Abstract
Surface modification of silver island films (SIFs) was carried out with Biotin-Poly (Ethylene-glycol)-Amine (BEA), which acts as a cross-linker between the silver surface and horse radish peroxidase (HRP) enzyme for optimum plasmon-enhanced enzymatic activity. SIFs-deposited blank glass slides and SIFs-deposited 3-Aminopropyltriethoxysilane(APTES)-coated glass slides were used as our plasmonic surfaces.In this regard, three different extent of loading of SIFs were also prepared (low, medium and high) on APTES-coated glass slides. Streptavidin-linked HRP enzyme was attached to SIFs-deposited blank glass slides and SIFs-deposited APTES-coated glass slides through the well-known biotin-streptavidin interactions. The characterization of these surfaces was done using optical absorption spectroscopy. The loading of SIFs on glass slides was observed to have significant effect on the efficiency of plasmon-enhanced enzymatic activity, where an enhancement of 200% in the enzymatic activity was observed when compared to our previously used strategies for enzyme immobilization in our preceding work[1]. In addition, SIFs-deposited on APTES-coated glass slides were found to be re-usable for plasmon-enhanced enzymatic reactions unlike SIFs deposited on to blank glass slides.
Keywords: Plasmon-enhanced enzymatic reactions, surface plasmons, surface plasmon resonance, silver island films, gold nanoparticles, horse radish peroxidase, enzymatic activity
1. Introduction
In the last two decades, several new plasmonic technologies that employ metal nanoparticles and planar thin films for the detection of biomolecular interactions have emerged: surface plasmon resonance analysis, surface enhanced Raman scattering and surface plasmon enhanced fluorescence spectroscopy[2, 3].In addition, the combined used of enzymes with plasmonic structures was shown to enable us design new hybrid enzyme-nanoparticle systems for applications in bio- and nanotechnology. It was previously shown that plasmonic nanoparticles enhance the electro-catalytic response of a glucose oxidase system (glucose biosensor)[4] and the detection of various analytes of interest, such as hydrogen peroxide and uric acid [5], thiocholine[6], carcinoembryonic antigen [7] and DNA [8]. More recently, Aslan Research Group recently presented a detailed investigation of the dependence of enzymatic activity on the nanoparticle-enzyme distance and nanoparticle loading on planar surfaces based on the new platform technology called plasmon-enhanced enzymatic reactions (PEER). In this regard, three different SIFs (low, medium and high loading) on APTES-coated glass slides and unsilvered (blank) APTES-coated glass slides (control samples) were used for the comparison of three different enzyme immobilization strategies for PEER. Three different strategies: 1) a biotin-avidin protein assay (strategy 1), 2) self-assembled monolayer of hexamethylenediamine (strategy 2) and 3) poly-l-lysine layer (strategy 3) were used to vary the distance of the enzyme from the silver surface. It was found that up to an 200% increase in enzymatic conversion of organic substrate by horse radish peroxidase (HRP) was observed from SIFs with high using strategy 1 compared to control samples, providing direct evidence that plasmon-enhanced enzymatic activity is highly dependent on the enzyme-nanoparticle distance and the extent of loading of silver nanoparticles.
In this paper, we used a compound (BEA) that has terminal biotin groups, polymer chain and an amine terminal group for the surface modification of SIFs to optimize plasmon-enhanced enzymatic activity of HRP on SIFs. Since most biologically active compounds, such as enzymes are known to lose activity when linked directly to a solid hydrophobic surface, we hypothesized that the use of a hydrophilic molecule (such as BEA) that has a spacer arm poly-ethylene glycol may shield the enzyme from denaturation and improve bioactivity [9-11]. Poly-ethylene glycol is routinely used to attach immunogens to a solid surface, where one can observe a reduction in non-specific enzyme absorption and increased enzyme stability. These factors are seen to be critical in optimizing the specificity and sensitivity of bioanalytical assays [10, 11]. Poly-ethylene glycol has also been used as a spacer molecule for the improvement of peptide bioactivity [11], and antigen detection in biosensor [12]. In this paper, the comparison between the use of SIFs-deposited blank glass slides and SIFs-deposited-APTES-coated glass slides for optimization of PEER is described.The plasmon-enhanced enzymatic activity was followed by the colorimetric measurement of the product produced as a result of enzymatic conversion of o-phenylenediamine (OPD) on silvered surfaces. In addition, the effect of loading of SIFs onto APTES-coated glass slides (low, medium and high) as an optimization parameter was carried out.
