Abstract
Rationale
MyomiRs miR-499, miR-208a and miR-208b direct cardiac myosin gene expression. Sequence complementarity between miRs and their mRNA targets determines miR effects, but the functional consequences of human myomiR sequence variants are unknown.
Objective
To identify and investigate mutations in human myomiRs in order to better understand how and to what extent naturally-occurring sequence variation can impact miR-mRNA targeting and end-organ function.
Methods and Results
Screening of ~2,600 individual DNAs for myomiR sequence variants identified a rare mutation of miR-499, u17c in the 3′ end, well outside the seed region thought to determine target recognition. In vitro luciferase reporter analysis showed that the 3′ miR-499 mutation altered suppression of a sub-set of artificial and natural mRNA targets. Cardiac-specific transgenic expression was used to compare consequences of wild-type and mutant miR-499. Both wild-type and mutant miR-499 induced heart failure in mice, but miR-499 c17 misdirected recruitment of a subset of miR-499 target mRNAs to cardiomyocyte RNA-induced silencing complexes, altering steady-state cardiac mRNA and protein makeup and favorably impacting cardiac function. In vitro analysis of miR-499 target site mutations and modeling of binding energies revealed abnormal miR-mRNA duplex configurations induced by the c17 mutation.
Conclusions
A naturally occurring miR-499 mutation outside the critical seed sequence modifies mRNA targeting and end-organ function. This first description of in vivo effects from a natural human miR mutation outside the seed sequence supports comprehensive studies of individual phenotypes or disease-modification conferred by miR mutations.
Keywords: microRNA, myomiR, gene mutation
Introduction
MicroRNAs (miRs) are small (18–25 nucleotide) non-coding RNAs that fine-tune protein expression by recruiting target mRNAs into macromolecular extra-nuclear complexes termed RISCs (RNA-induced silencing complexes) where protein translation is suppressed 1. DNA sequence analysis suggests that there are over 1,000 different human miRs. It is thought that one third of all mRNAs may be regulated by one or more human miRs 2. Because miRs recognize target mRNAs via Watson-Crick pairing of complementary bases, nucleotide sequence is a primary determinant of miR function. The prevailing view is that critical miR “seed sequences” (bases 2–8) largely determine miR-mRNA interactions,2, 3 although evidence is accumulating that extra-seed sequence pairing may play a discriminatory role in miR targeting.
Interactions between miRs and their target mRNAs are determined by miR abundance, mRNA target abundance, and miR-mRNA binding affinity; the latter is determined largely by binding site and miR sequence complementarity. There is tremendous variation in binding site sequence for a given miR within and between different mRNA targets. In contrast, nucleotide sequence for a given microRNA is highly conserved across species and between individuals 4. We postulated that naturally-occurring variation in mature microRNA sequences might have pathophysiological impact by creating new miR family members, thus altering suppression of target mRNAs with binding site nucleotides complementary either to the wild-type or mutant miR. In vivo data relating the effects of miR sequence variation on mRNA targeting function and end-organ phenotype are currently lacking.
To test the notion that miR mutations will alter in vivo heart function, we elected to study highly expressed cardiac miRs with well-established end-organ effects. The myomiR family of muscle-specific miRs coordinately regulate cardiac myosin isoform expression 5–7. MyomiRs are reportedly regulated in human heart failure as part of a broader genetic program involving dozens of miRs and mRNAs 8, 9,10, and forced cardiac-specific miR-499 expression at levels seen in human heart failure induces progressive cardiac enlargement and contractile dysfunction, i.e. dilated cardiomyopathy, in normal and pressure-overloaded hearts 10,11.
Here, we used pooled sequencing of a diverse cohort of human DNA samples to discover a novel, rare human miR-499 mutation that is within the mature miR sequence, but outside the 5′ seed region considered to be the major determinant of mRNA target recognition. Using in vitro assays of miR function to direct genome-wide in vivo molecular RNA proteomics analyses and computerized modeling of miR-mRNA duplex structures, we demonstrate that this 3′ extra-seed sequence miR-499 mutation conditionally changes miR-499 target mRNA recognition, thereby altering the effects of miR-499 on the cardiac transcriptome and proteome and modifying characteristics of the cardiomyopathy conferred by miR-499 overexpression. These findings strongly support further genome wide efforts for discovery and in vivo analysis of potentially pathological human microRNA sequence variants.
Materials and Methods
Pooled resequencing of human genomic DNA
Genomic regions containing miR precursors were analyzed in 1,742 individuals of Caucasian descent and 864 of African-American descent, using massively parallel sequencing of pooled samples, as previously described 12, 13.
Luciferase reporter studies
Three tandem copies of the nucleotide sequence perfectly complementary to wild-type miR-499, or mutant miR-499 c17 were cloned into pmiRGLO (Promega). Similar techniques were used to generate sequences containing single or double nucleotide changes in the binding site complementary to seed sequence positions 6 (Δ6) or 3 and 4 in combination (Δ3,4). miR-499 cDNA encoding the pri-miR was cloned from human DNA into pcDNA3.1 (Invitrogen) and mutations were engineered using site-directed mutagenesis (Stratagene QuikChange). Pri-miR and reporter constructs were transfected in sextuplicate into HEK293 cells using Fugene HD (Promega). Firefly 3′UTR-coupled luciferase activity was normalized to constitutive Renilla luciferase activity for each well. 3′ UTRs of Sox6, Pdcd4 and Skil were cloned into psiCheck2 (Promega) and assayed similarly. Saturation mutagenesis of the complementary miR-499 sequence was performed via ligation of double-stranded oligonucleotides directly into pmiRGLO.
