Abstract
Aggregation of Cu, Zn Superoxide Dismutase (SOD1) is often found in Amyotrophic Lateral Sclerosis (ALS) patients. The fibrillar aggregates formed by wildtype and various disease-associated mutants have recently been found to have distinct cores and morphologies. Previous computational and experimental studies of wildtype SOD1 suggest that the apo-monomer, highly aggregation-prone, displays substantial local unfolding dynamics. The residual folded structure of locally unfolded apoSOD1 corresponds to peptide segments forming the aggregation core as identified by a combination of proteolysis and mass spectroscopy. Therefore, we hypothesize that the destabilization of apoSOD1 caused by various mutations leads to distinct local unfolding dynamics. The partially unfolded structure, exposing the hydrophobic core and backbone hydrogen bond donors and acceptors, is prone to aggregate. The peptide segments in the residual folded structures form the “building block” for aggregation, which in turn determines the morphology of the aggregates. To test this hypothesis, we apply a multiscale simulation approach to study the aggregation of three typical SOD1 variants: wildtype, G37R, and I149T. Each of these SOD1 variants has distinct peptide segments forming the core structure and features different aggregate morphologies. We perform atomistic molecular dynamics simulations to study the conformational dynamics of apoSOD1 monomer, and coarse-grained molecular dynamics simulations to study the aggregation of partially unfolded SOD1 monomers. Our computational studies of monomer local unfolding and the aggregation of different SOD1 variants are consistent with experiments, supporting the hypothesis of the formation of aggregation “building blocks” via apo-monomer local unfolding as the mechanism of SOD1 fibrillar aggregation.
Keywords: SOD1 misfolding and aggregation, fibrillar aggregate, aggregation building block, molecular dynamics, multiscale modeling
Introduction
The misfolding and aggregation of Cu, Zn superoxide dismutase (SOD1) is associated with Amyotrophic Lateral Sclerosis (ALS)1; 2. More than 100 mutations in SOD1 have been identified in familial ALS patients. Both wildtype and mutant SOD1 can form insoluble fibrillar aggregates with a common cross-beta amyloid structure3; 4, as observed in many other amyloidogenic proteins with distinct primary, secondary, and tertiary structures5. Many of the research efforts in SOD1 misfolding have been focused on finding a general mechanism for how mutations promote SOD1 misfolding and aggregation, under the assumption of a common “mutation-independent” aggregation pathway and similar aggregate structures6. However, a recent study ofSOD1 aggregates formed by wildtype and various mutants revealed distinct fibrillar core compositions as well as aggregate morphologies7. Accordingly, phenotypic heterogeneity has been reported in familial ALS patients with different SOD1 mutations8. Increasing evidence suggests that the structures and morphologies of protein aggregates affect their respective disease phenotypes, and that polymorphism in protein aggregates associates with phenotypic heterogeneity9–12. Hence, uncovering the molecular mechanism governing the formation of polymorphic amyloid aggregates is important for gaining an understanding of ALS phenotypic heterogeneity.
SOD1 forms a stable dimer in solution, with each SOD1 monomer binding one copper and one zinc ion and forming one intra-monomer disulfide bond. Various biochemical and biophysical studies have suggested that wildtype SOD1 dimer is exceptionally stable because of the coordination of metal ions13. Mounting experimental and computational evidence suggests that apoSOD1 monomer is the most aggregation-prone species14–19. The loss of the coordinated metal ions destabilizes the protein with a significant population unfolded at physiological conditions14. Although apoSOD1 is native-like in the crystal structure20, the protein in solution features significant structural disorder and conformational flexibility21. Both experimental22; 23 and computational19 studies suggest that theapoSOD1 monomer features frequent local unfolding. In addition to the two long loops, the metal-coordinated (in the native state)strands 4,5, and 7feature a high level of local unfolding, and the N-terminal β-sheet is the most stable structural element (Fig. 1A). Interestingly, the same regions that are stable in the locally unfolded apoSOD1 also correspond to those regions that participate in the fibrillar core of wildtype SOD1 aggregates, having been identified as proteolysis-resistant peptides7. This observation is consistent with the generic aggregation mechanism proposed earlier24; 25, where the residual structural elements in the partially unfolded protein interact with each other and serve as “building blocks” for the formation of fibrillar amyloid aggregates. Recently, local unfolding induced by mutations has also been found to play an important role in the aggregation of gamma-crystallin in human cataracts26. Therefore, we hypothesize that the various disease-causative mutations in SOD1 have different impacts on apoSOD1 conformational dynamics, which in turn lead to distinct patterns of local unfolding and thus the varied morphologies of the resulted aggregates.
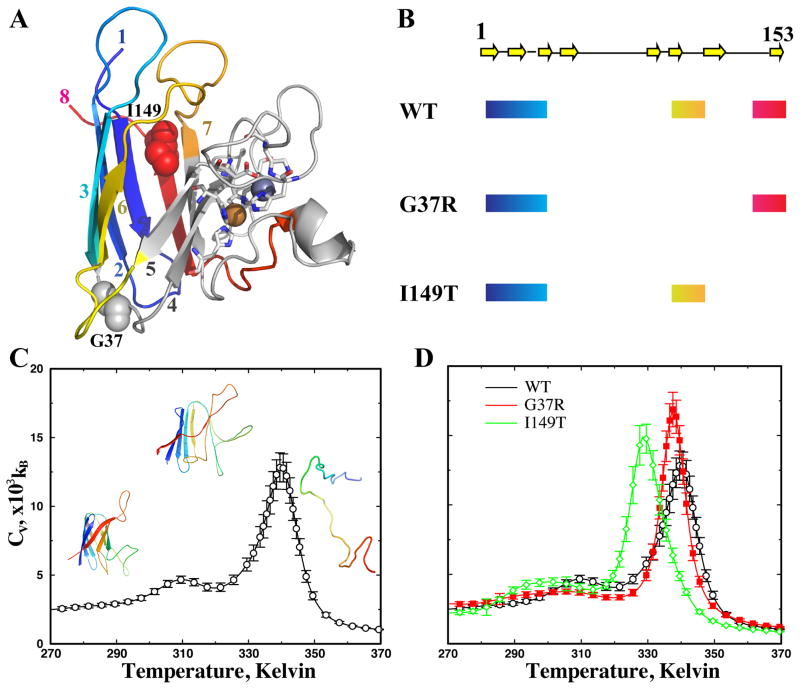
Figure 1. The thermodynamics of apoSOD1 monomer.
