Abstract
Formaldehyde-fixed, paraffin-embedded (FFPE) tissue repositories represent a valuable resource for the retrospective study of disease progression and response to therapy. However, the proteomic analysis of FFPE tissues has been hampered by formaldehyde-induced protein modifications, which reduce protein extraction efficiency and may lead to protein misidentification. Here, we demonstrate the use of heat augmented with high hydrostatic pressure (40,000 psi) as a novel method for the recovery of intact proteins from FFPE mouse liver. When FFPE mouse liver was extracted using heat and elevated pressure, there was a 4-fold increase in protein extraction efficiency, a 3-fold increase in the extraction of intact proteins, and up to a 30-fold increase in the number of nonredundant proteins identified by mass spectrometry, compared to matched tissue extracted with heat alone. More importantly, the number of nonredundant proteins identified in the FFPE tissue was nearly identical to that of matched fresh-frozen tissue.
Keywords: FFPE, formalin-fixed paraffin-embedded, high-pressure protein extraction, mouse liver, mass spectrometry, heat and pressure, proteomics
Introduction
Proteomics has emerged as a valuable tool for the identification of biomarkers associated with human disease1 including those resulting from very subtle changes in normal cell functions and signaling pathways. Proteomic technology has advanced to a state where thousands of proteins can be identified within complex samples,2−6 yet disease-based studies using fresh or frozen tissues are limited by a lack of available specimens for longitudinal clinical investigations. In contrast, there are millions of archival formalin-fixed, paraffin-embedded (FFPE) tissues for which the clinical course of disease and response to therapy has been established. FFPE tissue studies are affected greatly by sample collection, tissue processing, and archival time.7 Though these factors are difficult to control in archival samples, improvements in techniques such as mTRAQ have made relative quantitation of disease biomarkers in FFPE tissue possible.8,9 Improving protein extraction and detection of less abundant protein biomarkers is also of critical importance in proteomic analysis. Protein modifications by formaldehyde treatment and histological processing10,11 significantly limit the use FFPE tissues for proteomic analyses. This has prevented proteomic studies of the clinical course of diseases, such as prostate and breast cancer, that evolve slowly or where the time between treatment and recurrence is long. Coupling proteomic investigations with the retrospective pathology information available from archival FFPE tissues would produce a wealth of practical information on human diseases.
A variety of methods for profiling FFPE tissue have been employed recently. Some are practical for slide-mounted FFPE tissue, such as quantitative fluorescence imaging analysis (QFIA), which is reproducible and sensitive for specific standardized proteins,12−14 or MALDI-imaging mass spectrometry (MS).15−17 Other encouraging mass spectrometry (MS)-based proteomic studies of FFPE tissues have appeared in the recent literature;2−7,9,18−20 however, these investigations have typically been restricted to minute tissue specimens, such as those obtained by laser capture microdissection. Further, some studies report high rates of false-positive protein identification and are limited to the analysis of tryptic digests of FFPE tissues by liquid chromatography–MS (LC–MS).
Our laboratory has been studying the reactions of formaldehyde with proteins and ways to reverse these reactions.21 Using proteins in aqueous solution, we demonstrated that the majority of protein formaldehyde adducts and cross-links were consistently reversed with mild heating following the removal of excess formaldehyde by dialysis.22 We then developed a tissue surrogate, which consists of one or more proteins that form a gel-like plug when treated with formaldehyde at protein concentrations exceeding 75 mg/mL. These tissue surrogates have sufficient physical integrity to be processed using normal histological methods.23 A variety of extraction buffers and heating protocols were examined for their ability to recover proteins from tissue surrogates. Protein recovery was generally modest, and studies with multiprotein tissue surrogates revealed extraction bias, meaning that the composition of the solubilized proteins did not match that of the corresponding tissue surrogate. Subsequent studies showed that the ethanol dehydration step of histology caused most formaldehyde-treated proteins to adopt conformations enriched in β-sheets, leading to the formation of protein aggregates where the β-sheets form a dense network of intermolecular formaldehyde cross-links.11 We proposed that the difficulty of rehydrating these stabilized protein aggregates was the primary impediment to recovering proteins from FFPE tissue.
Pressure promotes water penetration into the inner core of proteins, causing denaturation, whereas heat alone causes protein unfolding followed by aggregation.24 Consequently, we hypothesized that the combined effects of heat and elevated pressure would facilitate the rehydration of the highly cross-linked protein aggregates in FFPE tissues. The increased exposure to water should greatly improve protein solubilization while simultaneously promoting the reversal of protein–formaldehyde adducts and cross-links. Initial physical studies on tissue surrogates supported this hypothesis25 and suggested that the effect of pressure was to reduce the size of protein aggregates through increased water penetration, rather than to increase the rate of reversal of protein–formaldehyde adducts and cross-links directly.26
Encouraged by these results, we extracted FFPE mouse liver tissues with heat augmented by elevated hydrostatic pressure, with the goal of reducing extraction bias, improving the recovery of intact proteins, and obtaining tryptic digests that more closely resemble those from matched fresh-frozen tissue. This manuscript describes these efforts.