Animals
miR-499 transgenic mice were generated by cloning a 530 bp fragment flanking mouse miR-499 (chromosome 2, intron 19 of Myh7b) into the cardiac-specific mouse Myh6 promoter construct. The wild-type miR-499 mouse (designated line 1, expressing at 16-fold normal levels) is described elsewhere10. The mutant miR-499 mouse (line 2, expressing at 23-fold normal levels) was chosen for comparative study because it expresses mature miR-499 in heart at levels similar to that of the wild-type miR-499 transgenic mouse, and to human heart disease 10. Mice were generated and housed according to procedures approved by the Washington University Institutional Animal Care and Use Committee.
Transcriptional and RISC mRNA profiling by high-throughput sequencing
Preparation of cDNA fragments from poly(A+)-selected cardiac RNA was as previously described 14. Conditions for Ago2 immunoprecipitation and isolation of bound RNA from mouse hearts were adapted from Karginov et al.15, as described in 14. Preparation of bar-coded Illumina sequencing libraries, sorting by barcode, and transcriptome mapping were performed as previously described 16.
Proteomics
3-color, 2D DiGE (differential in-gel electrophoresis) analyses of five paired hearts (miR-499 WT vs miR-499 mutant) were performed at Applied Biomics (Hayward, CA) as described 10.
Molecular modeling
Molecular modeling of miR-499 mRNA duplexes used RNAhybrid 17 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) to predict binding energies (minimum free energy of hybridization, mfe) associated with various miR-mRNA sequence mismatches and their corresponding multiply iterative structural conformations.
Results
A human miR-499 non-seed sequence mutation alters in vitro target mRNA suppression
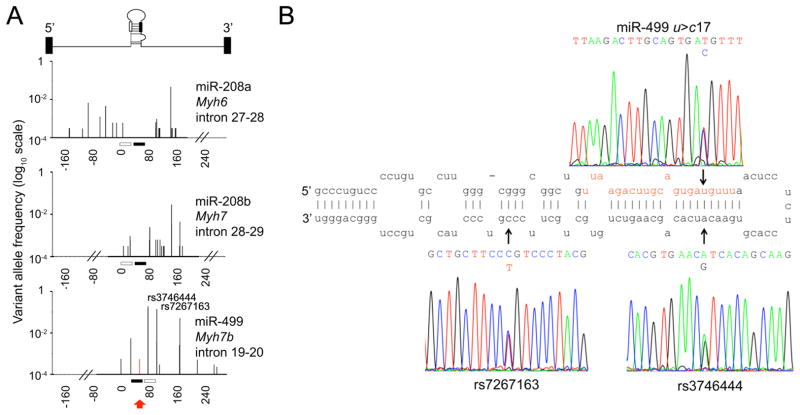
Some naturally-occurring miR sequence variations are predicted to impact mRNA targeting 18. While it is possible to explore the impact of binding site sequence variation on miR-mRNA interactions using mutagenesis, this approach lacks direct translational applicability to the human condition. On the other hand, little is currently known about the prevalence and pathophysiology of human miR sequence variation. Thus, we screened ~2,600 human DNA samples for sequence variation corresponding to the pre-miRs for each of the three human myomiRs, miR208a, miR-208b, and miR-499. Figure 1a graphically displays the position and frequency of sequence variations detected. MyomiR sequence variants were common in DNA corresponding to the primary intronic miRs and the precursor miR (the Drosha-processed stem-loop structure 1) (Online Table I), but only one myomiR sequence variant occurred within the mature miR, a rare C substitution for T at nucleotide (nt) 17 of miR-499 identified in two DNA samples (Figure 1a red arrow, Online Table I). Two previously reported common miR-499 precursor single nucleotide polymorphisms (SNPs) were also detected, rs3746444 and rs7267163 (Figures 1a, 1b). The former is located at nucleotide position 5 of miR-499* (aka miR-499-3p), the double-stranded complement to mature miR-499 (aka miR-499-5p), which is degraded during processing and has no known biological effect on the heart. All three of these miR-499 sequence variants were confirmed by Sanger sequencing (Figure 1b).
Figure 1. Pooled DNA sequencing detects a rare human nucleotide 17 miR-499 sequence variant.
(A) Allele frequencies (log10 scale) of myomiR sequence variants mapped to locations within their respective miRtrons, through screening 2,606 human subjects. One mature miR sequence variant (miR-499 u17c) was detected (red arrow, right panel). Nucleotide positions are numbered relative to the beginning of the pre-miR. (B) Sequence pherograms for the miR-499 u17c mutation and common single nucleotide polymorphisms rs3746444 and rs7267163, mapped to the stem-loop structure of pre-miR-499.
The novel miR-499 c17 mutation was detected in two of the 2,600 DNA samples. Non-seed sequence miR mutations are not generally considered to meaningfully impact miR function 2, 19. A genetic approach to determine whether miR-499 c17 affects the clinical status of the two human subjects in which it was identified was not possible because the number of affected individuals is too small to derive a statistically robust genotype-phenotype relationship; family pedigrees were not available. We considered, however, that a detailed functional and biological analysis of the miR-499 c17 mutation could establish whether this mutation has any effect on mRNA targeting, and test for the first time the conventional wisdom that non-seed sequence miR variation does not impact human health and/or disease.