(A) In SOD1 monomer, the two metal ions (spheres) are coordinated by residues (sticks) in strands 4, 5, and 7. The amyloid core of wildtype SOD1 is composed of N-terminal strands 1–3 (blue to cyan), middle strand 6 (yellow), and C-terminal strand 8 (red). Other regions are colored gray. The two mutated residues in this study, G37 and I149, are shown in sphere. (B) The regions forming the amyloid core of three typical SOD1 variants: wildtype (WT), G37R, and I149T. The arrows illustrate the strands along the primary sequence. The thick lines highlight the regions found in the proteolysis-resistant amyloid core for each of the three SOD1 variants. (C) The specific heat of the wildtype apoSOD1 as a function of temperature. The specific heat values and corresponding statistical uncertainties (shown as error bars) are computed using WHAM analysis of the replica exchange simulation trajectories (Methods). At temperatures bellow the low-temperature peak, the protein is native-like with the beta-barrel intact. At temperatures higher than the high-temperature peak, the protein is unfolded, without persistent secondary and tertiary structures. At the intermediate temperature between the two peaks, the protein is partially unfolded. (D) The specific heat curves for all three variants feature similar local unfolding and global unfolding thermodynamics.
To test this hypothesis, we apply a multiscale molecular dynamics approach to study the local unfolding of SOD1 monomer and the aggregation dynamics of multiple monomers. In the previous experimental study of SOD1 aggregates, Furukawa et al. discovered that three major regions comprise the fibril aggregate cores, including the N-terminal β-sheet (strands 1,2,3), the middle strand 6, and C-terminal strand 8 (Fig. 1B). The N-terminal strands are observed in the aggregates formed by all SOD1 variants. As a result, there are only three possible combinations of core-forming peptide patterns: all three segments, the N-terminal sheet plus middle strand, and the N-terminal sheet plus C-terminal strand. Therefore, beside the wildtype SOD1 whose aggregation core is composed of all three segments, we also include two mutants, G37R and I149T, having representative aggregation core compositions7. In the core of G37R and I149T, the C-terminal strand and middle strand 6 are observed, respectively, in addition to the N-terminal strands (Fig. 1B). In order to probe the monomer conformational dynamics, we perform atomistic discrete molecular dynamics (DMD) simulations19. In the atomistic simulations, we find that different mutations indeed result in different patterns of local unfolding, and that the residual structures in the locally unfolded states are consistent with the core-forming segments, supporting the “building block” aggregation mechanism24. We further perform coarse-grained DMD simulations to study SOD1 monomer aggregation for each of the three SOD1 variants. To promote the formation of partially unfolded structures, we develop a hybrid structure-based interaction model where the interactions between the core-forming residues are enhanced. The reconstructed model structure of the amyloid aggregates is consistent with the experimentally observed morphology. Therefore, our multiscale simulations suggest a molecular mechanism of mutation-dependent SOD1 aggregation polymorphism.
Results and Discussion
We use a multiscale molecular dynamics approach to study the misfolding and aggregation of apoSOD1. We use the atomistic DMD simulations27 to sample the conformational dynamics of the apoSOD1 monomer, the time scale of which is approximately μs to ms. For monomer aggregation, which spans hours and days, we use coarse-grained DMD simulations24 with experimental constraints to enhance sampling of protein aggregation.
Conformational dynamics of apoSOD1 monomer
We perform all-atom DMD simulations27 to sample the conformational dynamics of apoSOD1 monomers. The protein is in united-atom representation, with all heavy atoms and polar hydrogen explicitly modeled. The simulation is performed with implicit solvent, and inter-atomic interactions are modeled by a physical force field adapted from Medusa28, which includes Van der Waals, solvation29, and hydrogen bonding potentials. In addition to the previous version of the all-atom DMD force field27, we also introduce screened electrostatic interactions between charged residues (Methods). Atomistic DMD simulations allow efficient sampling of the large-scale conformational dynamics of proteins and protein complexes19; 30; 31.
In order to efficiently sample the conformational dynamics of apoSOD1, we utilize replica exchange32 DMD simulations, which allows enhanced sampling of the conformational space. We allocate twelve replicas for each SOD1 variant, with each replica running at different temperatures from low to high (Methods). Periodically, replicas with neighboring temperature values exchange their simulation temperatures stochastically. A temporarily trapped state in a replica can be rescued by simulating at a higher temperature, there by enhancing the sampling efficiency of DMD simulations. For each simulation, we start from the crystal structure conformation, with metal ions and the disulfide bond removed. The structures for wildtype (PDB: 1SPD) and G37R (PDB: 1AZV) are known. The starting structure of I149T is modeled by amino acid substitution and rotamer optimization using Eris33. In the simulations, cysteine residues do not form disulfide bond mimicking a reducing condition. The total simulation time of each replica is 100 ns. Therefore, we perform a total of 1.2 μs of DMD simulations for each SOD1 variant (Methods).
ApoSOD1 features frequent local unfolding before global unfolding
Based on replica exchange simulations, we use the weighted histogram analysis method (WHAM; Methods) to compute the thermodynamics of apoSOD1 unfolding. In WHAM analysis, the density of states is self-consistently determined from overlapping energy histograms for the replica exchange simulations34. To reduce structural relaxation artifacts from the starting crystallography structure, we exclude the first quarter of each simulation trajectory from the WHAM analysis. As a simple approach to test convergence of the simulations, we divide the rest of the simulations into halves, and then compute and compare the histogram of potential energies for each of the half. We find that the histograms from both halves are close to each other, suggesting the convergence of the simulations (Fig. S1). Given the density of states, thermodynamic parameters such as specific heat and average root-mean square deviation (RMSD) from the starting crystal structure can be calculated (details in the Methods).