Materials and Methods
Materials
Tissue Tek OCT compound was purchased from Sakura, USA, Amicon Ultra 3K centrifugal filters were purchased from Millipore, and 37% formaldehyde and Pierce Detergent Removal columns were obtained from Thermo Fisher. Precast NuPAGE Bis-Tris 4–12% gels, 2-(N-morpholino)ethanesulfonic acid–SDS running buffer, and the SilverQuest staining kit were purchased from Life Technologies. All other reagents were purchased from Sigma-Aldrich unless stated otherwise.
High-Pressure Instrumentation
Tissue extracts were heated under a pressure of 40,000 psi (276 MPa) using both home-built and commercial instruments. The home-built instrument consisted of a 2-mL capacity MS-1 stainless steel reaction vessel coupled to a manually operated high pressure piston screw pump available from High Pressure Equipment Company (Erie, PA). The temperature of the pressure vessel was regulated by a Eurotherm 2132 temperature controller (Leesburg, VA) connected to an aluminum heating collar surrounding the reaction vessel. The tissue extract (2 mL) was added directly to the reaction vessel using a syringe. The construction and operation of this pressure system has been described in detail previously.25,26 The commercial instrument was a model NEP 2320 barocycler (Pressure Biosciences) modified by the manufacturer to hold isobaric pressure and to provide temperature control up to 95 °C. The tissue extract (2 mL) was added to a FT500 sample tube, which was capped and placed in the pressure vessel of the Barocycler. The proteomic analyses were independent of which pressure instrument was used.
Mouse Liver Histology
The liver from a female BALB/c mouse was obtained under the secondary use provision from the Department of Laboratory Animal Medicine of the Armed Forces Institute of Pathology (USA). Half of the liver was immediately frozen in Tissue-Tek OCT compound, divided into several equal-sized pieces, and stored at −80 °C. The other half was fixed for 48 h at 4 °C in 10% formalin. The fixed liver tissue was washed for 30 min with distilled water and dehydrated through a graded series of alcohols (70, 85, and 100% by volume) and two changes of xylene, 30 min each. The tissue was then embedded in paraffin using established histology protocols.25,27 The FFPE tissue block was stored at room temperature. For protein recovery experiments, 10 μm sections of FFPE mouse liver were cleared of paraffin by incubating the sections through two changes of xylene for 10 min each. The sections were rehydrated through a series of graded alcohols for 10 min each—2 changes each of 100% ethanol, 85% ethanol, and 70% ethanol—and then incubated in distilled water for a minimum of 30 min, as described previously.25 Matched fresh-frozen and FFPE liver tissue sections were analyzed after 30 days, and again after 1 year, of storage.
Pressure-Assisted Extraction of FFPE Tissue
The ability of elevated hydrostatic pressure combined with heat to improve the recovery of proteins from FFPE mouse liver tissue was evaluated using two heat-based FFPE proteomic protocols recently reported in the literature.2,3 For each protocol, the experimental procedure was followed exactly as published, but with the heating step divided into two arms, or variations. In the first arm, the FFPE tissue extract was heated at the temperature and for the length of time reported in the original method. In the second experimental arm, the temperature and length of time was identical, but the experiment was performed under a pressure of 40,000 psi. The two experimental arms were then analyzed using identical gel electrophoresis and LC/MS conditions. The protein content of the tissue extracts was determined using a BCA protein assay kit (Pierce).
Protocol-1
Five 10 μm sections were cut from the mouse liver FFPE tissue block. Following deparaffinization and rehydration, the tissue was cut into small pieces and suspended in 3–6 mL of 50 mM Tris-HCl, pH 7, 2% SDS (extraction buffer 1, EB1) as described by Shi et al.2 The tissue suspension was homogenized with three 5-s cycles of sonication on ice using a probe-tip sonicator, and the resulting homogenate was split into two equal fractions. One fraction (protocol-1A) was incubated at 100 °C for 30 min followed by 80 °C for 2 h at atmospheric pressure (14.7 psi) in a sand bath. The second fraction (protocol-1P) was incubated at 100 °C for 30 min followed by 80 °C for 2 h at 40,000 psi using our hand-built pressure instrument. A 750 μm section (approximate thickness) of matched fresh-frozen liver was cut with a razor blade and homogenized in 22.5 mL of EB1 by sonication as described above. Proteins were extracted using two methods: incubation of the homogenate in an ice bath for 2.5 h (protocol-1FI) or incubation at 100 °C for 30 min followed by 80 °C for 2 h at atmospheric pressure using a sand bath (protocol-1FH). Protocol-2. Five 10 μm sections were cut from the mouse liver FFPE tissue block. Following deparaffinization and rehydration, the tissue was cut into small pieces and suspended in 3–6 mL of 100 mM Tris-HCl, pH 8, 100 mM DTT, 4% SDS (extraction buffer 2, EB2) as described by Ostasiewicz et al.3 and homogenized with three 5-s cycles of sonication on ice using a probe-tip sonicator. One fraction (protocol-2A) was incubated at 95 °C for 1 h at atmospheric pressure in a sand bath. The second fraction (protocol-2P) was incubated at 95 °C for 1 h at 40,000 psi using the Barocycler instrument. A 750 μm section of matched fresh-frozen liver was cut with a razor blade and homogenized in 22.5 mL of EB2 by sonication as described above. Proteins were extracted by incubating the homogenate at 95 °C for 3 min at atmospheric pressure using a sand bath (protocol-2FH).
