Abstract
Death domain (DD) superfamily members play a central role in apoptotic and inflammatory signaling through formation of oligomeric molecular scaffolds. These scaffolds promote the activation of proinflammatory and apoptotic initiator caspases, as well as Ser/Thr kinases. Interactions between DDs are facilitated by a conserved set of interaction surfaces, type I, type II, and type III. Recently structural information on a ternary complex containing the DDs of MyD88, IRAK4, and IRAK2 and a binary complex containing Fas and FADD DDs has become available. This review will focus on how the three DD interaction surfaces cooperate to facilitate the assembly of these oligomeric signaling complexes.
Introduction
Members of the death domain superfamily are critical components of apoptotic and inflammatory signaling [1]. This superfamily is composed of the Death Domain (DD), Caspase Recruitment Domain (CARD), Death Effector Domain (DED) [2], and Pyrin Domain (PYD) [3] subfamilies. Within each subfamily, members form homotypic interactions and facilitate the assembly of oligomeric signaling complexes.
A primary function of these signaling complexes is to promote the activation of pro-apoptotic initiator caspases. Upon release from the mitochondria cytochrome c binds to and triggers the oligomerization of Apaf-1 [4]. Apaf-1 is then able to bind to Caspase-9 through a CARD:CARD interaction and promote its activation via dimerization and subsequent autocatalytic cleavage. Upon ligand binding, the TNF receptor family member Fas forms a complex with FADD through DD:DD interactions. FADD then recruits Caspase-8 or Caspase-10 through their respective DEDs, leading to activation of their caspase activity. In the same manner, oligomerization of PIDD and RAIDD DDs leads to the formation of the PIDDosome and results in Caspase-2 activation [5]. Homotypic interactions between CARDs and PYDs also play an important role in the assembly of inflammasomes and their activation of pro-inflammatory caspases [6].
Assembly of DD signaling complexes can also promote the activation of Ser/Thr protein kinases. In D. melanogaster, Tube recruits the kinase Pelle through a DD:DD interaction, leading to phosphorylation and degradation of Cactus, an IκB homologue [14]. This results in the cessation of cytosolic sequestration of the transcription factor Dorsal, an NF-κB homologue [7]. In humans, DD containing homologues MyD88, IRAK4, and IRAK1/2 assemble into oligomeric signaling complexes to initiate a phosphorylation cascade that results in the degradation of IκB and translocation of NF-κB to the nucleus.
In this review, we will focus on the structures of several oligomeric death domain signaling complexes that have recently become available. Particular attention will be given to the recently solved structures of the ternary MyD88-IRAK4-IRAK2 DD complex, known as the Myddosome, and the Fas-FADD DD complex.
Death domains form oligomers using three conserved interaction types
Members of the DD superfamily share a common structural fold comprised of six antiparallel alpha helices arranged in a Greek key bundle (Figure 1a) [8,26]. Each subfamily within the DD superfamily has variations in sequence as well as the length and orientation of the helices that result in distinct structural characteristics [1]. In the DD subfamily, low sequence homology produces diverse interaction surfaces, enabling binding specificity within a subfamily. The DED subfamily contains a charged triad formed by the E/D-RxDL motif not conserved in other DD subfamilies [9]. In addition, the DED subfamily contains a conserved hydrophobic patch that facilitates DED:DED interactions [9,10]. H1 of the CARD subfamily is typically bent or broken into helices H1a and H1b [11, 30]. In the PYD subfamily, H3 is either shortened and preceded by a long loop [32,33], or replaced by a loop entirely [31].