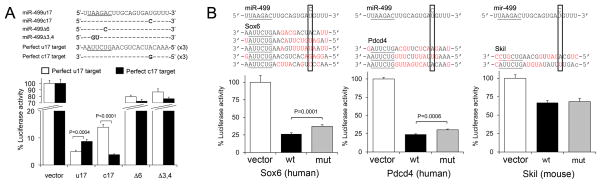
Initial functional studies were designed to test whether the miR-499 c17 nucleotide mutation has any functional impact under the most rigorous conditions, i.e. when all of the other miR-mRNA binding sites are perfectly matched. Accordingly, we designed luciferase reporter constructs with artificial binding site sequences exactly complementary to either the wild-type or mutant miR. This approach assays the potential for both loss-of-function (e.g. when the mutation introduces a binding pair mismatch at position 17) and gain-of-function (e.g. when the mutation corrects a mismatch at the same position). Overall luciferase suppression was 80–90% for both targets by wild-type (u17) and mutant (c17) miR-499, and each miR-499 suppressed its perfect complement to a significantly greater degree than it did the binding site with a single mismatch at position 17 (Figure 2a). By contrast, suppression of artificial mRNA target sites with single or double seed sequence mismatches (Δ6 and Δ3,4) was only ~20% by either c17 or u17 miR-499; there were no significant differences between wild-type and mutant miR-499 when binding site seed-sequence complementarity was disrupted (Figure 2a). The two miR-499 precursor polymorphisms that did not change mature miR-499 sequence had no effect on luciferase reporter activity (Online Figure I). These results show that the miR-499 seed region plays a major, but not complete, role in target recognition, and reveal that the miR-499 c17 mutation can conditionally affect mRNA target suppression.
Figure 2. The miR-499 nucleotide 17 mutation selectively changes in vitro mRNA reporter suppression.
(A) miR-499-u17, –c17, and miR-499 mutant Δ6 and Δ3,4 suppression of luciferase activity from dual-luciferase constructs with concatamerized sequences perfectly complementary to miR-499-u17 or -c17. miR-499 precursor and luciferase plasmids were co-transfected into HEK293 cells. (B) Human Sox6 and Pdcd4, and mouse Skil luciferase reporter suppression by wild-type (wt) and mutant miR-499. Predicted miR-499 binding sites are shown above; red nucleotides indicate sequence mismatches to wild-type miR-499. Rectangles indicate miR-499 nucleotide 17. White bars, vector treatment; black bars, addition of wild-type miR-499; gray bars, mutant miR-499 c17; n=12 per group.
Since perfect miR-mRNA sequence complementarity is exceedingly rare in animals 19, we examined the consequences of the miR-499 mutation on suppression of luciferase reporters containing partial 3′ untranslated regions of three bioinformatically-defined miR-499 target mRNAs. Because TargetScan relies heavily on seed sequence complementarity to predict target mRNAs, each of the predicted miR-499 target mRNAs, Sox6, Pdcd4, and Skil, had multiple binding sites with perfect seed sequences. Therefore, and as anticipated from the artificial reporter assays above, functional differences induced by the c17 mutation were modest. Nevertheless, Sox6 and Pdcd4 were more effectively suppressed by wild-type miR-499, whereas Skil exhibited no preference (Figure 2b). While in vitro reporter studies do not precisely reproduce in vivo miR targeting because of differences in mRNA structure and absence of co-regulatory miRs 20, 21, these results show that the miR-499 3′ mutation modifies suppression of a subset of target mRNAs.
miR-499 c17 mutation alters in vivo and cardiac function and mRNA targeting
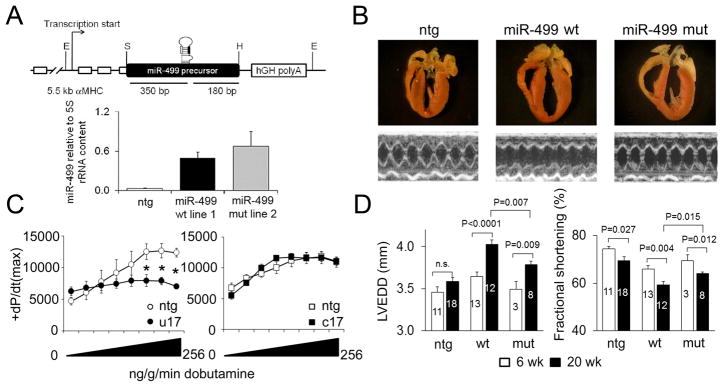
miR-499 is upregulated in human and experimental heart disease 9, 22 and confers a pathological phenotype when forcibly expressed in cardiac myocytes 10, 11. We created transgenic mice to explore whether the modest differences in suppression of some mRNA targets by mutant miR-499 c17 altered normal in vivo cardiac mRNA targeting, and therefore the cardiac phenotype. Based on previous results where unbiased RISC-sequencing identified cardiac miR-133 mRNA targets without perfect seed sequence complementarity 14, we posited that the 3′ miR-499 mutation might have greater impact on these “weaker” (and bioinformatically overlooked) mRNA targets. This notion was supported by our observation that, despite expressing at slightly greater levels in mouse myocardium than its wild type analog (Figure 3a), miR-499 c17 favorably impacted the characteristic progressive cardiomyopathy induced by miR-499 overexpression: Compared to wild-type miR-499 expressing hearts, those expressing mutant miR-499 developed less severe cardiac remodeling (Figure 3b) and impairment of ejection performance (Figures 3b, 3d), and exhibited preservation of the contractile response to β-adrenergic stimulation (Figure 3c).