Preparation of Samples for MS
All FFPE and matched fresh-frozen mouse liver tissue extracts (40 μg each) were separated by SDS-PAGE on precast NuPAGE Bis-Tris 4–12% gels using 2-(N-morpholino)ethanesulfonic acid–SDS running buffer. The gels were stained using the SilverQuest silver staining kit and documented using an Epson V500 photoscanner and annotated in Adobe Photoshop, version 7.1. Each gel lane was then divided into 10 bands and placed into microcentrifuge tubes. In-gel tryptic digestion was carried out as previously described.25
Mass Spectrometry and Data Analysis
Separation of the digested peptides was performed using nanocolumns prepared in-house (75 μm i.d. packed with Jupiter C18 particles, 5 μm, 300 Ǻ) connected to an Agilent 1100 nanoflow LC system, which was used to deliver binary gradient solvents A (0.1% formic acid (FA) in water) and B (0.1% FA in acetonitrile). Reversed-phase chromatography was performed by solubilizing the lyophilized tryptic peptides in 10 μL of 0.1% trifluoroacetic acid and injecting 7 μL of sample per analysis. After sample injection, a 20-min wash with 98% mobile phase A was used to remove any remaining salts from the sample. Peptide elution was accomplished using a linear gradient of 2% solvent B to 42% solvent B over 45 min at a constant flow rate of 250 nL/min.
The nanoflow reversed-phase LC column was coupled online to a linear ion trap mass spectrometer (LTQ, ThermoElectron) using the manufacturer’s nanoelectrospray source with an applied electrospray potential of 1.75 kV and a capillary transfer tube temperature of 185 °C. The LTQ-MS was operated in a data-dependent mode where each full MS scan was followed by seven tandem MS scans in which the seven most abundant peptide molecular ions detected were dynamically selected for MS/MS analysis using a normalized CID energy of 35%. A dynamic exclusion of 60-s was applied to reduce redundant selection of peptides.
The MS/MS spectra were analyzed using SEQUEST (ThermoElectron) against a combined UniProt nonredundant mouse proteome database containing 36,799 protein sequences. Only peptides with conventional tryptic termini (allowing for up to two internal missed cleavages) possessing delta-correlation scores (ΔCn) > 0.08 and charge state-dependent cross-correlation (Xcorr) criteria as follows were considered as legitimate identifications: >1.9 for +1 charged peptides, >2.2 for +2 charged peptides, and >3.1 for +3 charged peptides. A reverse-database search, performed using the respective databases, resulted in a calculated false-positive rate of <2% for all samples analyzed.
Results and Discussion
Extraction of FFPE Mouse Liver: Electrophoresis
FFPE and matched fresh-frozen mouse liver tissue stored for 30 days were extracted using protocol-1. The fresh liver tissue was completely solubilized in the extraction buffer (EB1) using extraction either on ice (protocol-1FI) or at elevated temperature (protocol-1FH). The quantity of protein solubilized in the buffer was designated as 100% protein recovery in Table 1. FFPE mouse liver tissue extracted at atmospheric pressure (protocol-1A) resulted in a protein recovery of only 17%, and a large plug of remaining tissue was observed in the extraction vial. In contrast, when the extraction was performed at 40,000 psi (protocol-1P), the solubilized protein increased to 77% and only a small amount of tissue residue remained in the vial. FFPE and matching fresh-frozen liver tissue stored for 1 year were extracted using protocol-2. The fresh liver tissue was completely solubilized in the extraction buffer (EB2) when extracted at elevated temperature (protocol-2FH). The quantity of protein solubilized in the buffer was designated as 100% protein recovery in Table 1. The advantage of using elevated pressure was again evident, as 79% of the FFPE liver protein was solubilized at elevated hydrostatic pressure (protocol-2P) while only 18% was recovered at ambient pressure (protocol-2A).
Table 1. MS Analysis for FFPE and Matched Fresh-Frozen Mouse Liver Tissue Extracted at Atmospheric (14.7 psi) or Elevated Hydrostatic Pressure (40,000 psi)a.
| tissue | pressure (psi) | protocolb | extraction conditions | %protein extractiond | unique peptide IDs | unique protein IDs |
|---|---|---|---|---|---|---|
| frozen, 30 days | 14.7 | 1FI | on ice, 2.5 h | 100% | 10237 | 4727 |
| frozen, 30 days | 14.7 | 1FH | 100 °C + 80 °Cb | 100% | 9964 | 4581 |
| FFPE, 30 days | 14.7 | 1A | 100 °C + 80 °Cb | 17% | 5565 | 3449 |
| FFPE, 30 days | 40,000 | 1P | 100 °C + 80 °Cb | 77% | 9621 | 5192 |
| frozen, 1 year | 14.7 | 2FH | 95 °C, 3 min | 100% | 5872 | 3415 |
| FFPE, 1 year | 14.7 | 2A | 95 °C, 1 h | 18% | 107 | 107 |
| FFPE, 1 year | 40,000 | 2P | 95 °C, 1 h | 79% | 5180 | 3492 |
FFPE mouse liver was homogenized in extraction buffer and heated with or without elevated pressure. Fresh-frozen tissue was extracted either at atmospheric pressure using the indicated extraction condition or on ice for 2.5 h.
