Abstract
Natural antimicrobial peptides (AMPs) are promising candidates for developing a generation of new antimicrobials to meet the challenge of antibiotic-resistant pathogens such as meticillin-resistant Staphylococcus aureus (MRSA). To facilitate the search for new candidates, we have utilised the Antimicrobial Peptide Database (APD), which contains natural AMPs from bacteria, fungi, plants and animals. This study demonstrates the identification of novel templates against MRSA by screening 30 peptides selected from the APD. These peptides are short (<25 residues), cysteine-free, cationic and represent candidates from different biological sources such as bacteria, insects, arachnids, tunicates, amphibians, fish and mammals. Six peptides, including ascaphin-8, database-screened antimicrobial peptide 1 (DASamP1), DASamP2, lycotoxin I, maculatin 1.3 and piscidin 1, were found to exert potent antimicrobial activity against an MRSA USA300 isolate. Although five of the six peptides showed broad-spectrum antibacterial activity, DASamP1 displayed killing of MRSA in vitro but not of Escherichia coli, Bacillus subtilis or Pseudomonas aeruginosa. In addition, DASamP1 suppressed early biofilm formation in a mouse model of catheter-associated MRSA infection. DASamP1 is a novel, short and potent peptide that will be a useful starting template for further developing novel anti-MRSA peptides.
Keywords: Antimicrobial peptides, Biofilms, Meticillin-resistant Staphylococcus aureus
1. Introduction
Meticillin-resistant Staphylococcus aureus (MRSA) USA300 represents a clade of genetically related strains that are a major cause of skin and soft-tissue infections in the hospital as well as the community settings in otherwise healthy individuals [1]. The annual frequency of deaths from MRSA is rapidly increasing and has surpassed those caused by human immunodeficiency virus/acquired immune deficiency syndrome (HIV/AIDS) [2–4]. [For US Centers for Disease Control and Prevention (CDC) estimated annual deaths from AIDS during 2001–2005 in the USA, refer to Table 7 at the following website: http://www.cdc.gov/hiv/surveillance/resources/reports/2005report/]. Therefore, there is an urgent need to develop new treatments against MRSA. Naturally occurring antimicrobial peptides (AMPs) are universal host defence molecules that have retained their potency throughout the years [5,6]. The Antimicrobial Peptide Database (APD) (http://aps.unmc.edu/AP/main.html) has collected more than 1900 AMPs from bacteria, fungi, plants and animals as of January 2012 [7]. These peptides display narrow- or broad-spectrum activity against bacteria, fungi, viruses and/or parasites. Some are also able to modulate immune responses. Because of multiple mechanisms of action, including membrane disruption, it is difficult and rare for bacteria to develop resistance to AMPs [5–7]. Thus, such peptides represent promising foundations for developing a new generation of antimicrobial agents [8].
To identify potent AMP templates against MRSA, a group of 30 peptide candidates (Table 1) from the APD [7,9] was screened for antimicrobial activity in vitro. One of the most promising peptides was also subject to in vivo testing. These peptides were chosen based on the following properties. First, the peptides are short (<25 amino acid residues) and do not contain cysteines, allowing cost-effective chemical synthesis. Second, they possess a net positive charge since cationic peptides are ideal to target bacteria with negatively charged surfaces. Third, these peptides are representative candidates across diverse biological sources, including bacteria, insects, arachnids, tunicates, amphibians, fish and mammals. To increase peptide efficacy, some variants were also generated by increasing positively charged amino acid residues in natural peptide templates. Fourth, and more importantly, their effects on MRSA USA300 have not yet been evaluated.
Table 1.