Figure 3. The miR-499 c17 mutation alters the cardiomyopathy phenotype induced by miR-499.
(A) A 530 bp mouse genomic fragment including the miR-499 precursor region was cloned into exon 3 of the murine Myh6/αMHC transgenic expression vector. E = EcoRI, S = SalI, H = HindIII restriction sites. B. miR-499 expression relative to 5S rRNA in independent mouse lines was assayed using the NCode miR RT-qPCR system (Invitrogen). Matching wild-type miR-499 (line 1) and mutant miR-499 c17 (line 2) were selected for comparative analyses. (B) Four-chamber cardiac sections (upper) and representative M-mode echocardiograms (lower) from 20 week-old wild-type miR-499 and mutant miR-499 c17 hearts. (C) Contractile response (peak +dP/dt) to β1-adrenergic agonist, dobutamine, in 20-week old wt miR-499 (left) and mutant miR-499 c17 (right) hearts compared to respective nontransgenic littermates (white) (2-way ANOVA, * P<0.05; n=4–5 per group). (D) Left-ventricular end-diastolic dimension (LVEDD) and fractional shortening assessed by M-mode echocardiography in 6 and 20 week-old wild-type miR-499 and mutant miR-499 c17 hearts; mean ± s.e.m. Numbers of mice used for each determination are shown within each column.
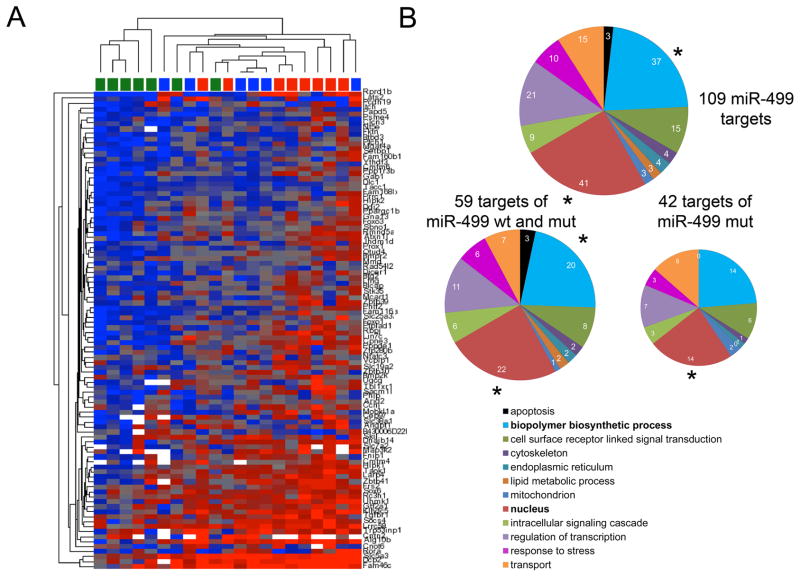
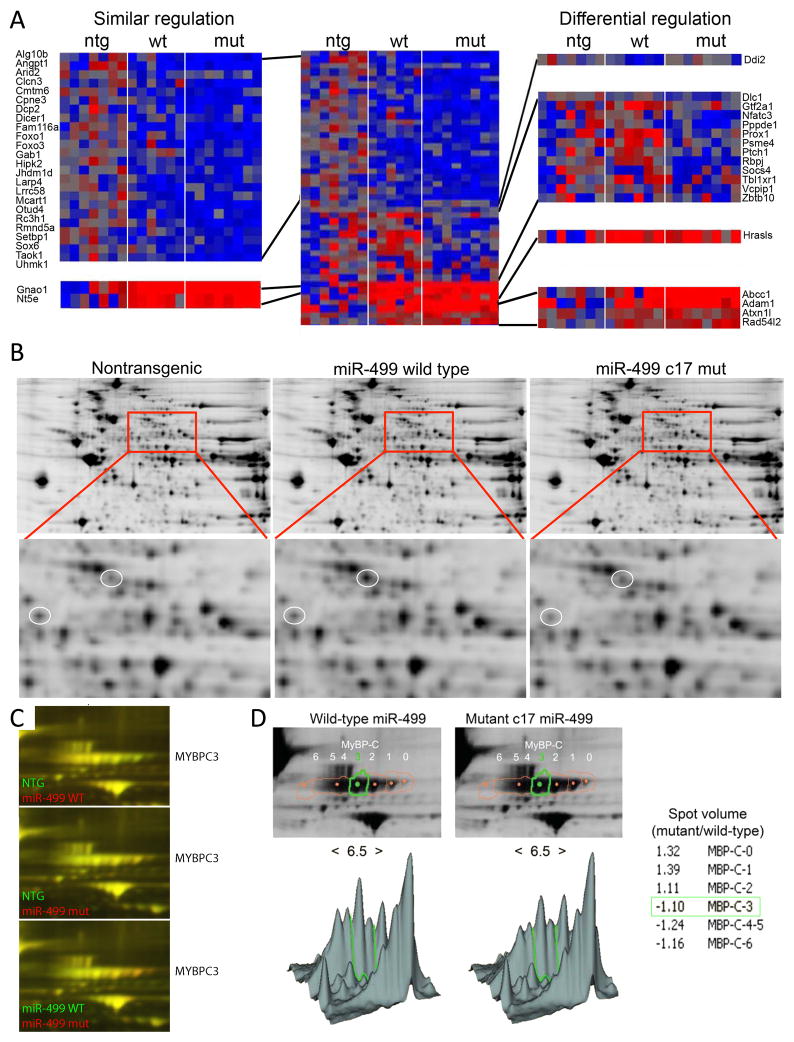
To better understand the relationship between phenotype and function in wild-type and mutant miR-499 mice, we used agnostic whole-genome approaches to compare cardiac mRNA targeting to the RISC, and to define primary and secondary effects of the miR-499 c17 mutant on steady-state cardiac mRNA levels 14, 23. The mRNA content of mouse heart RISCs genetically programmed with wild-type or c17 mutant miR-499 was assayed in 4 week old mice, at an age when heart function is still normal (thus avoiding potentially confounding effects of the age-dependent cardiomyopathy). A total of 109 mRNAs were enriched in miR-499 programmed cardiac RISCs, representing cardiac-expressed mRNAs targeted by miR-499. Of these, 42 were preferentially targeted by c17 mutant miR-499, 8 were preferentially targeted by wild-type miR-499, and 59 showed no preference (Online Table II). Unsupervised hierarchical clustering of RISC-enriched miR-499 target mRNAs was >80% accurate in grouping the mice according to genotype (Figure 4a). Gene-ontology analysis showed that both wild-type and mutant c17 miR-499 mRNA targets segregated into functional categories linked to cell signaling and transcriptional regulation (Figure 4b).