Protocol used for protein extraction (see Materials and Methods).
Tissue was heated at 100 °C for 30 min; then the temperature was lowered to 80 °C for 2 h.
The amount of protein extracted from fresh frozen tissue was set to 100%.
Previous studies using FFPE tissue surrogates containing several proteins23 indicated that failure to completely solubilize the tissue surrogate led to extraction bias, such that the composition of the solubilized protein solution differed significantly from that of the original surrogate. The extraction bias became less significant as the percentage of total protein solubilized from the tissue surrogate increased.23 Thus, the 4-fold improvement in protein solubilization realized when the extraction was performed at elevated pressure is likely to lead to a protein extract that more accurately represents the composition of the original FFPE tissue. Further, the increased amount of recovered protein allows a greater number of analytical techniques to be performed. While most published proteomic studies of FFPE tissue analyze only a few thousand cells from microdissected tissue,3−5,28 the use of elevated pressure has the advantage of improving protein extraction from whole tissue sections. This ability is particularly useful in instances where tissue microdissection is not practical or when a more global proteomic analysis is desired. Unbiased protein extraction and standardized protocols are particularly important with techniques such as reverse phase protein arrays (RPPAs), where the protein components of cell signaling pathways are quantified to direct clinical treatment of cancer.29
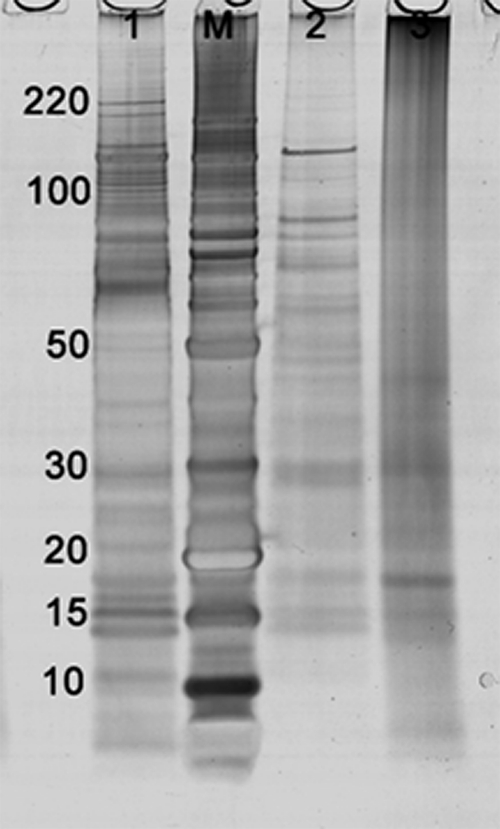
The 30-day-old FFPE mouse liver extracted at 40,000 psi (protocol-1P) exhibited a number of well resolved high and low molecular weight protein bands by SDS-PAGE, corresponding to ∼87% of those seen in the matched fresh-frozen tissue extracted on ice (Figure 1, lanes 2 and 1, respectively). The FFPE samples extracted at ambient pressure (protocol-1A) contained relatively few well-resolved protein bands equivalent to ∼25% of those seen in frozen mouse liver (Figure 1, lane 3). Similar results were seen with 1-year-old FFPE mouse liver extracted at 40,000 psi (protocol-2P) with well resolved high and low molecular weight protein bands corresponding to 74% of those seen in the matched fresh-frozen tissue (not shown). When the extraction was performed ambient pressure (protocol-2A) this value was reduced to 25%.
Figure 1.
1D SDS-PAGE of fresh-frozen and FFPE mouse liver extracts: lane 1, fresh-frozen tissue (protocol-1FI); lane M, molecular weight marker; lane 2, FFPE tissue extracted with heat at 40,000 psi (protocol-1P); lane 3, FFPE tissue extracted with heat alone (protocol-1A).
As top-down MS sequencing technology improves, the ability to extract and analyze intact proteins from FFPE tissue will become more important. Top-down sequencing facilitates the measurement of combinations of modifications, such as phosphorylation and glycosylation, and the direct quantitation of specific protein isoforms and splice variants. Few of these measurements are directly obtainable using bottom-up proteomic approaches in which proteins are digested into peptides. The ability to extract intact proteins from the seemingly inexhaustible source of FFPE tissues will increase the diagnostic and prognostic efficacy of proteomic-based biomarker discovery by allowing biomarker validation using orthogonal methods such as Western blotting, immunohistochemistry, immunoassays, and structural and interaction proteomics. In this context, the >3-fold increase in the recovery of intact proteins from FFPE tissue achieved by using elevated pressure represents a significant breakthrough.