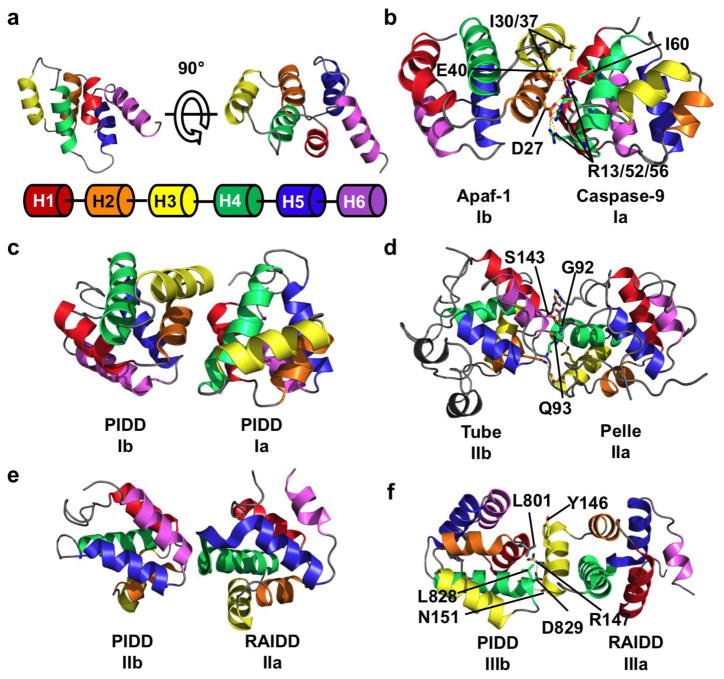
Figure 1.
Three types of asymmetric interactions mediate homotypic DD binding. (a) NMR structure of the DD of Fas, showing the six helical bundle domain architecture of the DD superfamily. Coloring scheme is shown below the structure and used throughout the figure. (b) Crystal structure of the heterodimer between the CARD domain of Apaf-1 and Caspase-9. In this type I interaction, negatively charged residues from H2 and H3 (interface Ib) of Apaf-1 interact with positively charged H1 and H4 (interface Ia) of Caspase-9. A hydrogen bond network formed by D27/E40 of Apaf-1 and R13/R52/R56 of Caspase-9 contributes to the interaction. Hydrophobic interactions between Apaf-1 I30/I37 and Caspase-9 I60 also contribute. (c) Type I interaction between two PIDD DDs. The interface architecture is highly similar to that of Apaf-1/Caspase-9. (d) Crystal structure of Tube and Pelle DD heterodimer showing a type II interface. The main chain amide of Tube S143 forms a pseudo α-helical hydrogen bond with the carbonyl of Pelle Q93. The side chain hydroxyl of S143 forms an addition hydrogen bond with the main chain carbonyl of Pelle G92. Several charge-charge interactions also contribute to binding. The interaction between the C-terminal tail of Tube and Pelle is not shown. (e) Type II interaction between PIDD and RAIDD DDs. The C-terminal end of RAIDD H4 and the H4–H5 loop (interface IIa) interacts with the N-terminal end of PIDD H6. (f) Type III interaction between PIDD and RAIDD DDs. H3 of RAIDD (interface IIIa) interacts with the H1–H2 and H3–H4 loops (interface IIIb) of PIDD. A salt-bridge between PIDD D829 and RAIDD R147, a hydrogen bond between PIDD L828 and RAIDD N151, and a hydrophobic interaction between PIDD L801 and RAIDD Y146.
The first structure of a DD complex was a heterodimer of the Apaf-1 and Caspase-9 CARD domains [11]. The CARD domains of Apaf-1 and Caspase-9 form an asymmetrical dimer through what is now known as a type I interaction (Figure 1b) [12]. Helices H1 and H4 of Caspase-9 form a positively charged type Ia surface that interacts with the negatively charged Ib surface formed by helices H2 and H3 of Apaf-1. This type I interaction buries a total surface area of 1,100 Å2. Contributing to the stability of this interaction is a hydrogen bond network between D27/E40 of Apaf-1 and R13/R52/R56 of Caspase-9, a second hydrogen bond network between S41 of Apaf-1 and R11 of Caspase-9 (not shown), and a hydrophobic interface formed by I30/I37 of Apaf-1 and I60 of Caspase-9 [11].