Antibacterial activity of 30 antimicrobial peptides against Staphylococcus aureus USA300 LAC and Escherichia coli K12
| Name (source)a | Peptide sequenceb,c | MIC (μM)
|
|
|---|---|---|---|
| S. aureus | E. coli | ||
| Apidaecin IA (insect) | GNNRPVYIPQPRPPHPRI | >100 | >100 |
| Ascaphin-8 (frog) | GFKDLLKGAAKALVKTVLF-NH2 | 3.1 | 12.5 |
| Brevinin-2-related (frog) | GIWDTIKSMGKVFAGKILQNL-NH2 | 25 | 25 |
| Buforin II (toad) | TRSSRAGLQFPVGRVHRLLRK | >71 | >71 |
| Clavanin-B (tunicate) | VFQFLGRIIHHVGNFVHGFSHVF | >100 | >100 |
| DASamP1 (synthetic)d | FFGKVLKLIRKIF-NH2 | 3.1 | >100 |
| DASamP2 (synthetic)e | IKWKKLLRAAKRIL-NH2 | 6.2 | 3.1 |
| Desertcolin 1 (frog) | GLADFLNKAVGKVVDFVKS-NH2 | >100 | >100 |
| Distinctin chain 1 C23R (synthetic)f | NLVSGLIEARKYLEQLHRKLKNRKV | >92 | >92 |
| Drosocin (insect)g | GKPRPYSPRPTSHPRPIRV | >100 | >100 |
| Hyposin-5 (frog) | FRPALIVRTKGTRL | >30 | >30 |
| Isracidin (cow) | RPKHPIKHQGLPQEVLNENLLRF | >100 | >100 |
| Latarcin 3a (spider) | SWKSMAKKLKEYMEKLKQRA | >100 | >100 |
| Lycotoxin I (spider) | IWLTALKFLGKHAAKHLAKQQLSKL | 3.1 | 25 |
| Maculatin 1.3 (frog) | GLLGLLGSVVSHVVPAIVGHF-NH2 | 6.2 | >100 |
| Mastoparan M (insect) | INLKAIAALAKKLL | >100 | 25 |
| Melectin (insect) | GFLSILKKVLPKVMAHMK-NH2 | 12.5–25 | 12.5–25 |
| Metalnikowin I (insect) | VDKPDYRPRPRPPNM | >100 | >100 |
| Misgurin (fish) | RQRVEELSKFSKKGAAARRRK | >84 | >84 |
| Parasin I (fish) | KGRGKQGGKVRAKAKTRSS | >84 | >84 |
| PGLa (frog) | GMASKAGAIAGKIAKVALKAL-NH2 | 25 | 25 |
| Piscidin 1 (fish) | FFHHIFRGIVHVGKTIHRLVTG | 3.1 | 12.5 |
| Plantaricin chain A (bacteria)h | GAWKNFWSSLRKGFYDGEAGRAIRR | >38.9 | >38.9 |
| Ponericin L2 (insect) | LLKELWTKIKGAGKAVLGKIKGLL | 85 | >25 |
| Pseudin-1 (frog) | GLNTLKKVFQGLHEAIKLINNHVQ | >100 | 100 |
| Ranatuerin 9 (frog) | FLFPLITSFLSKVL | 50 | >100 |
| Spinigerin (insect) | HVDKKVADKVLLLKQLRIMRLLTRL | >81 | >81 |
| Styelin A (tunicate) | GFGKAFHSVSNFAKKHKTA-NH2 | >100 | >100 |
| Temporin-LTc-3r (synthetic)i | SLSRFLRFLKIVYRRAF-NH2 | 12.5 | 25 |
| Uperin 7.1KRF (synthetic)j | GWFDVVKHIAKRF-NH2 | 50 | 12.5 |
Six peptides identified to be active against S. aureus and studied further are indicated in bold.
Peptide sequences were obtained from the Antimicrobial Peptide Database [7], and mutated residues are shown in bold.
C-terminal amidation is represented by NH2.
A peptide mutant of temporin-PTa with S4K, P10R and L13F mutations. In contrast to peptides from natural sources (e.g. frogs, fish), those labelled with ‘synthetic’ are man-designed based on natural templates.
A peptide mutant of polybia-MPI with the following mutations: D2K, D8R and Q12R.
The sequence of this peptide corresponds to chain A of distinctin with residue C23 changed to R.
Residue T11 is not O-glycosylated.
The sequence of this peptide corresponds to chain A of plantaricin JK.
A mutant of temporin-LTc with three mutations: S7R, P14R and P15R.
This peptide was obtained by changing the last three residues SAV of uperin 7.1 to KRF.