Extraction of FFPE Mouse Liver: LC-MS/MS Analysis
FFPE and matched fresh-frozen liver tissue stored for 30 days were extracted using protocol-1. FFPE tissue was extracted at ambient pressure (protocol-1A) or 40,000 psi (protocol-1P), while the matched fresh-frozen liver tissue was extracted using protocol-1FI and protocol-1FH. The solubilized proteins were separated by 1D-PAGE, and each gel lane was excised, digested with trypsin, desalted, and analyzed by LC-MS/MS. The total unique peptide and protein identifications for each tissue type and extraction condition are shown in Table 1. FFPE tissue extracted with heat alone resulted in the identification of 5565 unique peptides and 3449 unique proteins. The addition of elevated hydrostatic pressure significantly improved both the number of unique peptides (9621) and proteins (5192) identified. The number of proteins identified from the high pressure-extracted sample was comparable to the number of unique proteins identified from fresh-frozen tissue, which ranged from 4932 for tissue extracted on ice to 4451 for frozen tissue extracted with heat (Table 1).
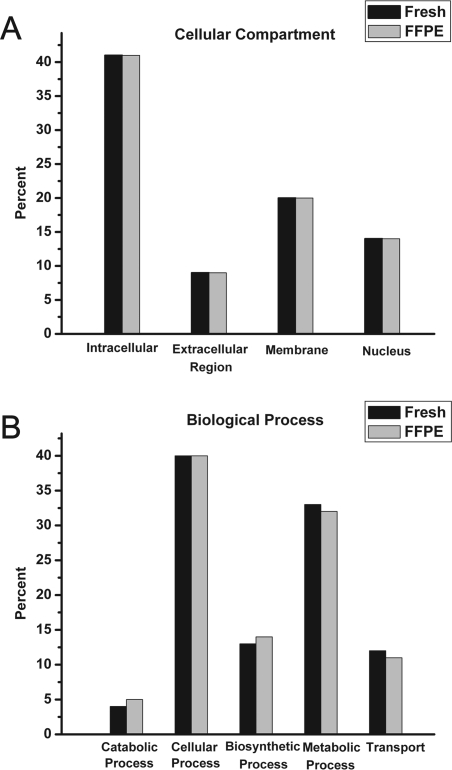
The MS results for the 30-day-old FFPE mouse liver extracted under elevated pressure (protocol-1P) and the matched fresh-frozen tissue extracted on ice (protocol-1FI) were searched using GOMiner, a gene ontology program. The identified proteins were categorized by their subcellular compartment and their biological function. The results of the gene ontology classification are shown in Figure 2. The percentages of nuclear, membrane, intracellular, and extracellular proteins identified in fresh-frozen and FFPE liver were virtually identical (Figure 2A), as were the results for classification by biological function (Figure 2B).
Figure 2.
Gene ontology analysis of proteins identified by LC-MS/MS. Proteins identified using fresh-frozen mouse liver (protocol-1FI) or FFPE liver extracted with heat and elevated pressure (protocol-1P) were categorized by subcellular localization (A) or biological process (B), using GoMiner gene ontology software.
To address the effect of long-term storage of the FFPE specimens, the mouse liver samples were investigated after an additional 11 months of storage (1-year-old sample). FFPE tissue was extracted at ambient pressure (protocol-2A) or 40,000 psi (protocol-2P), while the matched fresh-frozen liver tissue was extracted at high temperature (protocol-2FH). The solubilized proteins were separated by 1D-PAGE, and each gel lane was excised, digested with trypsin, desalted, and analyzed by LC-MS/MS. From the 1-D gel, we were able to identify 3492 nonredundant proteins in the 1-year-old FFPE liver extracted under elevated pressure, which was comparable to the 3415 nonredundant proteins identified in the matched fresh-frozen mouse liver extracted at high temperature. In contrast, only 107 unique proteins were identified in the FFPE tissue extracted at ambient pressure.