Shortly after the publication of the Apaf-1/Caspase-9 structure, the crystal structure of another heterodimeric complex between the DDs of Tube and Pelle was solved [7]. In D. melanogaster, the activation of the receptor Toll leads to translocation of Tube to the plasma membrane. Tube then recruits Pelle via homotypic DD interactions. Tube and Pelle interact in a manner distinct from that observed in the Apaf-1/Caspase-9 structure (Figure 1d). The C-terminal end of Pelle H4 and the H4–H5 loop form the type IIa surface. This surface interacts with the Tube type IIb surface consisting of a groove formed by the H1 and H2 corner on one side, with H6 and its preceding loop on the other. The C-terminus of Pelle H4 is aligned with the N-terminus of Tube H6 and forms a pseudo-helical hydrogen bond between the carbonyl of Pelle Q93 and the main chain amide of Tube S143 [7]. An additional hydrogen bond is formed between the Tube S143 side chain hydroxyl and the Pelle G92 carbonyl oxygen. Finally, there are several charge-charge interactions between the Tube H1/H2 corner and Pelle H1.
The structure of the PIDDosome demonstrated how these binary interactions cooperate to produce an oligomeric death domain assembly [13]. Cleavage of full length PIDD results in the generation of a death domain-containing 37 kDa C-terminal fragment. The PIDD DD interacts with the DD of RAIDD to function as an oligomeric platform for the activation of Caspase-2 through the CARD of RAIDD [5]. In the structure, five PIDD DDs and seven RAIDD DDs assemble into an asymmetric globular core that can be divided into three layers (Figure 2a–c). The lower layer contains five PIDD DD molecules (P1–P5), the middle layer contains five RAIDD DD molecules (R1–R5), followed by a top layer of 2 RAIDD DDs (R6 and R7). The interface between each PIDD DD and the RAIDD DD directly above (middle layer) is a type II interface. The C-terminus of RAIDD H4 and the following loop (IIa surface) interacts with PIDD H6 (IIb surface) in the same manner as Pelle and Tube (Figure 1e). This interaction is dependent on hydrogen bond networks and salt bridges. Type II interactions also exist between middle and upper layer RAIDD molecules. Five screw rotations are applied to this hypothetical subcomplex to obtain the first two layers of the PIDDosome (Figure 2a). There are two types of rotations applied, one rotating 84 degrees coupled with a downwards translation along the axis, and the other rotated 54 degrees with an upwards translation.
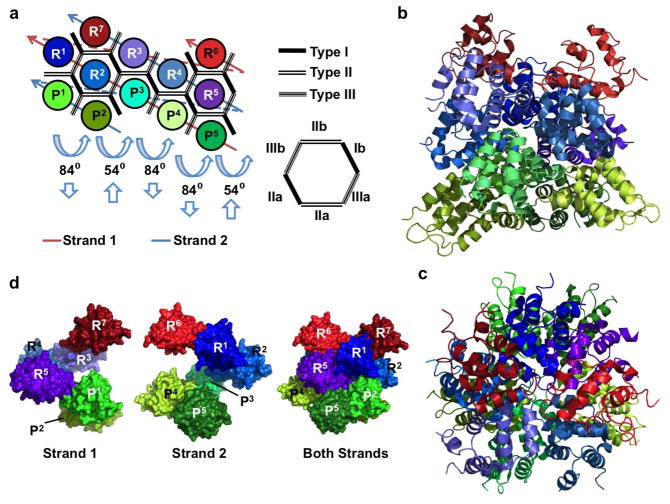
Figure 2.
Structure of the PIDDosome, a signaling complex containing PIDD and RAIDD DDs. (a) Planar schematic of the PIDD/RAIDD DD complex. The lower layer contains five PIDD DDs (P1–P5), the middle layer contains five RAIDD DDs (R1–R5), followed by a top layer of 2 RAIDD DDs (R6 and R7). Each DD interacts with between 3 and 6 adjacent DDs. Each hypothetical subcomplex consisting of a column of PIDD and RAIDD(s) (ex. P1 and R1 or P2, R2, and R7) is related to neighboring subcomplexes by two types of screw rotations, an 84° rotation coupled with downward translation, and a 54° rotation coupled with an upwards translation. (b) Crystal structure of the PIDDosome colored as in Figure 2a. Lower layer PIDD DDs are shown in greens, middle layer RAIDD DDs are in blues, and upper layer RAIDDs are shown in reds. (c) The PIDDosome viewed from above. (d) The PIDDosome is a double-stranded left-handed helix of DDs. Each strand contains 6 DDs connected via type III interactions (see (a) for the schematic of the strands). Shown are surface representations of strand 1 (left), strand 2 (middle) and both strands (right).