2. Materials and methods
2.1. Peptides
All peptides used in this study were chemically synthesised and purified to >95% (Genemed Synthesis Inc., San Antonio, TX), with peptide quality verified by reverse-phase high-performance liquid chromatography (HPLC) prior to use. Retention times of the peptides on a C8 column were also obtained from the HPLC chromatograms as detailed elsewhere [10]. Peptide concentrations were determined by ultraviolet spectroscopy.
2.2. Antimicrobial peptide activity in vitro
The four bacterial strains used in this study to determine AMP efficacy included the Gram-positive strains S. aureus USA300 LAC (a community-associated strain isolated from the Los Angeles County jail) [1] and Bacillus subtilis 168 as well as the Gram-negative isolates Escherichia coli K12 and Pseudomonas aeruginosa PAO1. The antimicrobial activity of peptides was evaluated using a standard broth microdilution protocol as described previously [11] and was repeated on different dates. In brief, logarithmic phase bacterial cultures [i.e. optical density at 600 nm (OD600) of ca. 0.5] were diluted to OD600 = 0.001 and were partitioned into a 96-well polystyrene microplate with ca. 105 colony-forming units (CFU)/well (90 μL aliquots). After treatment with 10 μL of peptide solution at various concentrations, microplates were incubated at 37 °C overnight and were read on a ChroMate®-4300 Microplate Reader (GMI, Ramsey, MN) at 630 nm. The minimal inhibitory concentration (MIC) was defined as the lowest peptide concentration that fully inhibited bacterial growth.
2.3. Haemolytic effects of peptides on human red blood cells in vitro
Haemolytic analysis of selected peptides was performed using an established protocol [12]. Briefly, blood cells [University of Nebraska Medical Center (UNMC) Blood Bank] were washed three times with phosphate-buffered saline (PBS) and were diluted to a 5% solution. Following peptide treatment, incubation at 37 °C for 1 h and centrifugation at 13 000 rpm, aliquots of the supernatant were transferred to a fresh 96-well microplate. To assess peptide cytotoxicity, the amount of haemoglobin released was measured at 545 nm. Percent lysis was calculated by assuming 100% release when human blood cells were treated with 2% Triton X-100 and 0% release when they were incubated with PBS buffer.
2.4. Staphylococcus aureus biofilm infection model
Biofilm infection was performed as described previously [13–15]. Male C57BL/6 mice (6–8 weeks old) were obtained from Charles River Laboratories (Frederick, MD). All animal procedures were in accordance with the National Institutes of Health ‘Guidelines for the care and use of laboratory animals’ and were approved by the UNMC Animal Care and Use Committee. Briefly, mice were anaesthetised with tribromoethanol (Avertin) and the skin was shaved and scrubbed with povidone–iodine. A small subcutaneous incision was made in the left flank and a blunt probe was used to create a pocket for insertion of a sterile 14-gauge Teflon catheter 1 cm in length (Exel International, St Petersburg, FL). The incision was sealed using Vetbond Tissue Adhesive (3M, St Paul, MN) and 1000 CFU of MRSA USA300 LAC::lux in 20 μL of sterile PBS was slowly injected through the skin into the infected catheter lumen. The health status of the mice was monitored throughout the infection procedure to ensure animal welfare, and mice found to be injured or moribund were immediately euthanised.
2.5. Peptide treatment of Staphylococcus aureus biofilms and bacterial enumeration
Animals were initially treated with 200 μg of peptide injected into the catheter at the time of infection (time 0), followed by 200 μg of peptide injected at four different sites surrounding the catheter at 24 h and 48 h post infection. Mice were euthanised by an overdose of inhaled isoflurane at Days 3 and 14 post infection to quantitate bacterial burdens associated with catheters and the surrounding tissues. Catheters were removed using aseptic technique, immediately placed in 1 mL of PBS and the biofilm was subsequently dissociated from the intraluminal and extraluminal surfaces using a sonicator. The tissue surrounding the catheter was also removed, weighed and homogenised in 500 μL of homogenisation buffer [25 mL of PBS supplemented with 100 μL of RNasin® (Promega, Madison, WI)] and one Roche Protease Inhibitor Tablet (Basel, Switzerland) using a Polytron® 1200E Tissuemizer® (Kinematica AG, Littau/Lucerne, Switzerland). Bacterial titres associated with catheters and surrounding tissues were quantified on tryptic soy agar plates supplemented with 5% sheep blood (HemoStat Laboratories, Dixon, CA). Previous evaluation of tissues surrounding infected catheters by scanning electron microscopy (SEM) revealed biofilm formation with tower structures that emulate those observed during in vitro biofilm growth conditions inside the catheter lumen [15]. Gram staining also revealed evidence of biofilm growth within the catheter lumen as well as at the catheter–tissue interface, and further examination by SEM verified a contiguous bacterial layer at the interface of the catheter and surrounding host tissue. Collectively, these findings demonstrate the ability to establish S. aureus biofilm infections in vivo. Significant differences between experimental groups were determined using an unpaired two-tailed student’s t-test in GraphPad Prism 4 (GraphPad Software Inc., La Jolla, CA). For all analyses, a P-value of <0.05 was considered statistically significant.