Figure 4. The miR-499 c17 mutation modifies in vivo RISC targeting of cardiac mRNAs.
(A) Unsupervised hierarchical clustering of non-transgenic, wild-type, and mutant miR-499 RISC-associated mRNAs. Each column represents a single mouse heart, each row a separate transcript (annotated on right margin). Unsupervised hierarchical clustering used Euclidean distance with average linkage. Non-transgenics are indicated by green squares at top, wild-type miR-499 by blue squares, and mutant miR-499 c17 by red squares. RISC score was 81% accurate in grouping the mice according to genotype, and 86% accurate in differentiating between wild-type miR-499 and mutant miR-499 c17-expressing hearts. Blue shading, lower RISC scores (RISCome content / transcriptome content); red shading, higher RISC scores. (B) Gene Ontology classification of miR-499 RISC-enriched mRNAs. Upper: 109 miR-499 mRNA targets. Bold type indicates over-represented categories. Lower: Functional classification of miR-499 target mRNAs that did not discriminate between wild-type miR-499 and mutant miR-499 (on the left) or that showed a preference for mutant miR-499 (on the right).
miRs act primarily by destabilizing their mRNA targets, thus decreasing steady-state mRNA levels 24, 25. Twelve target mRNAs were downregulated preferentially by mutant miR-499 c17, 1 mRNA was downregulated preferentially by wild-type miR-499, and twenty-four miR-499 target mRNAs were decreased by both (Figure 5a). A small number of miR-499 RISC-targeted mRNAs showed increased steady-state levels, as described for atypical miR-target interactions 26 (Figure 5a). Concordance of mRNA downregulation and RISC enrichment by the respective miR-499 was 68% for wild-type miR-499 and 94% for miR-499 c17 hearts.
Figure 5. The c17 miR-499 mutation alters cardiac mRNA and protein content.
(A) Heatmap of cardiac transcriptome mRNA levels for 114 miR-499 target mRNAs. Left: expanded view of 24 downregulated mRNAs and 2 upregulated mRNAs in both wild-type and mutant miR-499 c17 hearts. Upper right: mRNAs downregulated only by wild-type miR-499 or mutant miR-499 c17; lower right: mRNAs upregulated only by wild-type miR-499 or mutant miR-499 c17. (B) Representative (of 5) 2D DiGE (2D differential in-gel electrophoresis) images showing staining for nontransgenic and wild-type miR-499 cardiac protein (left); nontransgenic and mutant miR-499 c17, right; expanded view shows regulation of specific protein spots. (C) Expanded view of MyBP-C protein species. Highly phosphorylated MyBP-C migrates to the left (lower pH). (D) Location of MyBP-C spots quantitated using DeCyder software (GE Healthcare). Lower panels show the density of each spot in 3D view, with low pH (greater phosphorylation) to the left and high pH to the right (pH 6.5 at the center). Ratios of spot volume show that less MyBP-C phosphorylation is present in mutant miR-499 c17 hearts.
miR-499 c17 mutation alters myocardial protein levels
The major function of myomiRs is regulating myosin isoform levels in response to stress or hormonal factors 6, 7. Yet, neither of the myomiR-regulated myosin heavy chain mRNAs is a direct target of any myomiR. This is an example of indirect targeting, and reflects secondary and tertiary consequences of miRs acting their direct mRNA targets 20. We used two-dimensional differential in-gel electrophoresis (DiGE) 27 of mouse myocardium to examine the proteomes of ~8 week old wild-type and mutant c17 miR-499 expressing hearts, in comparison to non-transgenic controls. Although DiGE is an agnostic measure of cardiac protein content, it preferentially examines the more highly expressed portion of the proteome and cannot detect poorly-expressed direct miR targets 20. Of 56 regulated spots identified by mass spectroscopy, 41 proteins were co-regulated by wild-type and mutant miR-499, 9 proteins were preferentially regulated by wild-type miR-499, and 6 proteins were preferentially regulated by mutant c17 miR-499 (Figure 5b, Online Table III).