Three types of type I interactions are present, PIDD:PIDD, RAIDD:PIDD, and RAIDD:RAIDD. Each of these is mediated by the type Ia surface formed by H1 and H4 of one DD and the type IIa surface formed by H2 and H3 of the adjacent DD, similar to the interface between Apaf-1 and Caspase-9 CARD domains (Figure 1c). In addition, a new type III interface was found in the PIDDosome (Figure 1f). This is formed by the interaction of H3 (Type IIIa surface) of the first DD with a groove formed by the H1–H2 and H3–H4 loops on the second DD. Like the type I interaction, three subtypes of the type III interaction exist between PIDD:PIDD, RAIDD:PIDD, and RAIDD:RAIDD. The Type III interface contains hydrophobic, charged, and polar interactions. The Type III interface between RAIDD and PIDD contains a hydrophobic interaction between PIDD L801 and RAIDD Y146, a salt bridge between PIDD D829 and RAIDD R147, and a hydrogen bond between PIDD L828 and RAIDD N151. Certain DDs in the PIDDosome use type Ia, Ib, IIa, IIb, IIIa and IIIb interaction surfaces simultaneously, forming interactions with six neighboring DDs.
The PIDDosome can also be thought of as a double stranded left-handed helical oligomer. Strand 1 contains P3–5, R1–2, and R6, while strand 2 contains DDs P1–2, R3–5, and R7 (Figure 2a). Sequential DDs in a strand are mediated by type III interactions. Surface types I and II are responsible for interstrand interactions. Shown are surface representations of each strand, as well as the complete PIDDosome for comparison (Figure 2d).
Structure of the MyD88-IRAK4-IRAK2 death domain complex
Members of the Toll-Like Receptor (TLR) family recognize pathogen associated molecular patterns and trigger signaling cascades to activate immune responses [14]. They share a common cytoplasmic domain with the IL-1 and IL-18 receptors, known as a Toll/IL-1 Receptor (TIR) domain [15]. These receptor TIR domains bind to the C-terminal TIR of the adapter protein MyD88, which performs a critical role in signal propagation from both IL-1 and IL-18 receptors, as well as all TLRs excluding TLR3 [16]. The N-terminal death domain of MyD88 then binds IRAK family members, which contain N-terminal DDs and C-terminal kinase domains [17]. Formation of an oligomeric DD signaling complex containing MyD88, IRAK4, and IRAK1/2 results in auto- and cross-phosphorylation followed by downstream activation of transcription factors, including NF-κB and Interferon Regulatory Factors (IRFs) [16–18].
The structure of a ternary death domain complex containing DDs from MyD88, IRAK4, and IRAK2 was recently solved [19]. The DDs in the structure are arranged in a tower of approximately four layers, with 6 MyD88 forming the bottom two layers, 4 IRAK4 forming the middle layer, and 4 IRAK2 in the top layer (Figure 3a). The tower is formed from a single left-handed helix of DDs, beginning with MyD88 (M1–M6), followed by IRAK4 (I41–I44), and finally IRAK2 (I21–I24). In this helix, the type IIIb surface of the nth DD interacts with the type IIIa surface of the adjacent (n+1)th DD. Each successive DD is approximately rotated 98° and translated 6 Å along the helical axis. Interaction types I and II are responsible for contact between layers of the helix. Interfaces Ib and IIb of the nth DD interact with surface Ia of the (n+3)th DD and surface IIa of the (n+4)th DD. The various interaction subtypes (i.e. Type II between MyD88:MyD88, MyD88:IRAK4, and IRAK4:IRAK2) are similar and superimpose well. The final dimensions of the tower are 110 Å in height and 70 Å in diameter.