Materials and Methods
Materials
Biotin-poly(ethylene-glycol)-amine (BEA) was obtained from Laysan Bio Inc. Sodium phosphate monobasic anhydrous was obtained from Fisher Scientific. Sodium hydroxide (99.99%), streptavidin-peroxidase from streptamycesavidinii (HRP-streptavidin), D-glucose, 3-Aminopropyltriethoxysilane (APTES) (99%), ammonium hydroxide (30%) and nitric acid (A.C.S reagent grade) were all obtained from Sigma-Aldrich. Culture well chambered cover glass (2 wells, 15 mm diameter, 2.0 mm deep) used for this work, were obtained from Electron Microscopy Sciences (Hatfield, PA). O-phe nylenediaminedihydrochloride (OPD) was obtained from Thermo-Scientific.Silver nitrate (A.C.S reagent grade) was purchased from Spectrum Chemical Corporation. Citric acid (analytical reagent grade) was purchased from Mallinckrodt Chemical Works. Sulfuric acid (A.C.S reagent grade) was purchased from Pharmco product Inc.All solutions were prepared using deionized water obtained from Millipore Direct Q3 system.
Methods
(1)Deposition of Silver Island Films (SIFs) on Glass Slides
The glass slides were first cleaned in piranha solution (70% sulfuric acid, 30% hydrogen peroxide). The slides were rinsed with deionized water, then kept to dry at room temperature. The cleaned glass slides were later immersed in an ethanolic solution containing 2% (3-aminopropyltriethoxysilane) APTES for 1 hour. The APTES-coated glass slides were rinsed with ethanol and allowed to dry at room temperature. The preparation and deposition of SIFs on to blank glass slides and to the APTES-coated glass slides at different loading (low, medium and high) was carried out as in the previously described procedure[1]. The characterization of the slides was done using UV-Visible spectrophotometer, where absorbance spectra of SIFs-deposited glass slide and SIFs-deposited APTES-coated glass slides were recorded, indicating the relative amount of silver nanoparticles to the surface of the glass slides.
(2)Preparation of Biotin-Poly(Ethylene-glycol)-Amine assay on SIFs and SIFs-deposited APTES-coated glass slides
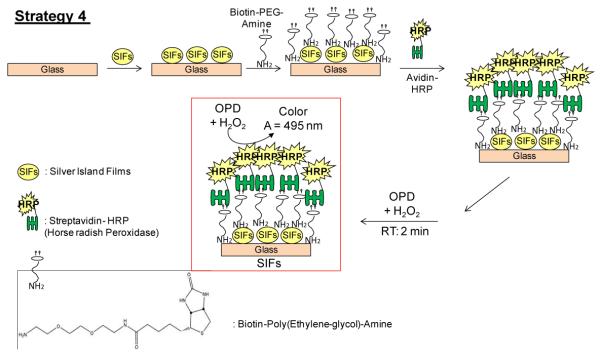
BEA, which has a biotin group attached to an ethylene-glycol core and a terminal amine surface group, was used for surface modification of SIFs surfaces, as shown in Scheme 1. BEA was prepared at a concentration of 0.5 mM using sodium phosphate monobasic buffer solution at pH 7.Culture well chambered cover glass was used to cover the SIFs-deposited glass slide and SIFs-deposited APTES-coated glass slides before the addition of 200 μL of BEA solution in the well chambers, the incubation was kept for 30 minutes. The slides were rinsed with deionized water to remove the unbounded BEA [Note: Incubation longer than 30 minutes will result in the rapid loss of SIFs on the surfaces of both glass slides].HRP-streptavidin (~200 μL) solution was subsequently added into the wells containing BEA-coated with SIFs-deposited glass slide and SIFs-deposited APTES-coated glass slides. The preparation of HRP-streptavidin was done as described previously [1]. The slides were then rinsed with deionized water to remove the unbounded HRP-streptavidin. HRP-streptavidin was used with BEA for the preparation of the monolayer by specific interaction with biotin group.
Scheme. 1.
Schematic depiction of Surface modification of SIFs with Biotin-PEG-Amine: HRP enzyme is deposited onto SIFs via Biotin-PEG-Amine / streptavidin interactions. Two different glass slides were used for the preparation of SIFs: blank glass slides and glass slides modified with 3-aminopropyltriethoxysilane, which contains terminal amine groups.