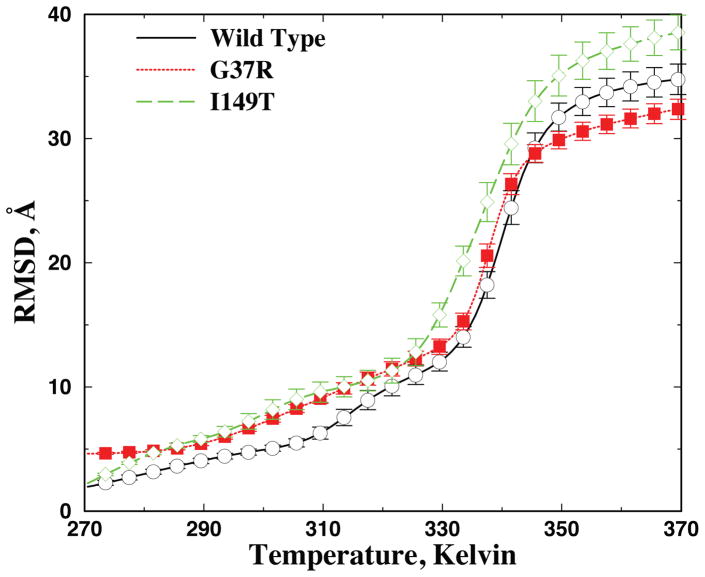
The specific heat of wildtype apoSOD1 monomer features a major peak at Tf~330 Kelvin (Fig. 1C), which corresponds to the global unfolding or melting transition of the protein. Above the transition temperature T>Tf, the protein is random coil-like with large RMSD (Fig. 2). At lower temperatures, the protein is folded or at least partially folded with lower RMSD. Interestingly, a minor peak appears at a lower temperature Tp ~ 310 Kelvin, which corresponds to a partial unfolding transition. Such a weak structural transition has also been observed in earlier computational studies, although the global unfolding transition was the main focus of these studies, using the melting temperature as the measure of thermal stability19. In an earlier work by Zhou and Karplus35, a similar transition was termed surface-molten to solid transition. The lower value of the local unfolding temperature as compared to the global unfolding temperature suggests that the apo-protein undergoes significant local unfolding before it becomes globally unfolded (Fig. 2), which is consistent with the NMR study of the wildtype apoSOD1 monomer in solution21.
Figure 2. Average RMSD as a function of temperature.
Average RMSD is computed using WHAM analysis. Based on the self-consistently determined density of states ρ(E) and the conditional probability to observe a structure with RMSD of R at a given energy E, P(R|E), the average RMSD and corresponding error bar can be computed accordingly (Methods).
We similarly compute the specific heat of the two SOD1 mutants (Fig. 1D). Both mutants have a major and a minor peak in the specific heat, similar to that of the wildtype. Because of the vast conformational space of SOD1, we do not expect DMD simulations to reach folding/unfolding equilibrium as observed in simulations of small fast-folding proteins27. For example, the melting temperature of wildtype apoSOD1 monomer in DMD simulations (~330K) is higher than that measured in experiments (~315K)36. However, starting from the crystal structure, we expect that that our μs-long replica exchange simulations would be able to sufficiently sample conformations in the folded and partially folded states (e.g., the convergence of simulations illustrated in Fig. S1 and Fig. 2), and the computed unfolding temperatures can be used as a qualitative measure of a protein’s thermostability. Compared to the wildtype, the I149T mutant has a lower melting temperature, and thus a weaker thermal stability, while G37R has a similar or slightly higher melting temperature. Interestingly, the two mutants have lower local unfolding temperatures (the low temperature peak in Fig. 1D) than that of the wildtype, suggesting that mutations enhance the local unfolding of apoSOD1.
Mutations lead to different local unfolding dynamics
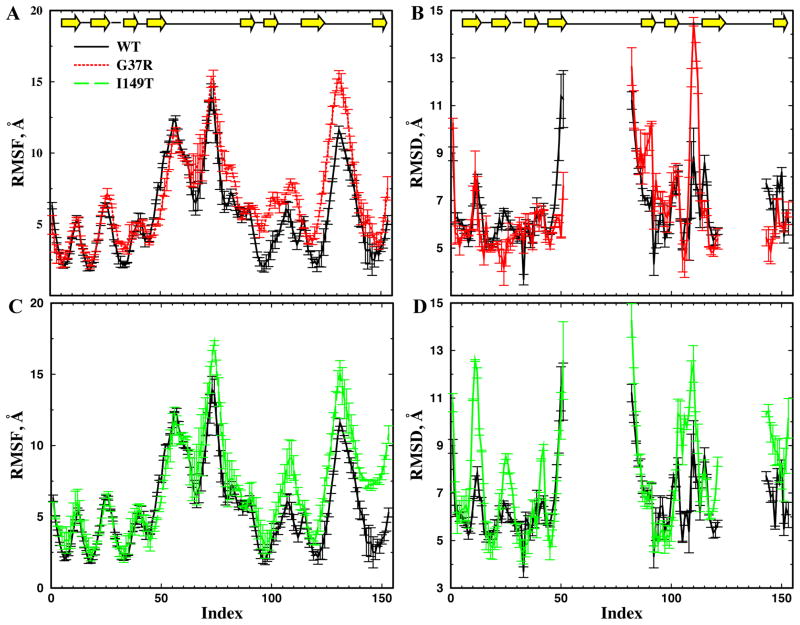
In order to characterize the conformational dynamics of different SOD1 variants in the partially unfolded states, we reconstruct the conformational ensemble from the replica exchange simulation trajectories (see Methods) based on the RMSD ranges identified from WHAM calculations (Fig. 2). Given the partially unfolded structural ensemble for each SOD1 variant, we compute the root-mean-square fluctuation (RMSF) of each residue around its average positions (Fig. 3A,C). A larger RMSF value denotes higher conformational flexibility of the residue in the locally unfolded state. We also compute the average root-mean-square deviation (RMSD) of each residue from the corresponding native structure (Fig. 3B,D). In the RMSD per residue calculation, we exclude the two major loops, residues 51–80 and 121–141, which are coordinated by metal ions in the crystal structure and are highly disordered without metals bound. For the wildtype, the three N-terminals trands, the middle strand near residue 100, and the C-terminal strand all feature low RMSD and RMSF values. These results are consistent with previous experimental22 and computational6; 19 studies of local unfolding in the apoSOD1 monomer. These same segments were also found to participate in the fibrillar core by wildtype apoSOD17.