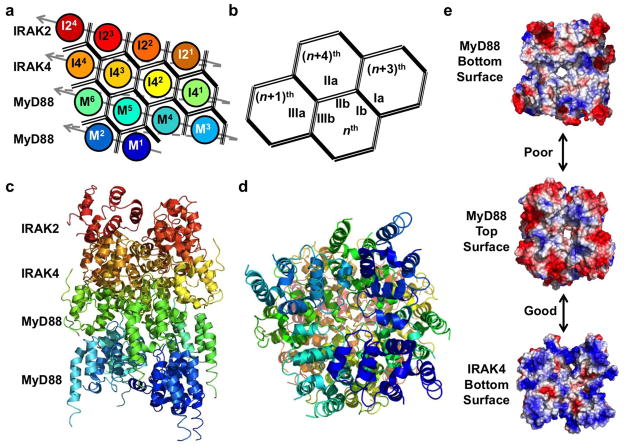
Figure 3.
Structure of the Myddosome, a complex of MyD88, IRAK4, and IRAK2 DDs. (a) Planar schematic of the MyD88-IRAK4-IRAK2 complex. The Myddosome is a four layered tower formed from a single stranded left-handed helical DD oligomer. The strand begins with 6 MyD88 DDs (M1–M6), followed by IRAK4 DDs (I41–I44), and finally IRAK2 DDs (I21–I24). Interactions between successive DDs are type III. (b) The nth DD uses its IIIb surface to interact with the IIIa surface of the (n+1)th DD. Interactions between strands are mediated by type I and type II interfaces. The nth DD uses its Ib and IIb surfaces to associate with the Ia surface of the (n+3)th DD and the IIa surface of the (n+4)th DD. (c) The crystal structure of the MyD88-IRAK4-IRAK2 oligomer colored as in (a). (d) Myddosome viewed from below. (e) Surface electrostatic potential map showing poor charge complementarity between MyD88 top and bottom surfaces (top) and good charge complementarity between MyD88 top and IRAK4 bottom surfaces (bottom).
While the three DDs in the Myddosome share the canonical DD superfamily fold, MyD88 has features that differentiate itself including a long H1–H2 loop and an extended H6. The long H1–H2 loop forms interactions with IRAK4 at their shared interface. This interaction is not present between IRAK4 and IRAK2. In addition, the charge and shape complementarity varies between the layers. The top and bottom surfaces of MyD88 are only weakly complimentary, preventing stable interactions between layers (Figure 3e). The top and bottom surfaces of the IRAK2 layer are also poorly complimentary [19]. In contrast, the bottom surface of IRAK4 is complimentary with the top surface of MyD88 (Figure 3e). Additionally, the top surface of IRAK4 is complimentary with the bottom surface of IRAK2 [19]. These features explain the assembly specificity and order of assembly of the various DDs present in the Myddosome.
Structure of the Fas-FADD death domain complex
The expression of FasL is responsible for much of the cytotoxic activity of CD8+ T lymphocytes and natural killer cells [20,23]. Ligation of FasL to the receptor Fas results in the formation of the death-inducing signaling complex (DISC) [21] containing Fas, FADD, and Caspase-8/10. Fas contains a cytoplasmic DD, while FADD is an adapter that containing an N-terminal DED and a C-terminal DD. The interactions between Fas and FADD are mediated by DDs [22], while the FADD DED associates with the tandem DEDs of Caspase-8/10 [24]. The formation of the DISC triggers both a dimerization induced conformational change and autoproteolytic processing of Caspase-8 that result in its activation.