(3) Preparation of o-phenylenediaminedihydrochlor ide (OPD) solution
The preparation of OPD solution was carried outas described previously[1].The wavelength of the product obtained after the interaction of OPD with HRP-streptavidin was at 495 nm for all experiments.
Instrumentation
Cary 50 Bio UV-Visible spectrophoto-meter (Varian, Inc.) was used to obtain all absorption measurements. Real-color photographs of SIFs before and after use were taken by a 5 MP digital camera.
Results and Discussion
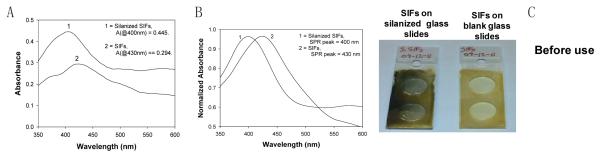
SIFs were deposited on blank glass slides and APTES-coated glass slides by using the method as described in our previous work[1]. Theseplasmonic surfaces were later characterized by measuring their absorbance spectrum. Fig. 1 shows the absorbance and normalized absorbance spectrum of the SIFs-deposited glass slide and SIFs-deposited APTES-coated glass slides. The maximum absorbance and wavelength recorded for SIFs-deposited glass slide and SIFs-deposited APTES-coated glass slides were (0.294, @430 nm) and (0.445, @400 nm) respectively(Fig. 1A). The normalized absorbance spectrumfor SIFs on silanized glass slides shows a blue shiftin the SPR peak and an increase in the surface plasmon resonance peak (SPR)despitethe fact that identical preparationprocedure was carried out for both surfaces (Fig. 1B). Fig. 1C shows the real color photograph of SIFs-deposited glass slides and SIFs-deposited APTES-coated glass slides.
Fig. 1.
(A): Absorption spectrum for glass slides modified with 3-aminopropyltriethoxysilane (SIFs-deposited APTES-coated glass slide) and SIFs-deposited blank glass slides. (B): Normalized Absorbance for both surfaces. (C): Real-color photographs of both surfaces.
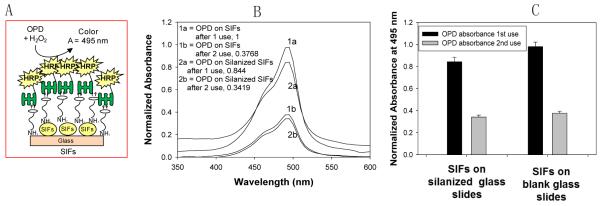
It is important to discuss the reasons for the selection of the surface modification strategy in this work before the presentation of the data. The selected molecule, BEA, is a water-soluble compound that contains a terminal primary amine, biotin and polyethylene glycol chain, which gives the BEA a long and flexible connection to minimize steric hindrance involved with binding to avidin molecules [13-15]. The terminal amine group is used for the chemisorption of BEA to SIFs, while the biotin group is used to attach HRP-streptavidin to SIFs surface. This assembly scheme places HRP~6 nm away from the silver surface (Fig. 2A). Since streptavidin is attached to the SIFs surface via the biotin groups on BEA, HRP is placed as the top layer of the assembly and involve in enzymatic conversion of OPD without steric hindrance from the molecules within the assembly.
Fig. 2.
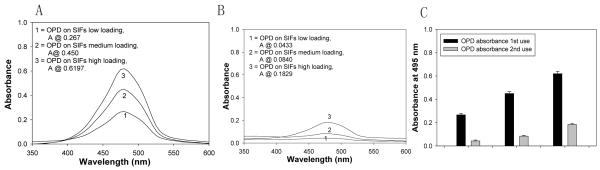
Schematic depiction of the new enzyme immobilization strategy employed in this study (A). Normalized absorbance spectrum of OPD at 495 nm (B) and (C) absorption of OPD on SIFs-deposited APTES-coated glass slide, and SIFs-deposited glass slide after 1st and 2nd use.