miR-499 has been implicated as an indirect regulator of protein phosphorylation by suppressing the phosphatase calcineurin 28. We did not find direct or indirect regulation of calcineurin mRNA in either wild-type or mutant miR-499 programmed hearts. However, we did find evidence of indirect effects on myocardial phosphoprotien content, providing mechanistic insight into the salutary effects of the c17 mutation on the cardiomyopathy induced by increased miR-499 expression. Myosin binding protein C (MyBP-C) is a potent modifier of cardiac contractility; its effects are regulated through phosphorylation by several cardiac protein kinases 29. MyBP-C phosphorylation, indicated as a leftward shift toward lower pH 27, is increased in wild-type miR-499 expressing hearts, but not in mutant c17 miR-499 expressing hearts (Figures 5c and 5d). Increased MyBP-C phosphorylation is a biochemical signature of cardiac pathology 27. While protein kinases and phosphatases were not identified as direct miR-499 targets, the alpha subunits of G13 and Go and other cell signaling factors are (Online Table II), and modulate downstream protein kinase activity 30. Thus, differential effects of mutant c17 miR-499 on its direct and indirect mRNA targets can explain observed differences in protein expression, processing, and on end-organ phenotype.
The structural basis for differential mRNA targeting by wild-type and c17 mutant miR-499
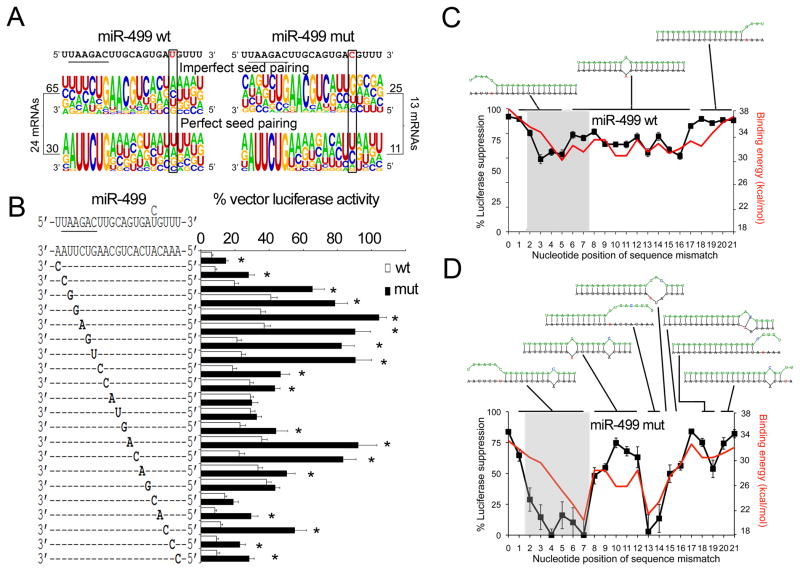
To better grasp the consequences of miR sequence variation on miR-mRNA interactions, we defined the characteristics of miR-499 binding sites within its antithetically regulated mRNA targets. Approximately one third of these sites exhibited perfect miR-499 seed sequence complementarity (Figure 6a); the influence of nt17 on miR-499 target binding was greater in the majority of mRNA sites wherein seed sequence pairing was imperfect 3, 31, with g corresponding to nt17 in 56% of miR-499 c17 target sites, versus 20% of wild-type miR-499 u17 target sites (P=0.002; Figure 6a, top row). By comparison, nt17 had little impact on target sites having perfect seed sequence complementarity (Figure 6a, bottom row). Thus, the probability of miR-499/mRNA pairing is interactively determined by sequence complementarity at both ends of the duplex.
Figure 6. miR-499 c17 mutation uncovers interactive intra-molecular sequence determinants of mRNA targeting.
(A) Motif plot of proportional nucleotide composition in miR-499 binding sites of 24 non-preferential miR-499 target mRNAs (left), comprising 95 sites, and 13 preferential miR-499 c17 mRNAs (right), comprising 36 sites. miR-499 sequences are at top, with seed sequences underlined. Sites were segregated into sites with imperfect seed sequence pairing (top) and perfect seed sequence pairing (bottom). Rectangles indicate nucleotide 17. (B) Luciferase reporter activities of miR-499 binding site mutations. Suppression by wild-type miR-499 (white bars) or mutant miR-499 c17 (black bars), is shown relative to control transfection for each binding site mutation. Mutations were engineered as a↔ c or g↔ u to avoid g:u base-pairing. * denotes significant difference between wild-type and mutant miR-499 suppression. (C, D) Molecular structures of miR-499 and target mRNA duplexes determined by modeling RNAhybrid binding energy predictions (red line, right axes) to observed luciferase reporter suppression (black line, left axes). Luciferase suppression (the inverse of data in panel B) is plotted over a range of 0–100%, and minimum free energy of hybridization (mfe) between −18 and −38 kcal/mol, both as a function of target site mutation for wild-type miR-499 (u17; left) or mutant miR-499 (c17, right); gray shading indicates miR-499 seed sequence. Optimal miR-499-mRNA duplex structures for target site mutants are schematically depicted above the plots; the green strand is miR-499 and black strand is mRNA target.