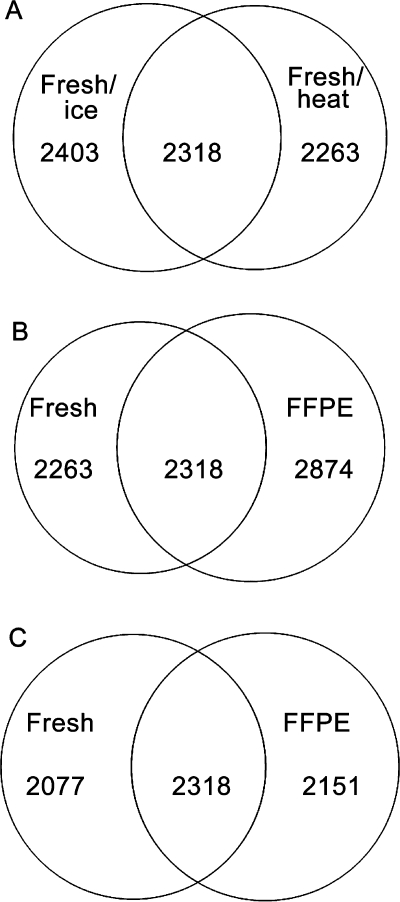
Figure 3 shows Venn diagrams of the unique and common (overlapping) proteins identified in mouse liver tissue extracts prepared by different methods. Figure 3A compares 30-day-old fresh-frozen liver tissue extracted on ice (protocol-1FI) versus 100 °C for 30 min followed by 80 °C for 2 h (protocol-1FH). Figure 3B compares 30-day-old FFPE liver tissue extracted under pressure (protocol-1P) versus matched fresh-frozen liver extracted at elevated temperature (protocol-1FH). Figure 3C compares 1-year-old FFPE liver tissue extracted under pressure (protocol-2P) versus matched fresh-frozen liver extracted at elevated temperature (protocol-2FH). Notably, the common proteins, expressed as a percentage of either the FFPE or matched fresh-frozen mouse liver tissue, were ∼50% for all three tissue pairs.
Figure 3.
Venn diagrams showing the number of unique and common proteins identified using LC MS/MS analysis: panel A, fresh-frozen tissue extracted on ice (protocol-1FI) or with heat (protocol-1FH); panel B, fresh-frozen tissue extracted with heat (protocol-1FH) and FFPE mouse liver extracted with elevated pressure (protocol-1P); panel C, fresh-frozen tissue extracted with heat (protocol-2FH) and FFPE mouse liver extracted with elevated pressure (protocol-2P).
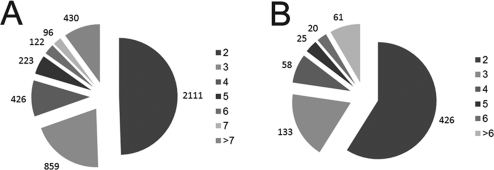
Figure 4A is a pie chart showing the number of unique proteins identified by two or more fully tryptic peptides for 30-day-old FFPE mouse liver extracted under pressure (protocol-1P). The results reveal that 49% of the proteins were identified by two peptides, 51% were identified by three or more peptides, and 21% of the proteins were identified by five or more peptides. Figure 4B shows similar results for 1-year-old FFPE mouse liver extracted under pressure (protocol-2P), with 57% of the proteins identified by two peptides, 41% by three or more peptides, and 15% identified by five or more peptides. These results are similar to those for the matched fresh-frozen mouse liver tissue (not shown). The complete list of peptides identified in the fresh-frozen and high-pressure-recovered FFPE tissue, with their corresponding Xcorr values, can be found online (Supporting Information Table 1).
Figure 4.
Total number of unique proteins identified using LC-MS/MS by two or more unique, fully tryptic peptides in FFPE mouse tissue extracted with heat and elevated pressure (40,000 psi): A, 30-day-old FFPE mouse liver (protocol-1P); B, 1-year-old FFPE mouse liver (protocol-2P).
Conclusion
Virtually all protocols reported in the literature for the extraction of proteins from FFPE tissue use a variation of the heat-induced antigen retrieval technique developed by Shi and Taylor30 for the recovery of antigenicity in immunohistochemical studies. This method involves exposing FFPE tissue sections to a buffer solution containing a detergent and/or protein denaturant and elevated temperatures of 90–120 °C for a period of 10–30 min.31 Optimal antigen recovery varies with the host FFPE tissue, with each type requiring different buffers, pH values, buffer additives, and incubation temperatures.32,33 Although the FFPE proteomic literature is still quite limited, it is not unreasonable to propose that the same situation applies to the proteomic analysis of FFPE tissues. Elevated hydrostatic pressure is not a technique unto itself but rather an adjuvant method that can be applied to existing or future FFPE protein extraction protocols. While it remains to be shown that this method is useful with every tissue fixation and protein extraction protocol, elevated pressure acts through purely physical means26 and should be compatible with FFPE protein extraction buffers of any pH and containing any detergent, protein denaturant, or other additive.25 This should allow its integration into a wide range of protein extraction protocols for MS-based proteomics with little to no alteration to downstream sample preparation and analysis. This was demonstrated in this report by the successful application of elevated pressure to two very different published FFPE protein extraction protocols. An increase in pressure to 40,000 psi, to augment heat treatment, improved protein extraction efficiency from FFPE mouse liver tissue by approximately 4-fold and increased the number of unique proteins identified by up to 30-fold over the published methods used at ambient pressure. Further, the tryptic digests of these pressure-extracted tissues resulted in protein profiles that more closely resembled those from matched fresh-frozen tissue when analyzed by LC/MS than did those extracted with heat alone, while maintaining a false-identification rate of <2%. The ability of elevated pressure to significantly improve the recovery of intact proteins from FFPE tissues over the use of heat alone has great potential for broad application to top-down proteomic studies for the identification of disease biomarkers.
Acknowledgments
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, the Department of Defense, or the Veterans Health Administration, nor does the mention of trade names, commercial products, or organization(s) imply endorsement by the United States Government. The authors wish to thank Pressure Biosciences, Inc. for making a modified NEP 2320 barocycler available for these studies.