3. Results
3.1. Minimal inhibitory concentrations of antimicrobial peptides
For initial in vitro screens, the antimicrobial activities of 30 selected peptides against S. aureus USA300 and E. coli were evaluated in this study (Table 1). Six peptides were identified to be active against S. aureus with MICs ranging from 3.1 μM to 6.2 μM. Only two of these six peptides (DASamP1 and maculatin 1.3) failed to kill E. coli up to a concentration of 100 μM. Lycotoxin I has known activity against Gram-negative E. coli and yeast [16]; however, here we demonstrate activity of this peptide against Gram-positive S. aureus. Prior to this study, there were no antibacterial data available for DASamP1 (database-screened AMP1), its wild-type peptide temporin-PTa [17] or DASamP2.
Therefore, the in vitro activity of these six peptides against B. subtilis and P. aeruginosa was also evaluated (Table 2). Whilst ascaphin-8, DASamP2, lycotoxin I and piscidin 1 showed activity both against B. subtilis and P. aeruginosa, maculatin 1.3 was active against B. subtilis but not P. aeruginosa. Interestingly, DASamP1 was not active against B. subtilis or P. aeruginosa. Taken together, we conclude that ascaphin-8, DASamP2, lycotoxin I and piscidin 1 are broad-spectrum AMPs that are active against all of the Gram-positive and Gram-negative organisms tested here. In contrast, maculatin 1.3 is active against the two Gram-positive but not the two Gram-negative bacterial strains tested, whilst DASamP1 is active only against S. aureus but not other bacteria examined in this study (Tables 1 and 2).
Table 2.
Antimicrobial and haemolytic activities of the six anti-meticillin-resistant Staphylococcus aureus peptides identified in this study
| Peptide | MIC (μM)
|
HL50 (μM)a | CSb | Net chargec | Pho%c | RT (min)d | |
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | Pseudomonas aeruginosa | ||||||
| Ascaphin-8 | 6.25 | 12.5 | 45 | 15 | +4 | 55 | 10.08 |
| DASamP1 | >100 | >100 | 25 | 8 | +5 | 61 | 12.83 |
| DASamP2 | 12.5 | 6.25 | 75 | 12 | +7 | 57 | 10.73 |
| Lycotoxin I | 6.25 | 25 | 125 | 40 | +7 | 50 | 10.94 |
| Maculatin 1.3 | 25 | >100 | 25 | 4 | +3 | 54 | 15.15 |
| Piscidin 1 | 12.5 | 50 | 20 | 7 | +7 | 43 | 14.68 |
MIC, minimal inhibitory concentration.
Concentration at which 50% of red blood cells were lysed.
Cell selectivity (CS) of the peptide against S. aureus USA300 LAC was calculated as HL50/MIC (MICs in Table 1).
Net charge and hydrophobic residues% (pho%) were calculated using the Antimicrobial Peptide Database [7].
High-performance liquid chromatography retention time (RT) measured as described previously [10].