Figure 3. The local unfolding conformational dynamics of apoSOD1 monomer.
The locally unfolded conformational ensemble for each SOD1 variant is reconstructed from the simulation trajectories (Methods). The corresponding conformational dynamic parameters, including RMSF(A,C) and RMSD (B,D) per residue are computed based on the reconstructed structural ensemble. In order to estimate the error bars, we split the simulation trajectory into halves and compute the dynamic parameters independently. To avoid overlapping lines, we compare the RMSF and RMSD between WT and G37R (A,C), and between WT and I149T (B,D), separately. The average RMSD per residue is computed with respect to the native states. Due to their large conformational flexibility, we do not include the loops in the RMSD calculation.
Compared to the wildtype, G37R displays larger RMSD and RMSF in the middle strand near residue 100, while the N- and C-terminal strands of G37R have low RMSD and RMSF, similar to that of the wildtype (Fig. 3A,B). In the I149T mutant, the C-terminal strand has larger RMSF and RMSD values than those of the wildtype. The N-terminal and middle strands of I149T feature low RMSD and RMSF as is found in the wildtype, although the RMSD of the turns in I149T is large (Fig. 3C,D). These regions with low RMSD and RMSF values in mutant SOD1 correspond to the core-forming peptides in the respective aggregates7 (Fig. 1). Therefore, there is a correlation between the computationally identified stable, native-like regions in the locally unfolded apoSOD1 monomer and the proteolysis-resistant peptide segments in the corresponding amyloid core. Taken together, these results suggest that mutations affect the local unfolding dynamics and result in different patterns of local unfolding. The peptide segments in the well-defined residual structure in the locally unfolded apoSOD1 can serve as a “building block” for fibril aggregates, which interact with each other to form the fibrillar core.
Aggregation of apoSOD1 monomers
The aggregation of SOD1 is a long time-scale process, which takes days, weeks, and months in the quiescent condition37 and hours in the agitated condition7. Modeling this process with high-resolution DMD simulations is computationally challenging. Instead, we use a coarse-grained two-bead protein model in DMD simulations24 to study the aggregation of apoSOD1 monomers, where a structure-based interaction potential for aggregation24; 38 is used (Methods). To promote the formation of partially unfolded SOD1 as observed in both computation and experiments7, we assign stronger attractive interactions between residues forming the amyloid core. The core-forming residues are identified from proteolysis and mass spectroscopy of the aggregates of SOD1 variants (Methods; Fig. 4).
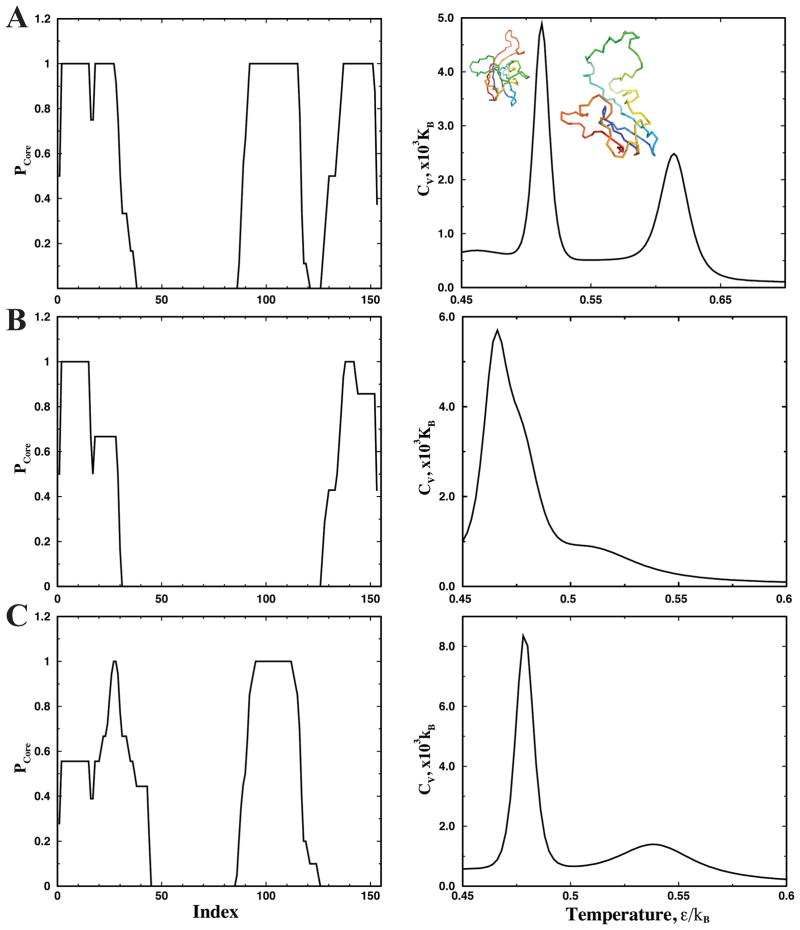
Figure 4. Thermodynamics of coarse-grained SOD1 monomer.
(A) WT, (B) G37R, and (C) I149T. In the left column, the probability of each residue to form the amyloid core (Pcore) is computed from the experimentally identified, proteolysis-resistant peptides in the amyloid core (Methods). In the right column, the specific heat of the coarse-grained SOD1 monomer is computed from replica exchange simulations of monomer folding. Due to the experimentally-based bias potential, the protein features a stable partially folded intermediate state.