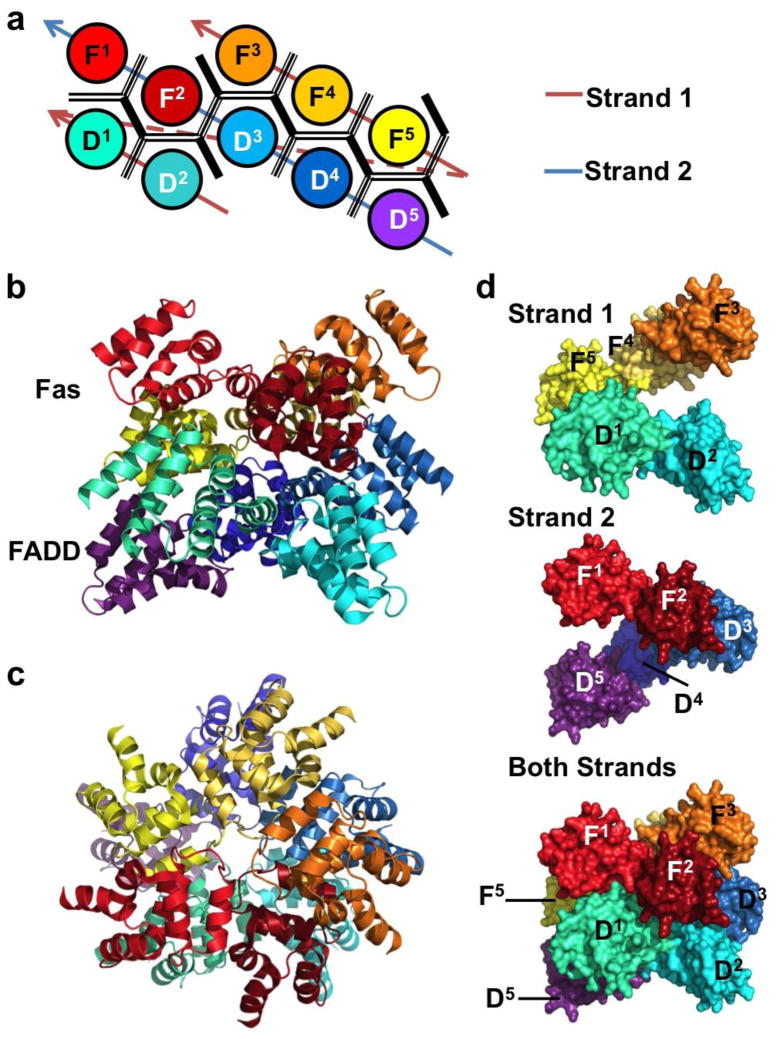
The structure of a signaling complex containing the DDs of both Fas and FADD has recently been elucidated [25]. Five Fas DDs and five FADD DDs are arranged in an asymmetric globular core similar to that observed in the PIDD-RAIDD structure. The lower layer contains the five FADD DDs (D1–5) while the upper layer contains five Fas DDs (F1–5). Similarly, a hypothetical subcomplex of one FADD DD and the Fas DD directly above it can be defined for the convenience of discussion. At the interface of this subcomplex is a type II interface, formed by the IIa surface of Fas and the IIb surface of FADD. This subcomplex can then be rotated and translated along a screw axis five times to obtain the two layered Fas-FADD complex. Several types of type I and type III interactions mediate the interactions between different subcomplexes. Mutations on the interfacial residues between Fas and FADD had an effect on complex formation in vitro and in vivo. A semi-quantitative relationship was observed between mutational severity and the incidence frequency of the surface used in the complex. Like the PIDD-RAIDD oligomer, the Fas-FADD DISC can be interpreted as a double-stranded left-handed helical oligomer (Figure 4d). The first strand consists of D1–2 and F3–5, while the second strand contains D3–5 and F1–2 (Figure 4a). Like the PIDDosome and Myddosome, intrastrand contacts between sequential DDs are mediated by type III interactions, while types I and II are responsible for interstrand contacts.
Figure 4.
The structure of the DISC, a complex of Fas and FADD DDs. (a) Planar schematic of the Fas-FADD DD complex. The DDs form a two layered oligomeric core. The lower layer is comprised of five FADD DDs (D1–5), and five Fas DDs (F1–5) form the upper layer. (b) The crystal structure of the FAS-FADD DD oligomer colored as in (a). (c) DISC viewed from above. (d) Like the PIDDosome, the DISC is a double-stranded left-handed helix. Each strand consists of five DDs joined through type III interactions. Shown are surface representations of strand 1 (top), strand 2 (middle) and both strands (bottom).