3.2. Haemolytic effects of anti-meticillin-resistant Staphylococcus aureus inhibitory peptides
The cytotoxicity of the six peptides selected (ascaphin-8, DASamP1, DASamP2, lycotoxin I, maculatin 1.3 and piscidin 1) was determined and reported as the concentration causing 50% blood cell lysis (HL50). The haemolytic activity of these six peptides was ranked in the following order: piscidin 1 > DASamP1 ~ maculatin 1.3 > ascaphin-8 > DASamP2 > lycotoxin I. The selectivity indexes of the six peptides were calculated by taking the ratio between HL50 and the MIC of the peptide against S. aureus USA300 LAC (Table 2). Lycotoxin I was observed to have the highest selectivity index of 40. Ascaphin-8, DASamP1, DASamP2 and piscidin 1 had a selectivity index of ca. 10. The selective index of lycotoxin I is comparable with the best reported in a recent article [18].
3.3. Correlation of peptide activity with physical properties
To understand better the sequence–activity relationships, peptide parameters were obtained from the APD [7] and HPLC retention times were measured [10] for the six selected peptides (Table 2). Retention time provides one approach for the measurement of peptide hydrophobicity [10]. The correlation between activity (MICs for B. subtilis, E. coli, P. aeruginosa, S. aureus or haemolytic HL50) of the six peptides (Tables 1 and 2) and their properties [net charge, hydrophobic content (pho%) or HPLC retention times] (Table 2). Whilst poor correlations were found in most instances (correlation coefficients 0.003–0.42), the antibacterial activity of the six peptides against P. aeruginosa displayed a moderate correlation with HPLC retention times (correlation coefficient 0.6).
3.4. DASamP1 impacts Staphylococcus aureus biofilm establishment in vivo
DASamP1 was chosen for further efficacy testing in vivo because this novel peptide is short, easy to synthesise and displayed bacteria species-specific activity. Bacterial specificity is an important consideration in the search for novel antimicrobials to prevent unwanted cidal activity against probiotic bacteria. The in vivo efficacy of DAPamP1 against MRSA USA300 LAC was examined using a mouse model of catheter-associated biofilm infection as previously described [13–15]. Animals were initially treated at the time of infection (time 0) followed by additional peptide administration at 24 h and 48 h post infection. Bacterial titres associated with the catheter and surrounding host tissue were evaluated at Day 3 (Fig. 1A,B) and Day 14 post infection (Fig. 1C,D) to determine the impact of peptide treatment on bacterial burdens. Mice treated with DASamP1 exhibited a significant decrease in bacterial burdens associated with biofilm-infected catheters (Fig. 1A,C) as well as in surrounding tissues (Fig. 1B,D) compared with vehicle-treated mice. Importantly, early peptide treatment was key to preventing S. aureus biofilm establishment, since minimal bacterial growth was detected at Day 14 following infection even though the last dosing of DASamP1 occurred at 48 h (Fig. 1C,D).
Fig. 1.
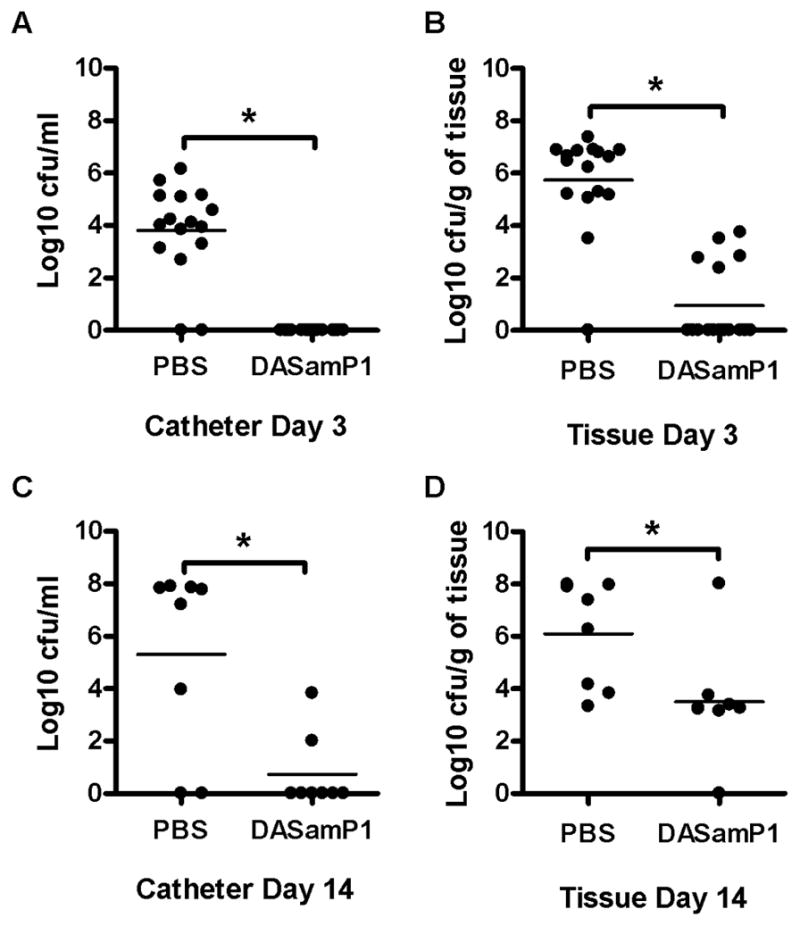