Experimental constraints promote partial unfolding in SOD1 monomers
We first characterize the coarse-grained monomer folding dynamics of the three SOD1 variants. For each variant, we perform replica exchange DMD simulations of the monomer and compute the specific heat using WHAM (Methods). Due to the two types of interactions (structure-based interactions and experimentally-derived core interactions), the coarse-grained SOD1 monomers feature three-state folding dynamics with two distinct peaks in specific heat, which corresponds to folded-intermediate, and intermediate-unfolded transitions respectively39 (Fig. 4A). Strong attractions between the experimentally-determined core-forming residues stabilize the partially unfolded intermediates, the structures of which are consistent the experimentally-derived input constraints with the core-forming segments folded(Fig. 4). Although the specific heat plots between coarse-grained and atomistic simulations of corresponding proteins are different (Fig. 1 and S1), which is expected due to different models and also different types of interaction potentials, they all display two peaks featuring a partially unfolded intermediate. To model the aggregation of SOD1 via the association of the partially unfolded intermediates, we perform equilibrium simulations of multiple SOD1 monomers at the average temperature between the two transitions.
Formation of amyloid-like SOD1 aggregates
For each SOD1 variant, we perform DMD simulations with eight SOD1 monomers in a cubic box with dimensions of 227 Å, corresponding to a high concentration of approximately 1mM. At a temperature that promotes local unfolding, each isolated monomer remains in the intermediate state. For example, in wildtype SOD1 monomers, the N-terminal strands (blue strands in Fig. 5A), central strand 6 (yellow strand in Fig. 5A), and C-terminal strand (red strand in Fig. 5A) are all folded. The monomers associate with each other in simulations and form amyloid-like oligomers (Fig. 5,6), where the residual strands in each monomer assemble into beta sheets via inter- and intra-monomer hydrogen bonds. These beta-sheets face each other to form high-order “cross-beta” structures as shown by the computed fibrillar diffraction pattern (Fig. 6). There are strands from the same protein incorporated into neighboring sheets, which further stabilize the aggregates in addition to the side chain-side chain interactions. Our aggregation simulations of three SOD1 variants demonstrate the formation of amyloid-like aggregates by association of the partially unfolded SOD1 monomers. The amyloid-like aggregate structure of SOD1 is not a simple stacking of the residual folded structure, but requires a major rearrangement of each monomer.
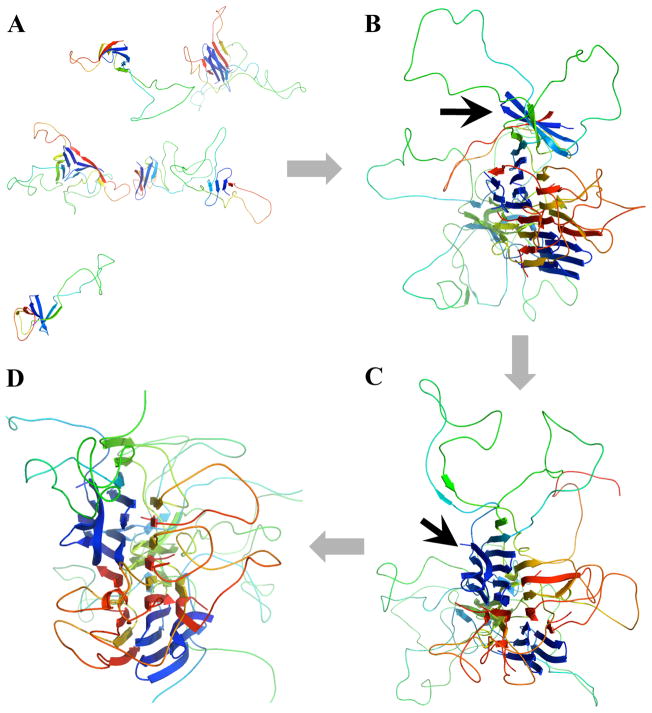
Figure 5. The aggregation process of wildtype SOD1 monomers.
(A) The initial configuration of eight SOD1 monomers in the simulation box. The monomer concentration is as high as 1mM. The cartoon representation is assigned and illustrated by using MolScript47 and PyMol, respectively.(B) One monomer (indicated by the arrow) associates with the end of the nascent amyloid-like aggregate and forms the cross-beta core. (C) The associated monomer is incorporated into the aggregate after structural rearrangement. (D) The resulting stable aggregate structure with well-formed cross-beta core.
Figure 6. The fibrillar aggregates of SOD1.
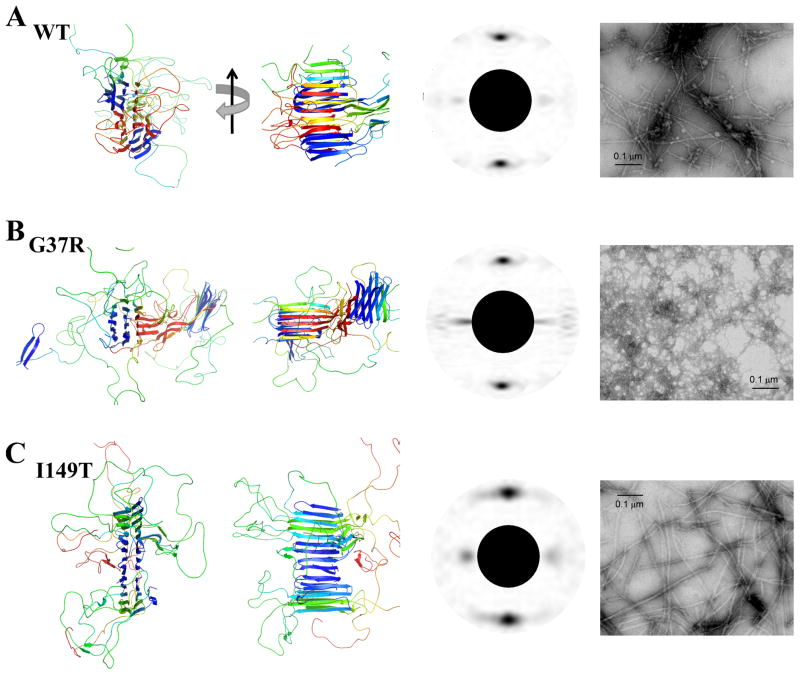
(A)WT, (B)G37R and (C) I149T. The first and second columns correspond to the aggregates formed in simulations. Two views are shown by 90° rotation along the axis of the amyloid fibril. The aggregate of G37R contains two cores. The computed fibril diffraction patterns of the aggregates24 feature the typical “cross-beta” characteristics (third column). The peak along the fibril axis corresponds to the hydrogen bonds between strands, and the peak perpendicular to the axis corresponds to the separation between the adjacent beta-sheet. The fourth column corresponds to the electron microscopic images of aggregates formed in vitro. The SOD1 fibrils were prepared by shaking disulfide-reduced apo-SOD1 at 37 °C, 1,200 rpm for 50 hours (Methods). A bar in each panel represents 0.1 μm.