Supporting Information Available
Complete list of peptides identified in the fresh-frozen and high-pressure-recovered FFPE tissue, with their corresponding Xcorr values. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under grant 1R21 CA134359 (C.B.F.), and contract NO1-CO-12400 (T.D.V.), and the Veterans Health Administration under a Merit Review award (J.T.M. and T.J.O.). Carol B Fowler, Jeffrey T Mason, and Timothy J O'Leary are named as lead contributors in a patent application filed with the USPTO by the Veterans Health Administration and the Armed Forces Institute of Pathology.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Mallick P.; Kuster B. Proteomics: a pragmatic perspective. Nat. Biotechnol. 2010, 28 (7), 695–709. [DOI] [PubMed] [Google Scholar]
- Shi S.-R.; Liu C.; Balgley B. M.; Lee C.; Taylor C. R. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J. Histochem. Cytochem. 2006, 54 (6), 739–43. [DOI] [PubMed] [Google Scholar]
- Ostasiewicz P.; Zielinska D. F.; Mann M.; Wisniewski J. R. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J. Proteome Res. 2010, 9 (7), 3688–700. [DOI] [PubMed] [Google Scholar]
- Hood B. L.; Darfler M. M.; Guiel T. G.; Furusato B.; Lucas D. A.; Ringeisen B. R.; Sesterhenn I. A.; Conrads T. P.; Veenstra T. D.; Krizman D. B. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell. Proteomics 2005, 4 (11), 1741–53. [DOI] [PubMed] [Google Scholar]
- Patel V.; Hood B. L.; Molinolo A. A.; Lee N. H.; Conrads T. P.; Braisted J. C.; Krizman D. B.; Veenstra T. D.; Gutkind J. S. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin. Cancer Res. 2008, 14 (4), 1002–14. [DOI] [PubMed] [Google Scholar]
- Azimzadeh O.; Barjaktarovic Z.; Aubele M.; Calzada-Wack J.; Sarioglu H.; Atkinson M.; Tapio S. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J. Proteome Res. 2010, 9, 4710–4720. [DOI] [PubMed] [Google Scholar]
- Balgley B. M.; Guo T.; Zhao K.; Fang X.; Tavassoli F. A.; Lee C. S. Evaluation of archival time on shotgun proteomics of formalin-fixed and paraffin-embedded tissues. J. Proteome Res. 2009, 8 (2), 917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSouza L. V.; Krakovska O.; Darfler M. M.; Krizman D. B.; Romaschin A. D.; Colgan T. J.; Siu K. W. mTRAQ-based quantification of potential endometrial carcinoma biomarkers from archived formalin-fixed paraffin-embedded tissues. Proteomics 2010, 10 (17), 3108–16. [DOI] [PubMed] [Google Scholar]
- Xiao Z.; Li G.; Chen Y.; Li M.; Peng F.; Li C.; Li F.; Yu Y.; Ouyang Y.; Xiao Z.; Chen Z. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J. Histochem. Cytochem. 2010, 58 (6), 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonder J.; Veenstra T. D. Clinical proteomic applications of formalin-fixed paraffin-embedded tissues. Clin. Lab. Med. 2009, 29 (1), 101–13. [DOI] [PubMed] [Google Scholar]
- Fowler C. B.; O’Leary T. J.; Mason J. T. Modeling formalin fixation and histological processing with bovine ribonuclease A: Effects of ethanol dehydration on reversal of formaldehyde-induced cross-links. Lab. Invest. 2008, 88 (7), 785–91. [DOI] [PubMed] [Google Scholar]
- Huang D.; Casale G. P.; Tian J.; Lele S. M.; Pisarev V. M.; Simpson M. A.; Hemstreet G. P. III. Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer. Int. J. Cancer 2010, 126 (2), 315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.; Casale G. P.; Tian J.; Wehbi N. K.; Abrahams N. A.; Kaleem Z.; Smith L. M.; Johansson S. L.; Elkahwaji J. E.; Hemstreet G. P. III. Quantitative fluorescence imaging analysis for cancer biomarker discovery: application to beta-catenin in archived prostate specimens. Cancer Epidemiol. Biomarkers Prev. 2007, 16 (7), 1371–81. [DOI] [PubMed] [Google Scholar]
- Richardson T.; McCanse W.; Casale G. P.; Huang D.; Tian J.; Elkahwaji J. E.; Lele S.; Hemstreet G. P. Tissue-based quantification of 8-hydroxy-2′-deoxyguanosine in human prostate biopsies using quantitative fluorescence imaging analysis. Urology 2009, 74 (5), 1174–9. [DOI] [PubMed] [Google Scholar]
- Casadonte R.; Caprioli R. M. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 2011, 6 (11), 1695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronci M.; Bonanno E.; Colantoni A.; Pieroni L.; Di I. C.; Spagnoli L. G.; Federici G.; Urbani A. Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: application to MALDI-TOF imaging MS investigations. Proteomics 2008, 8 (18), 3702–14. [DOI] [PubMed] [Google Scholar]