Database-screened antimicrobial peptide 1 (DASamP1) impacts Staphylococcus aureus biofilms in vivo. C57BL/6 mice (Day 3, n = 16/group; Day 14, n = 8/group) were infected with 103 colony-forming units (CFU) of MRSA USA300 LAC::lux in the lumen of surgically implanted catheters to establish biofilm infection. Animals received DASamP1 injections at 0, 24 and 48 h following infection and were sacrificed at Day 3 (A,B) or Day 14 (C,D) following S. aureus exposure, whereupon catheters (A,C) and surrounding host tissues (B,D) were recovered to quantitate bacterial burdens. Results are expressed as the number of CFU/mL for catheters or CFU/mg tissue in order to correct for differences in tissue sampling size. * Significant difference in bacterial burden between phosphate-buffered saline (PBS)- and peptide-treated mice (P < 0.05). Results presented from individual animals in two independent experiments, with bars representing the mean of each group.
4. Discussion
To date, S. aureus remains one of the major causes both of healthcare- and community-associated infections [1,3]. With the emergence of drug-resistant strains in the 1960s, primarily MRSA, this ubiquitous pathogen has become an even greater therapeutic challenge and, at present, MRSA strains account for >50% of all S. aureus strains causing nosocomial infections. Accordingly, the interest and need for novel therapeutic modalities against MRSA has been growing worldwide. Host defence peptides such as AMPs are naturally occurring universal signalling and effector molecules important during the host immune response, and engineered peptides derived from these molecules have the potential to overcome the increasing antibiotic resistance seen in many pathogens [5,6,8].
In the current study, 30 peptides were selected from the APD [7] for their ease of chemical synthesis, net positive charge and source diversity and were tested against the pathogens S. aureus and E. coli. Antibacterial assays of the 30 peptides here led to the identification of six potent anti-MRSA peptides (Table 2). The range of MICs of these six AMPs varied from 3.1 μM to 6.2 μM against S. aureus and from 3.1 μM to >100 μM against E. coli. These six peptides with the greatest efficacy against these pathogens only showed significant haemolysis at concentrations higher than those required to kill S. aureus effectively, indicating that these peptides are capable of selectively targeting bacterial membranes without lysing mammalian cells. These same six peptides also showed varied efficacy against B. subtilis and P. aeruginosa. The peptide DASamP1 was the most promising, with a selective ability to kill S. aureus but not E. coli, B. subtilis or P. aeruginosa or to lyse mammalian erythrocytes. Furthermore, it demonstrated effective antimicrobial capabilities and antibiofilm activity in vivo. Collectively, these findings demonstrate that DASamP1 provides an effective means to prophylactically reduce MRSA colonisation and biofilm formation on artificial surfaces in vivo. The growing prevalence of nosocomial infections, increased use of procedures that can give rise to biofilm development, and the difficulties of treating/controlling S. aureus within a biofilm support the use of AMPs and improved analogues as a means to treat biofilm infections.