The ends of each fibril core expose unsatisfied hydrogen bond donors and acceptors, which allow for further fibril growth. For instance, in Fig. 5B,C,D we illustrate one monomer incorporation event from the wildtype simulation. A SOD1 monomer initially associates with one end of the amyloid-like aggregate by diffusion (Fig. 5B), and undergoes structural rearrangement in order to be incorporated into the ordered aggregate (Fig. 5C,D). In the final aggregate structures, we notice that both wildtype (Fig. 5D) and the I149T mutant (Fig. 6C) form a single-core aggregate, while G37R forms an aggregate with two cores during the course of DMD simulations (Fig. 6B). This result is consistent with the experimentally observed morphology of G37R fibrillar aggregate, which is much thinner, branched, and less ordered compared to he fibrils formed by wildtype and I149T(Fig. 6). Therefore, we postulate that the abundance of flexible loops in partially unfolded G37R results in the formation of many smaller beta-rich aggregates and inhibits the formation of the long and discrete fibrillar aggregates observed in wildtype and I149T.
In summary, we performed multiscale molecular dynamics simulations describing monomer conformational dynamics and oligomer formation of apo SOD1 in wildtype and two mutants, G37R and I149T, each of which has distinct aggregation morphology and peptide segments forming the fibrillar core. Our simulation results suggested a generic SOD1 aggregation mechanism. After the loss of the stabilizing metal ions and disulfide bond, apoSOD1 monomer undergoes significant local unfolding before global unfolding. Mutations affect the conformational dynamics and result in distinct local unfolding patterns. The residual structure of the partially unfolded protein exhibits an exposed hydrophobic core and unsatisfied hydrogen bond donors and acceptors, which is prone to aggregate. At aggregation-favoring conditions, including high concentration and temperatures that promote local unfolding, the partially unfolded apoSOD1 monomers associate into amyloid-like aggregates with “cross-beta” characteristics. The aggregate structure is not formed by a simple stacking of the persistent residual structure of the monomer, but rather requires structural rearrangement to form the ordered structure. We also found a correlation between the aggregate structures in simulations and the mesoscopic aggregate morphologies observed in experiments. Mutant G37R, which forms less ordered fibrillar aggregates in vitro, is found to form two smaller amyloid-like cores due to the large portion of unstructured segments inhibiting the formation of a single ordered “cross-beta” core as observed in the two other variants (Fig. 5). Therefore, mutations affect the residual structure of the locally unfolded apoSOD1, where the peptide segments serve as the “building block” of SOD1 aggregation24; 25. The structured as well as the unstructured regions of the partially folded state determine the morphology of the aggregates.
Materials and Methods
Atomistic discrete molecular dynamics simulations
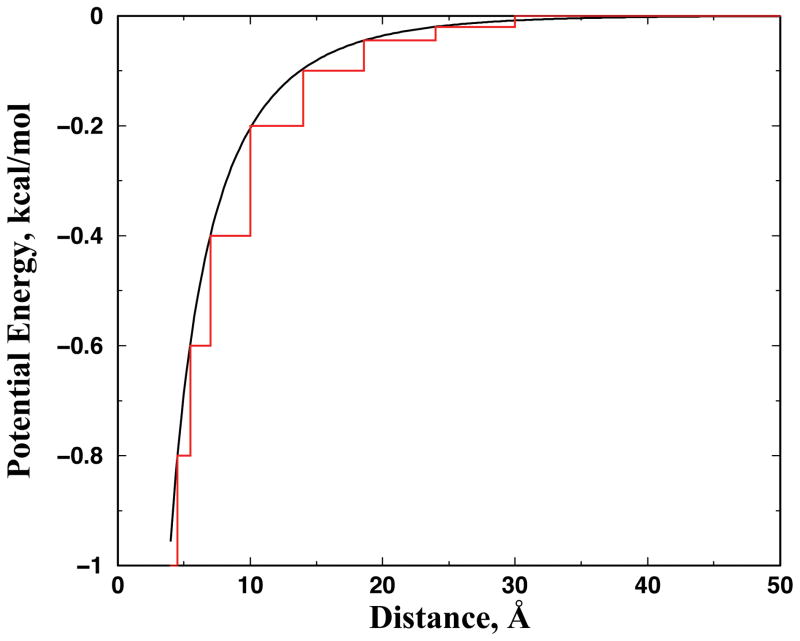
DMD is a special type of molecular dynamics simulation where pairwise interaction potentials are modeled with discontinuous functions40. The algorithm for DMD can be found in Refs.41; 42 We use an atomistic DMD force field introduced in Ref.27 to study apoSOD1 monomer dynamics. Briefly, we use the united-atom model to represent the protein, where all heavy-atoms and polar hydrogen atoms are explicitly modeled. The bonded interactions include covalent bonds, bond angles, and dihedrals. We include Van der Waals, solvation, and environment-dependent hydrogen bonding interactions in the non-bonded interactions. The solvation energy is modeled using the Lazaridis-Karplus implicit solvation model with the fully-solvated conformation as the reference state29. The hydrogen bond interaction is modeled using a reaction algorithm43. In addition to the previous version of the atomistic DMD force field27, we also add electrostatic interactions between charged residues, including the basic and acidic residues. We assign integer charges to the central atoms of charged groups: CZ for Arginine, NZ for Lysine, CG for Aspartic acid, and CD for Glutamic acid. We use the Debye-Hückel approximation to model the screened charge-charge interactions. The Debye length is set at approximately 10 Å by assuming water relative permittivity of 80, and a monovalent electrolyte concentration of 0.1 mM. We discretize the continuous electrostatic interaction potential with an interaction range of 30 Å, where the screened potential approaches zero (Fig. 7). We use the constant volume DMD simulations with period boundary conditions and control the simulation temperature using the Anderson thermostat 44.