Concluding remarks
The asymmetric type I, II, and III interactions between DDs are conserved in all current structures of oligomeric DD signaling complexes. These interactions likely represent the predominant mechanism of DD polymerization. While the overall architecture of each interaction is conserved, variations in residues that form each interaction surface contribute to the binding specificity between DDs. The use of helical oligomerization in signal transduction has several potential benefits. In the case of Fas and MyD88, extracellular ligand binding triggers receptor clustering. This allows for the transmission of the signal across the plasma membrane by inducing oligomerization of resident cytosolic DD proteins. The requirement for a nucleation event protects cells from errant activation of irreversible signaling pathways. This is of paramount importance as the downstream effects of these DD oligomerization events are largely bimodal. Activation of apoptotic initiator caspases invariably results in the activation of effector caspases and apoptosis [27]. Also, NF-κB activation has recently been shown to respond digitally to TNF-α [28] and signaling through the T cell receptor [29] on a single cell level. The helical nature of DD signaling complexes allows for interaction with a variable number of binding partners. It also allows for the conversion from an analog signal in the form of extracellular ligand concentration (or in the case of PIDD, cellular stress) to an intracellular digital one by setting a threshold for signal propagation.
HIGHLIGHTS.
Death domain oligomerization is facilitated by conserved interaction surfaces.
Known death domain oligomers possess helical symmetry.
Helical oligomerization provides a versatile digital signal propagation mechanism.
Acknowledgments
This work was supported by the National Institute of Health (AI50872 and AI076927 to H.W.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valmiki MG, Ramos JW. Death effector domain-containing proteins. Cell Mol Life Sci. 2009;66:814–830. doi: 10.1007/s00018-008-8489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohl A, Grütter MG. Fire and death: the pyrin domain joins the death-domain superfamily. C R Biol. 2004;327:1077–1086. doi: 10.1016/j.crvi.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 5.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 6.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steward A, McDowell GS, Clarke J. Topology is the principal determinant in the folding of a complex all-alpha Greek key death domain from human FADD. J Mol Biol. 2009;389:425–437. doi: 10.1016/j.jmb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ, Wu H. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L, Lenardo MJ, Fesik SW. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 11.Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 12.Weber CH, Vincenz C. A docking model of key components of the DISC complex: death domain superfamily interactions redefined. FEBS Lett. 2001;492:171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- 13.Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 15.O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol Rev. 2008;226:10–18. doi: 10.1111/j.1600-065X.2008.00701.x. [DOI] [PubMed] [Google Scholar]
- 16.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki N, Suzuki S, Yeh WC. IRAK-4 as the central TIR signaling mediator in innate immunity. Trends Immunol. 2002;23:503–506. doi: 10.1016/s1471-4906(02)02298-6. [DOI] [PubMed] [Google Scholar]
- 18.Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. The crystal structure of the Myddosome reveals elegant and versatile helical symmetry and hierarchical DD assembly, providing a structural framework for understanding the mechanism of auto- and cross-phosphorylation of IRAK family kinases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kägi D, Vignaux F, Ledermann B, Bürki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 21.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 23.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrington PE, Sandu C, Wei Y, Hill JM, Morisawa G, Huang T, Gavathiotis E, Wei Y, Werner MH. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 25••.Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. The crystal structure of the Fas-FADD DD complex provides an explanation for the requirement for hexameric or membrane-bound FasL. When superimposed with the structure for full-length FADD [24], a model for Caspase-8/10 recruitment and activation is proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 27.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 28•.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. In this paper, the authors make single cell measurements of NF-κB nuclear localization and create time dependent target gene expression profiles in response to TNF-α stimulation. They then develop a mathematical model that describes bimodal activation of NF-κB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-κB. J Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaughn DE, Rodriguez J, Lazebnik Y, Joshua-Tor L. Crystal structure of Apaf-1 caspase recruitment domain: an α-helical Greek key fold for apoptotic signaling. J Mol Biol. 1999;293:439–447. doi: 10.1006/jmbi.1999.3177. [DOI] [PubMed] [Google Scholar]
- 31.Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grütter MG, Wüthrich K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure. 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Bae JY, Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]