To understand the sequence determinants of miR-499 target recognition we engineered a complete series of miR-499 target site mutations and compared efficiency of target suppression by wild-type and c17 mutant miR-499 (Figure 6b). Target suppression by wild-type miR-499 was most compromised by nucleotide mismatches at the 5′ end of the miR, affecting nt 3 through 5 of the seed sequence. Single target site mismatches corresponding to nt 6–16 in the mid-region of the miR-mRNA duplex retained ~70% of wild-type suppression, and target site mutations complementary to the 5′ end of miR, between nt 17 and 21, had little effect on target suppression (Figure 6b, white bars). In contrast, target site mismatches corresponding to nt 2 through 7 severely impaired or completely abrogated target suppression by the c17 miR-499 mutant, revealing functional cooperativity between 5′ and 3′ nucleotides in the miR-mRNA duplex. Target site mismatches complementary to miR-499 nt 13 and 14 also eliminated target suppression by the c17 miR-mutant (Figure 6b, black bars). By comparison, sequence mismatches in the mid-region of the duplex, between nt 8 and 12, and those corresponding to miR-499 nt 18 through 21, retained a high level of target suppression by mutant miR-499 c17 (Figure 6b, black bars). Co-expression of wild-type and mutant c17 miR-499, which simulates heterozygosity in the human subjects wherein the miR-499 mutant was detected, induced intermediate functional phenotypes (Online Figure II).
Molecular modeling of miR-mRNA hybrid binding energy to target suppression revealed structural diversity of the RNA duplexes that explains differential mRNA targeting by the miR-499 c17 mutant (Figures 6c–d and Online Figure III). Single base-pair mismatches within 4 nucleotides of either end of the wild-type miR-499-mRNA duplex disrupt one end of the miR-mRNA hybrid, effectively truncating the miR while otherwise maintaining continuous miR-mRNA binding (Figure 6c); single base-pair mismatches at any position between nucleotides 6 and 17 produce a localized bulge in the duplex (Figure 6c). The c17 miR-499 mutation altered the miR-mRNA interaction in two ways: The cooperative interaction between 3′ and 5′ base pairs within the duplex exaggerates disruption of miR-mRNA binding when there is a target site mismatch corresponding to nt 2–7 (Figure 6d), and target site mismatches within four nt 5′ or two nt 3′ of the mutant base produce atypical miR-mRNA duplex configurations that adversely impact target suppression (Figure 6d).
Discussion
Here, we provide evidence that naturally-occurring miR sequence variation can broadly affect miR function, even when the nucleotide mismatch is located far away from the critical 5′ seed sequence generally considered to drive mRNA target recognition and suppression. Using high-throughput next-generation sequencing, we identified a human mutation in mature miR-499 that alters its mRNA targeting profile and end-organ effects, enhancing contractile function. By comparing myocardial protein levels on two dimensional gels, we identified numerous changes in protein expression induced by both wild-type and mutant miR-499 and uncovered differences in protein mobility/phosphorylation induced by the mutant that may reflect its favorable impact on cardiac contraction. Finally, we took advantage of this naturally occurring mutation to interrogate the structural determinants of miR-mRNA pairing. To our knowledge these studies are the first to establish a functional consequence of 3′ miR sequence variation and to demonstrate (at least in the mouse model) disease modification by a non-seed sequence miR mutation.
The most important aspect of this work is the finding that mature miR sequence variation outside of the “seed” sequence can have meaningful biological effects. Our results show that nucleotide variation in the 3′ end of a miR (or of the corresponding mRNA binding site) has less impact when miR and binding site seed sequence complementarity are perfect or nearly so. There is increasing evidence for non-seed sequence miR binding (through “centered pairing” sites for example32), and our results show that only approximately one third of in vivo miR-499 mRNA targets have perfect seed sequences. Thus, 3′ miR sequence variants have the potential to modify the effects of a miR, while retaining its primary functional characteristics. This observation may deserve consideration in human genetic screening as well as when designing long or short miR-targeting molecules.
The conventional approaches of defining mRNA targets of miRs reasonably uses bioinformatics to define a population of candidate mRNA targets, based on sequence complementarity. We found that this approach was not useful to direct comparative studies of wild-type and c17 mutant miR-499 because their seed sequences are identical, and current miR target analysis software emphasizes seed sequence complementarity. Indeed, analysis with the popular TargetScan platform did not predict any differences in wild-type and mutant miR-499 targets. Because our previous RISC-enrichment studies of mouse hearts programmed with miR-133 had revealed a large number of miR-targets lacking perfect seed sequences that were not identified as targets by bioinformatics 33, here we used unbiased RISC profiling to compare the direct mRNA targets of wild-type and mutant miR-499. Whereas many mRNAs were recruited to cardiac RISCs by both miR-499s, a minority of RISC-enriched mRNAs showed a preference for one or the other miR-499. Post-hoc analysis of complementary sequences in the 3′UTRs of these biologically-defined preferential targets revealed the role of binding site complementation corresponding to the mutant miR nucleotide in selective targeting.
Another novel aspect of these studies is incorporation RNA duplex structure information into the data analysis. Our results show how miR-mRNA binding energy, mRNA suppression, and miR-mRNA duplex structure are inter-related. The interactions between 5′ seed and 3′ non-seed sequence mismatches we identified were not predicted by RNA hybrid software, and were identified only by modeling structure and binding energy to luciferase suppression data. Translating these types of in vitro reporter data to the in vivo condition will require not only extrapolation of information about RNA duplex configuration, but also on secondary mRNA 3′UTR structure that may either limit or facilitate binding of individual mRNA binding sites to miR/RISCs. These considerations are particularly well illustrated by the 3′UTR structure of miR-499 binding domains within the thoroughly validated mRNA target, Sox-6 (Online Figure IV).