Figure 7. Screened Debye-Hückel potential function between two opposite single charges.
The continuous potential has a Debye length of ~10 Å, assuming the relative permittivity of water 80 and a monovalent salt concentration of 0.1 mM. The step-function in red is the discretized step function utilized in DMD simulations. For two atoms of the same charge, we change the sign of the potential to indicate a repulsive.
Replica Exchange simulations and WHAM analysis
We use the replica exchange method to perform simulations of multiple copies of the same system in parallel at various temperatures. At given time intervals, replicas with neighboring temperatures exchange temperature values according to a Metropolis-based stochastic algorithm. We set the temperature exchange interval as 50ps. Exchange between replicas increases the sampling efficiency, in that energetic barriers can be overcome more easily, and in less time, with exposure to higher temperatures. In our simulations, we allocate 12 replicas with temperatures of 270, 282, 294, 303, 312, 321, 330, 339, 345, 352.5, 365, and 377.5 K for the simulations of apoSOD1 monomers.
We perform Weighted Histogram Analysis Method (WHAM) analysis using the trajectories from the replica exchange simulations. The WHAM method utilizes multiple simulation trajectories with overlapping sampling along the reaction coordinates to self-consistently compute the density of states ρ(E) by combining histograms from different simulation trajectories34. Given the density of states, the folding specific heat (Cv) can be computed at various temperatures according to the partition function, . Here, kB is the Boltzmann constant. To compute the average RMSD as a function of temperature, we compute the conditional probability P(A|E) of observing a structure with a RMSD of A at given energy E, evaluated from all simulation trajectories. The average RMSD as a function of temperature can be computed as 〈A(T)〉 = 1/Z ∫ A·P(A|E)ρ(E)exp(− E/kBT)dEdA. We also estimate the error bars as statistical uncertainty45 in the WHAM estimation of specific heat and average RMSD. The temporal correlation in sequentially generated configurations is obtained by autocorrelation analysis.
Reconstruction of the locally unfolded states
We use a RMSD range [Dmin, Dmax] to identify the locally unfolded structures from the trajectories. In order to identify the cutoff values, we compute average RMSD as a function of temperature using WHAM analysis (Fig. 2). From the specific heat, we identify the transition temperature at low temperatures corresponding to local unfolding, Tl. We assign the average RMSD at Tl as Dmin for each of the three SOD1 variants. For all three SOD1 variants, the melting or global unfolding starts from a state with an average RMSD of approximately 10 Å (Fig. 2). Therefore, we use a Dmax of 10 Å for all three variants.
Coarse-grained DMD simulations
We use a two-bead protein model to study apoSOD1 aggregation24; 46. In the two-bead model, each amino acid is represented by only the alpha-carbon (backbone) and beta-carbon (side chain). The bonded interactions between neighboring atoms along the peptide chain are assigned so as to mimic peptide geometry46. We use a structure-based potential to model the side chain-side chain packing interactions, where native interactions in the native state as observed in the crystal structure are favored. Two interacting residues can form either intra- or inter-monomer contacts, in order to promote protein-protein association24; 38. The attractions between beta-carbons are assigned with a hardcore distance of 3 Å and interaction range of 7.5 Å. We also model the backbone-backbone hydrogen bond interaction as in Ref.24
To stabilize the partially unfolded state, where the amyloid core-forming residues remain folded, we assign a strong attraction between the core residues. In the proteolysis/mass spectroscopy study of aggregates, there are overlapping peptides observed for each core-forming region (e.g., the N-terminal region for wildtype SOD1)7. For all of the overlapping peptides observed experimentally, we assume that each peptide has an equal chance to participate in the amyloid core. Therefore, we can compute for each residue in a given region the probability to be observed in the amyloid core, PCore(i) (Fig. S2). By combing all regions, we obtain the core-forming probability for all residues. We therefore introduce an experimentally-derived bias potential to the structure-based potential:
Here, Δij is the native interaction matrix, which equals one if the two residues i and j are in contact in the native state and zero otherwise. εGō and εCore are the energy scales for the structure-based and experimentally-determined bias potential, correspondingly. In our simulations, we assign εGō =0.5ε0 and εCore=1.0 ε0 with ε0 as the energy unit. The hydrogen bond interaction strength is 2.0 ε0.
Electron microscopic observation of in vitro SOD1 fibrils
Preparation and observation of SOD1 fibrils (WT, G37R, I149T) were performed as described previously7. Briefly, following constant agitation of 100 mM SOD1 at 37 °C, 1,200 rpm for 50 hours, insoluble aggregates were collected by ultracentrifugation at 110,000 g for 30 min. In order to avoid shearing fibrillar structures during agitation, a seeding reaction was further performed by adding 10 mM SOD1 aggregates (monomer-based) to 100 mM soluble SOD1 and incubating the mixture at 37 °C for 3 days in the absence of agitation. Insoluble pellets were again collected by ultracentrifugation, re-suspended in pure water, and adsorbed on 400-mesh grids coated by a glow-charged supporting membrane. After negative staining with 1% uranyl acetate, images were obtained using an electron microscope (1200EX, JEOL).
Supplementary Material
Highlights.
ApoSOD1 monomer features significant local unfolding.
Different ALS mutations induce distinct local unfolding dynamics of apoSOD1.
Residual folded structures in the locally unfolded apoSOD1 form the aggregation building block.
The local unfolding dynamics of apoSOD1 determines the morphology of the fibrillar aggregate.