Here we demonstrate that DASamP1, an easily produced synthetic AMP, provides a convenient and inexpensive yet effective way to prophylactically reduce bacterial burdens in S. aureus biofilms associated with catheters and other artificial implants. Bacteria within a biofilm are not responsive to conventional antibiotic treatment and are physically protected from the cellular and molecular components of host immunity by the biofilm matrix, making immunotherapy/vaccines problematic. As a result, the only therapeutic option for dealing with biofilms developing around an artificial device is a cycle of removal and replacement—an inconvenient, ineffective and undesirable option. These difficulties of biofilm treatment/control are underscored even further when it is associated with more permanent artificial implants such as hips, knees and heart valves. The fact that DASamP1 was demonstrated to suppress biofilm formation in vivo provides a possible treatment in those implanted areas.
5. Conclusions
This study demonstrates that potent anti-MRSA peptides can be identified based on database screening. By evaluating 30 peptides, 6 potent anti-MRSA candidates were found, including ascaphin-8, DASamP1, DASamP2, lycotoxin I, maculatin 1.3 and piscidin 3. These six compounds (Table 2) represent excellent first templates for developing a new generation of antimicrobials against MRSA USA300 isolates that are problematic both in the hospital and community settings. In particular, DASamP1 is a novel peptide that is easy to synthesise and displayed potent antimicrobial activity against MRSA both in vitro and in vivo, opening the door to an alternative treatment for implant patients.
Acknowledgments
Funding
This work was supported by the Most Promising New Invention Award from UNeMED of the University of Nebraska Medical Center (Omaha, NE) and by R56AI081975 from the National Institute of Allergy and Infectious Disease (National Institutes of Health) to GW, and P01 AI083211 Project 4 to TK.
Footnotes
Competing interests
None declared.
Ethical approval
This study was approved by the University of Nebraska Medical Center Animal Care and Use Committee (Omaha, NE).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105:1327–32. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bancroft EA. Antimicrobial resistance: it’s not just for hospitals. JAMA. 2007;298:1803–4. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- 3.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Stein R. The Washington Post. Oct 17, 2007. Drug-resistant staph germ’s toll is higher than thought. [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellullar organisms. Nature. 2002;415:359–65. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–8. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37(Database issue):D933–7. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G. Antimicrobial peptides: discovery, design and novel therapeutic strategies. Wallingford, UK: CABI; 2010. [Google Scholar]
- 9.Wang G, Watson KM, Peterkofsky A, Buckheit RW., Jr Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob Agents Chemother. 2010;54:1343–6. doi: 10.1128/AAC.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Elliott M, Cogen AL, Ezell EL, Gallo RL, Hancock REW. Structure, dynamics, and antimicrobial and immune modulating activities of human LL-23 and its single residue variants mutated based on homologous primate cathelicidins. Biochemistry. 2012;51:653–64. doi: 10.1021/bi2016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Li Y, Li X. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J Biol Chem. 2005;280:5803–11. doi: 10.1074/jbc.M410116200. [DOI] [PubMed] [Google Scholar]
- 12.Malmsten M, Kasetty G, Pasupuleti M, Alenfall J, Schmidtchen A. Highly selective end-tagged antimicrobial peptides derived from PRELP. PLoS One. 2011;6:e16400. doi: 10.1371/journal.pone.0016400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassat JE, Lee CY, Smeltzer MS. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol. 2007;391:127–44. doi: 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–32. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–96. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L, Adams M. Lycotoxins, antimicrobial peptides from venom of the wolf spider Lycosa carolinensis. J Biol Chem. 1998;273:2059–66. doi: 10.1074/jbc.273.4.2059. [DOI] [PubMed] [Google Scholar]
- 17.Conlon JM, Kolodziejek J, Nowotny N, Leprince J, Vaudry H, Coquet L, et al. Characterization of antimicrobial peptides from the skin secretions of the Malaysian frogs, Odorrana hosii and Hylarana picturata (Anura:Ranidae) Toxicon. 2008;52:465–73. doi: 10.1016/j.toxicon.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Dawson RM, Fox MA, Atkins HS, Liu CQ. Potent antimicrobial peptides with selectivity for Bacillus anthracis over human erythrocytes. Int J Antimicrob Agents. 2011;38:237–42. doi: 10.1016/j.ijantimicag.2011.05.006. [DOI] [PubMed] [Google Scholar]