In conclusion, we describe mechanisms for, and consequences of, altered mRNA targeting by a rare 3′ human miR-499 mutant. Because miR-499 is highly expressed in cardiac tissue, is upregulated in human heart failure, and is sufficient to cause heart failure in mice, we postulated that misregulation of even a fraction of normal miR-499 targets by the c17 mutant might significantly impact cardiac function. The only known prior example linking functional miR mutations to a human disorder are seed-sequence mutations of miR-96 in familial hearing loss 34. The current results suggest that non-seed sequence miR mutations may act as disease modifiers by altering the pattern of mRNA suppression for targets lacking perfectly complementary seed sequences. Our findings emphasize the need to comprehensively and agnostically define miR-mRNA interactions and miR-regulated protein expression in proper biological context, accounting for endogenous mRNA expression patterns and cooperativity with other regulatory factors. The insights derived from in vivo studies of miR-mRNA interactions in relevant tissues, and from integrating molecular modeling with biological data, provide a different perspective on the consequences of miR mutations and suggest opportunities for better defining the individual genetic determinants of disease and future miR-based therapeutics 1, 35.
Supplementary Material
Novelty and Significance.
What is known?
MicroRNAs suppress their target mRNAs by recruiting them to RNA-induced silencing complexes outside the nucleus.
MicroRNAs recognize their mRNA targets through complementary nucleotide sequences in critical “seed” regions (nucleotides 2–8).
Human microRNA sequence variation is rare, suggesting evolutionary suppression.
What new information does this article contribute?
We discovered a rare human miR-499 mutation at nucleotide 17 (of 21), well outside of the seed region thought to determine miR-mRNA pairing.
Compared to wild-type miR-499, the mutant reduced in vitro suppression of known miR-499 targets, altered in vivo recruitment of cardiac mRNAs to RNA-induced silencing complexes, and modified the cardiac phenotype.
The structural basis for dysfunction induced by the miR-499 mutation is unstable miR-mRNA duplex configurations.
Summary.
Here, we describe and characterize a new mutation in the cardiac-expressed microRNA, miR-499. MicroRNA mutations are rare in available human genome databases. We postulated that this rarity relates to the primary function of microRNAs, which is to bind to specific target mRNAs and orchestrate their destruction. Altered nucleotide sequence would alter miR-mRNA binding, and alter the profile of mRNA targeting. Since microRNAs target many different mRNAs, and each of these mRNA products can have secondary and tertiary end-organ effects, the consequences of microRNA mutations are potentially quite broad. On the other hand, if miR-mRNA interactions are driven largely by 5′ seed sequences (nt 2–8) (as is commonly believed), then only seed sequence mutations should have significant effects. The current studies provide the first example of a non-seed sequence human microRNA mutation that alters normal microRNA function. We show that the substitution of wild-type u17 for c17 alters in vivo cardiac mRNA recognition, we reveal that differential mRNA recognition is driven by binding site complementarity corresponding to miR nt17, and we describe the broad and unpredictable consequences of this mutation. These findings should stimulate further discovery of human miR mutations and define a comprehensive platform for their analysis.
Acknowledgments
Sources of Funding
Supported by NIH/NHLBI R01 HL108943, RC2 HL102222, and by a Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Program.
Non-standard abbreviations and acronyms
- miR
microRNA
- RISC
RNA-induced silencing complex
- DiGE
differential in-gel electrophoresis
Footnotes
Disclosures
The authors declare that they have no conflicts of interest relating to this manuscript.
References
- 1.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorn GW., 2nd Decoding the Cardiac Message. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.111.256768. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 8.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal Regulation of Myocardial microRNAs and Messenger RNA in Human Cardiomyopathy and Reversal of the microRNA Signature by Biomechanical Support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE, Dorn GW., 2nd Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.112.265736. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PLos One. 2011;6:e19481. doi: 10.1371/journal.pone.0019481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappola TP, Matkovich SJ, Wang W, van Booven D, Li M, Wang X, Qu L, Sweitzer NK, Fang JC, Reilly MP, Hakonarson H, Nerbonne JM, Dorn GW., 2nd Loss-of-function DNA sequence variant in the CLCNKA chloride channel implicates the cardio-renal axis in interindividual heart failure risk variation. Proc Natl Acad Sci USA. 2011;108:2456–2461. doi: 10.1073/pnas.1017494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., 2nd Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW., 2nd Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq. Circ Res. 2010;106:1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 20.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol. 2010;30:1937–1945. doi: 10.1128/MCB.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW, 2nd, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 26.Buchan JR, Parker R. Molecular biology. The two faces of miRNA. Science. 2007;318:1877–1878. doi: 10.1126/science.1152623. [DOI] [PubMed] [Google Scholar]
- 27.Kang MY, Zhang Y, Matkovich SJ, Diwan A, Chishti AH, Dorn GW., 2nd Receptor-independent cardiac protein kinase Calpha activation by calpain-mediated truncation of regulatory domains. Circ Res. 2010;107:903–912. doi: 10.1161/CIRCRESAHA.110.220772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 29.James J, Robbins J. Signaling and myosin-binding protein C. J Biol Chem. 2011;286:9913–9919. doi: 10.1074/jbc.R110.171801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu J, Vaiskunaite R, Suzuki N, Kozasa T, Carr DW, Dulin N, Voyno-Yasenetskaya TA. Interaction of heterotrimeric G13 protein with an A-kinase-anchoring protein 110 (AKAP110) mediates cAMP–independent PKA activation. Curr Biol. 2001;11:1686–1690. doi: 10.1016/s0960-9822(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang WC, Juan AH, Panebra A, Liggett SB. MicroRNA let-7 establishes expression of beta2-adrenergic receptors and dynamically down-regulates agonist-promoted down-regulation. Proc Natl Acad Sci USA. 2011;108:6246–6251. doi: 10.1073/pnas.1101439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW., 2nd MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 35.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.