Acknowledgments
We thank Elizabeth A. Proctor and Rachel Redler for helpful discussions and critical reading of the manuscript. Calculations are performed on the Topsailhigh-performance computing cluster at the University of North Carolina at Chapel Hill. This work was supported by the National Institutes of Health grant R01 GM080742.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copy editing, type setting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 3.Furukawa Y, Kaneko K, Yamanaka K, O’Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2008;283:24167–76. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oztug Durer ZA, Cohlberg JA, Dinh P, Padua S, Ehrenclou K, Downes S, Tan JK, Nakano Y, Bowman CJ, Hoskins JL, Kwon C, Mason AZ, Rodriguez JA, Doucette PA, Shaw BF, Selverstone Valentine J. Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One. 2009;4:e5004. doi: 10.1371/journal.pone.0005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–27. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci U S A. 2006;103:3147–52. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa Y, Kaneko K, Yamanaka K, Nukina N. Mutation-dependent polymorphism of Cu, Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2010;285:22221–31. doi: 10.1074/jbc.M110.113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen PM, Nilsson P, Keranen ML, Forsgren L, Hagglund J, Karlsborg M, Ronnevi LO, Gredal O, Marklund SL. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120 (Pt 10):1723–37. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 9.Bruce ME, Fraser H. Scrapie strain variation and its implications. Curr Top Microbiol Immunol. 1991;172:125–38. doi: 10.1007/978-3-642-76540-7_8. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 11.Jones EM, Surewicz WK. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 12.Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc Natl Acad Sci U S A. 2009;106:9679–84. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw BF, Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Kayatekin C, Zitzewitz JA, Matthews CR. Disulfide-reduced ALS variants of Cu, Zn superoxide dismutase exhibit increased populations of unfolded species. J Mol Biol. 2010;398:320–31. doi: 10.1016/j.jmb.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2004;101:15094–9. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnesano F, Banci L, Bertini I, Martinelli M, Furukawa Y, O’Halloran TV. The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–8003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 17.Hough MA, Grossmann JG, Antonyuk SV, Strange RW, Doucette PA, Rodriguez JA, Whitson LJ, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc Natl Acad Sci U S A. 2004;101:5976–81. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu, Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 19.Ding F, Dokholyan NV. Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc Natl Acad Sci U S A. 2008;105:19696–701. doi: 10.1073/pnas.0803266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, Hart PJ. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–7. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 21.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu, Zn superoxide dismutase: role of metal ions in protein folding. Biochemistry. 2003;42:9543–53. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 22.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc Natl Acad Sci U S A. 2006;103:10218–23. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw BF, Durazo A, Nersissian AM, Whitelegge JP, Faull KF, Valentine JS. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–76. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- 24.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J Mol Biol. 2002;324:851–7. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- 25.Sinha N, Tsai CJ, Nussinov R. A proposed structural model for amyloid fibril elongation: domain swapping forms an interdigitating beta-structure polymer. Protein Eng. 2001;14:93–103. doi: 10.1093/protein/14.2.93. [DOI] [PubMed] [Google Scholar]
- 26.Das P, King JA, Zhou R. Aggregation of gamma-crystallins associated with human cataracts via domain swapping at the C-terminal beta-strands. Proc Natl Acad Sci U S A. 2011;108:10514–9. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–8. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazaridis T, Karplus M. Effective energy functions for protein structure prediction. Curr Opin Struct Biol. 2000;10:139–45. doi: 10.1016/s0959-440x(00)00063-4. [DOI] [PubMed] [Google Scholar]
- 30.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28:743–7. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proctor EA, Ding F, Dokholyan NV. Structural and Thermodynamic Effects of Post-translational Modifications in Mutant and Wild Type Cu, Zn Superoxide Dismutase. J Mol Biol. 2011;408:555–67. doi: 10.1016/j.jmb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314:141–151. [Google Scholar]
- 33.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–7. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 35.Zhou Y, Karplus M. Folding thermodynamics of a model three-helix-bundle protein. Proc Natl Acad Sci U S A. 1997;94:14429–32. doi: 10.1073/pnas.94.26.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez JA, Shaw BF, Durazo A, Sohn SH, Doucette PA, Nersissian AM, Faull KF, Eggers DK, Tiwari A, Hayward LJ, Valentine JS. Destabilization of apoprotein is insufficient to explain Cu, Zn-superoxide dismutase-linked ALS pathogenesis. Proc Natl Acad Sci U S A. 2005;102:10516–21. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassall KA, Stubbs HR, Primmer HA, Tong MS, Sullivan SM, Sobering R, Srinivasan S, Briere LA, Dunn SD, Colon W, Meiering EM. Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS. Proc Natl Acad Sci U S A. 2011;108:2210–5. doi: 10.1073/pnas.0913021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Cho SS, Levy Y, Cheung MS, Levine H, Wolynes PG, Onuchic JN. Domain swapping is a consequence of minimal frustration. Proc Natl Acad Sci U S A. 2004;101:13786–91. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khare SD, Ding F, Dokholyan NV. Folding of Cu, Zn superoxide dismutase and familial amyotrophic lateral sclerosis. J Mol Biol. 2003;334:515–25. doi: 10.1016/j.jmb.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 40.Rapaport DC. Molecular-Dynamics Simulation of Polymer-Chains with Excluded Volume. Journal of Physics a-Mathematical and General. 1978;11:L213–L217. [Google Scholar]
- 41.Allen MP, Tildersley DJ. Computer simulation of liquids. Clarendon Press; New York, NY, USA: 1989. [Google Scholar]
- 42.Rapapport DC. The art of molecular dynamics simulation. Cambridge University Press; Cambridge: 1997. [Google Scholar]
- 43.Ding F, Borreguero JM, Buldyrey SV, Stanley HE, Dokholyan NV. Mechanism for the alpha-helix to beta-hairpin transition. Proteins. 2003;53:220–8. doi: 10.1002/prot.10468. [DOI] [PubMed] [Google Scholar]
- 44.Andersen HC. Molecular-dynamics simulations at constant pressure and-or temperature. J Chem Phys. 1980;72:2384–2393. [Google Scholar]
- 45.Chodera JD, Swope WC, Pitera JW, Seok C, Dill KA. Use of the weighted histogram analysis method for the analysis of simulated and parallel tempering simulations. Journal of Chemical Theory and Computation. 2007;3:26–41. doi: 10.1021/ct0502864. [DOI] [PubMed] [Google Scholar]
- 46.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Direct molecular dynamics observation of protein folding transition state ensemble. Biophys J. 2002;83:3525–32. doi: 10.1016/S0006-3495(02)75352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraulis PJ. MOLCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.