- Wisztorski M.; Franck J.; Salzet M.; Fournier I. MALDI direct analysis and imaging of frozen versus FFPE tissues: what strategy for which sample?. Methods Mol. Biol. 2010, 656:303–22, 303–22. [DOI] [PubMed] [Google Scholar]
- Addis M. F.; Tanca A.; Pagnozzi D.; Crobu S.; Fanciulli G.; Cossu-Rocca P.; Uzzau S. Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics 2009, 9 (15), 3815–23. [DOI] [PubMed] [Google Scholar]
- Cheung W.; Darfler M.; Alvarez H.; Hood B. L.; Conrads T. P.; Habbe N.; Krizman D. B.; Mollenhaur J.; Feldmann G.; Maitra A. Application of a global proteomic approach to archival precursor lesions: Deleted in malignant brain tumors 1 and tissue transglutaminase 2 are upregulated in pancreatic cancers. Pancreatology 2008, 8, 608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto D. A.; Hood B. L.; Darfler M. M.; Guiel T. G.; Lucas D. A.; Conrads T. P.; Veenstra T. D.; Krizman D. B. Liquid Tissue: proteomic profiling of formalin-fixed tissues. Biotechniques 2005, 38 (Suppl), 32–5. [DOI] [PubMed] [Google Scholar]
- O’Leary T. J.; Fowler C. B.; Evers D. L.; Mason J. T. Protein fixation and antigen retrieval: chemical studies. Biotechnol. Histochem. 2009, 84 (5), 217–21. [DOI] [PubMed] [Google Scholar]
- Rait V. K.; O’Leary T. J.; Mason J. T. Modeling formalin fixation and antigen retrieval with bovine pancreatic ribonuclease A: I-structural and functional alterations. Lab. Invest. 2004, 84 (3), 292–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C. B.; Cunningham R. E.; O’Leary T. J.; Mason J. T. ″Tissue surrogates″ as a model for archival formalin-fixed paraffin-embedded tissues. Lab. Invest. 2007, 87 (8), 836–46. [DOI] [PubMed] [Google Scholar]
- Balny C.; Masson P.; Heremans K. High pressure effects on biological macromolecules: from structural changes to alteration of cellular processes. Biochim. Biophys. Acta 2002, 1595 (1–2), 3–10. [DOI] [PubMed] [Google Scholar]
- Fowler C. B.; Cunningham R. E.; Waybright T. J.; Blonder J.; Veenstra T. D.; O’Leary T. J.; Mason J. T. Elevated hydrostatic pressure promotes protein recovery from formalin-fixed, paraffin-embedded tissue surrogates. Lab. Invest. 2008, 88 (2), 185–95. [DOI] [PubMed] [Google Scholar]
- Fowler C. B.; Chesnick I. E.; Moore C. D.; O’Leary T. J.; Mason J. T. Elevated pressure improves the extraction and identification of proteins recovered from formalin-fixed, paraffin-embedded tissue surrogates. PLoS One 2010, 5 (12), e14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratthauer G. L. Processing of tissue specimens. Methods Mol. Biol. 2010, 588, 93–102. [DOI] [PubMed] [Google Scholar]
- Guo T.; Wang W.; Rudnick P. A.; Song T.; Li J.; Zhuang Z.; Weil R. J.; DeVoe D. L.; Lee C. S.; Balgley B. M. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J. Histochem. Cytochem. 2007, 55 (7), 763–72. [DOI] [PubMed] [Google Scholar]
- Berg D.; Hipp S.; Malinowsky K.; Bollner C.; Becker K. F. Molecular profiling of signalling pathways in formalin-fixed and paraffin-embedded cancer tissues. Eur. J. Cancer 2010, 46 (1), 47–55. [DOI] [PubMed] [Google Scholar]
- Shi S. R.; Key M. E.; Kalra K. L. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 1991, 39 (6), 741–8. [DOI] [PubMed] [Google Scholar]
- Mason J. T.; Fowler C. B.; O’Leary T. J.. Study of Formalin-Fixation and Heat-Induced Antigen Retrieval. In Immunohistochemistry & Antigen Retrieval Technique Based Research & Diagnostics; Taylor C., Shi S.-R., Eds.; Wiley-Blackwell: Hoboken, NJ, 2010; pp 253–286. [Google Scholar]
- Pileri S. A.; Roncador G.; Ceccarelli C.; Piccioli M.; Briskomatis A.; Sabattini E.; Ascani S.; Santini D.; Piccaluga P. P.; Leone O.; Damiani S.; Ercolessi C.; Sandri F.; Pieri F.; Leoncini L.; Falini B. Antigen retrieval techniques in immunohistochemistry: comparison of different methods. J. Pathol. 1997, 183 (1), 116–23. [DOI] [PubMed] [Google Scholar]
- D’Amico F.; Skarmoutsou E.; Stivala F. State of the art in antigen retrieval for immunohistochemistry. J. Immunol. Methods 2009, 341 (1–2), 1–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.