The enzymatic activity was studied by placing a solution of OPD on the enzyme-modified surfaces of the SIFs-deposited blank glass slides and SIFs-deposited APTES-coated glass slides. The conversion of OPD by HRP into a colored product (from colorless to light brown) was followed by measuring absorption spectrum of the initial and final OPD solution. In this regard, the colored solution was removed after ~2 mins of incubation and placed in a glass vial containing sulfuric acid to stop the conversion of OPD. The absorbance spectrum of the colored solution was then taken after two uses of the same surface. The absorption spectrum of OPD solution after 1st and 2nd use on both SIFs-deposited blank glass slides and SIFs-deposited APTES-coated glass slides is shown in Fig. 2B. The comparison of the enzymatic activity by both surfaces was carried out by plotting absorbance value at 495 nm after 1st and 2nd use (Fig. 2C), which shows a similar extent of enzymatic activity after 1st and 2nd use. During these experiments, we visually observed a significant loss of silver nanoparticles from both surfaces and carried out further experiments to determine the extent of loss of nanoparticles.
Fig. 3 shows the real-color photographs of SIFs-deposited blank glass slides and SIFs-deposited APTES-coated glass slides after the 1st and 2nd use, and Fig. 3C shows the corresponding absorbance values at their surface resonance peaks for both surfaces before use, after 1st and 2nd use. The decrease in absorbance from SIFson blank glass slides after two uses (>50%)implies thatthese surfaces are unlikely to be re-usable for further repeated use. In contrast, there was a relativelysmaller loss of silver nanoparticles from the SIFs-deposited APTES-coated glass slides (40%). Subsequently, further investigation for optimum enzymatic activity was carried out using SIFs-deposited APTES-coated glass slides, where the extent of silver nanoparticles was varied: low, medium and high loading.
Fig. 3.
Real-color photographs of SIFs used in surface modification of SIFs with Biotin-PEG-Amine: SIFs-deposited APTES-coated glass slides and SIFs-deposited blank glass slide, (A) after 1st use, and (B) after 2nd use. (C) Corresponding absorbance values of SIFs.
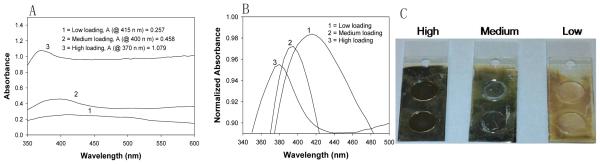
Fig. 4A, Fig. 4B shows absorbance and normalized absorbance for SIFs with low, medium and high loading on APTES-coated glass slides. As observed and described previously [1], the surface resonance peak for SIFs show a blue shift as the loading of silver nanoparticles is increased. The real-color photographs of SIFs with low, medium and high loading on APTES-coated glass slides visually corroborate the increase in nanoparticle loading on SIFs.
Fig. 4.
(A) Absorption spectrum for SIFs-deposited APTES-coated glass slides: low, medium and high loading, (B) Normalized absorbance spectrum. (C) Real-color photographs.
Fig. 5 shows the absorbance spectrum of the colored solution obtained after the interaction of OPD and HRP-streptavidin for the different loading after 1st and 2nd use. The absorbance value at 495 nm for OPD measured for the SIFs with differentloading after 1st use was increased as the loading of silver nanoparticles was increased: low loading (A495 nm=0.265), medium loading (0.450), and high loading (0.6197). The increase in absorbance of the different loading indicates that Biotin-PEG-Amine, which is used for the optimization of enzymatic activity on SIFs appears to improve the enzymatic activity as the loading is increased. When compared to our previous work where we used three different strategies for enzyme immobilization (Biotin-avidin, self-assembled monolayer hexamethylenediamine, and Poly-L-lysine monolayer[1]) to study the nanoparticle-enzyme distance- and nanoparticle loading-dependent enzymatic activity, the use of Biotin-PEG-Amine further improved the enzymatic activity. After the 2nd use of the SIFs with different loading (Fig. 5B), we observed a decrease in the OPD absorbance spectrum, which indicates either the loss of enzyme activity and/or loss of SIFs from the glass surface due to multiple washing steps taking place after each step of the assay. Fig. 5C shows the absorbance of the colored solution at 495 nm after 1st and 2nd use for all loading, where 80%, 80% and 70% decrease in absorbance value for SIFs with low, medium and high loading, respectively. To further investigate whether the decrease in OPD absorbance was due to the potential detachment of SIFs from the surfaces as a result of extensive washing and/ or loss of enzymatic activity, the absorbance spectrum and real-color photographs of SIFs after 1st and 2nd use were taken and shown in Fig. 6. Fig. 6 clearly shows that there was only a slight detachment of SIFs with low and medium loading from the surface after the 2 runs (<10%), which implies that the decrease in OPD absorbance on these surfaces was mainly due to the loss of enzymatic activity (and/or enzyme detachment). On the other hand, SIFs with high loading showed >60% decrease in absorbance for the surface plasmon resonance peak of SIFs, which implies that the decrease in OPD absorbance on these surfaces was mainly due to the detachment of SIFs from surfaces. Although not determined, it is thought that the loss of enzymatic activity also contributed to the decrease in the OPD absorbance. Based on the comparison of the data presented here with the data generated for other enzyme immobilization strategies in our previous work [1], we can conclude that the immobilization of enzyme onto SIFs using BEA as the linker yields the highest enzyme activity to date. Further investigation of other enzymes for plasmon-enhanced enzymatic reactions is ongoing in our laboratory and will be reported in due course.
