, Jin H, Chen Y, Lin W, Wang C, Chen Z, Han N, Bian H, Zhu M, Wang J. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2011 doi: 10.1111/j.1469-8137.2011.03867.x. This study demonstrates that a member of the Cid/Lrg protein family is important in chloroplast development, carbon partitioning and leaf senescence.
- 34.Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol. 2009;191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton TG, Yang SJ, Bayles KW. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol Microbiol. 2006;59:1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x. [DOI] [PubMed] [Google Scholar]
- 37.Rice KC, Nelson JB, Patton TG, Yang SJ, Bayles KW. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J Bacteriol. 2005;187:813–821. doi: 10.1128/JB.187.3.813-821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCleary WR, Stock JB, Ninfa AJ. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 41.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun. 2003;71:4724–4732. doi: 10.1128/IAI.71.8.4724-4732.2003. In this work, it was demonstrated that carbohydrate metabolism has an important role the growth characteristics and virulence factor production of S. aureus.
- 43.Yang SJ, Rice KC, Brown RJ, Patton TG, Liou LE, Park YH, Bayles KW. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J Bacteriol. 2005;187:5893–5900. doi: 10.1128/JB.187.17.5893-5900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 45.Henikoff S, Haughn GW, Calvo JM, Wallace JC. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol. 2003;328:555–566. doi: 10.1016/s0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 47.Rice KC, Patton T, Yang SJ, Dumoulin A, Bischoff M, Bayles KW. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J Bacteriol. 2004;186:3029–3037. doi: 10.1128/JB.186.10.3029-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang SJ, Dunman PM, Projan SJ, Bayles KW. Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol Microbiol. 2006;60:458–468. doi: 10.1111/j.1365-2958.2006.05105.x. [DOI] [PubMed] [Google Scholar]
- 49.Patton TG, Rice KC, Foster MK, Bayles KW. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol Microbiol. 2005;56:1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 50.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Beer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. Relation between the arrangement of structural components of biofilm, oxygen distribution and mass transfer.
- 52.Sternberg C, Christensen BB, Johansen T, Toftgaard Nielsen A, Andersen JB, Givskov M, Molin S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. This study investigated the role of oxygen availability in determining the spatial physiological activity of P. aeruginosa growing in biofilms.
- 54.von Ohle C, Gieseke A, Nistico L, Decker EM, DeBeer D, Stoodley P. Real-time microsensor measurement of local metabolic activities in ex vivo dental biofilms exposed to sucrose and treated with chlorhexidine. Appl Environ Microbiol. 2010;76:2326–2334. doi: 10.1128/AEM.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189:4223–4233. doi: 10.1128/JB.00107-07. In this work, spatial patterns of DNA replication and protein synthetic activity in staphylococcal biofilms were imaged and quantified. The results of this work suggest that staphylococcal biofilms contain cells in at least four distinct states: growing aerobically, growing fermentatively, dead, and dormant.
- 56.Thomas VC, Hancock LE. Suicide and fratricide in bacterial biofilms. Int J Artif Organs. 2009;32:537–544. doi: 10.1177/039139880903200902. [DOI] [PubMed] [Google Scholar]
- 57. Crabtree HG. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23:536–545. doi: 10.1042/bj0230536. The first report on glucose-mediated inhibition of cellular respiration and its relationship to tumor development.
- 58. McLaughlin SG, Dilger JP. Transport of protons across membranes by weak acids. Physiol Rev. 1980;60:825–863. doi: 10.1152/physrev.1980.60.3.825. This review summarizes the possible molecular mechanisms by which weak acids move protons across artificial bilayers.
- 59.Baronofsky JJ, Schreurs WJ, Kashket ER. Uncoupling by Acetic Acid Limits Growth of and Acetogenesis by Clostridium thermoaceticum. Appl Environ Microbiol. 1984;48:1134–1139. doi: 10.1128/aem.48.6.1134-1139.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Axe DD, Bailey JE. Transport of lactate and acetate through the energized cytoplasmic membrane of Escherichia coli. Biotechnol Bioeng. 1995;47:8–19. doi: 10.1002/bit.260470103. [DOI] [PubMed] [Google Scholar]
- 61.Zhu T, Lou Q, Wu Y, Hu J, Yu F, Qu D. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. BMC Microbiol. 2010;10:287. doi: 10.1186/1471-2180-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]