Fig. 5.
Absorbance of OPD at 495 nm (A and B) and (C) Absorption spectrum of OPD on SIFs-deposited APTES-coated glass slide (low, medium and high loading) after 1st and 2nd use.
Fig. 6.
Real-color photographs of SIFs used in surface modification of SIFs with Biotin-PEG-Amine: low, medium and high loading (A) after 1stuse, (B) after 2nd use. (C) Corresponding absorbance values of these SIFs at their relative surface plasmon resonance peaks.
Conclusions
In this work, surface modification of SIFs was carried out using Biotin-PEG-Amine for the optimization of the plasmon-enhanced enzymatic activity. The enzyme (HRP) activity on several SIFs surfaces was assessed by absorption measurements. It was demonstrated that the enzymatic activity can be enhanced using SIFs-deposited blank glass slides and SIFs-deposited APTES-coated glass slides. However, significant losses of silver nanoparticles from SIFs-deposited blank glass slides were observed. Subsequently, we employed SIFs-deposited APTES-coated glass slides with different loading (low, medium and high) for further investigation.The enzymatic activity was also shown to be increased as the loading of SIFs on APTES-coated glass slides was increased. Biotin-PEG-Amine, which has a spacer arm (PEG) that places the enzyme 6 nm away from the silver surface, was shown to be the best option for the immobilization of enzymes on SIFs to date, which are employed in plasmon-enhanced enzymatic reactions.
Acknowledgements
The project described was supported by Award Number 5-K25EB007565-05 from the National Institute of Biomedical Imaging and Bioengineering. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health.
Footnotes
Citation: Biebele Abel, et al. Plasmon-Enhanced Enzymatic Reactions 2:Optimization of Enzyme Activity by Surface Modification of Silver Island Films with Biotin-Poly (Ethylene-glycol)-Amine. Nano Biomed. Eng. 2012, 4(1), 23-28. DOI: 10.5101/nbe.v4i1.p23-28.
References
- 1.Abel B, et al. Plasmon-Enhanced Enzymatic Reactions: A Study of Nanoparticle-Enzyme Distance- and Nanoparticle Loading-Dependent Enzymatic Activity. Nano Biomed Eng. 2011;3(3):184–191. doi: 10.5101/nbe.v3i3.p184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salamon Z, Macleod HA, Tollin G. Surface plasmon resonance spectroscopy as a tool for investigating the biochemical and biophysical properties of membrane protein systems. II: Applications to biological systems. Biochim Biophys Acta. 1997;1331(2):131–52. doi: 10.1016/s0304-4157(97)00003-8. [DOI] [PubMed] [Google Scholar]
- 3.Wegner GJ, Lee HJ, Corn RM. Characterization and optimization of peptide arrays for the study of epitope-antibody interactions using surface plasmon resonance imaging. Analytical Chemistry. 2002;74(20):5161–8. doi: 10.1021/ac025922u. http://dx.doi.org/10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 4.Ren ML, et al. Using silver nanoparticle to enhance current response of biosensor. Biosensors & Bioelectronics. 2005;21(3):433–437. doi: 10.1016/j.bios.2004.08.052. http://dx.doi.org/10.1016/j.bios.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 5.Raj CR, Jena BK. Enzyme integrated silicate-Pt nanoparticle architecture: A versatile biosensing platform. Biosensors & Bioelectronics. 2011;26(6):2960–2966. doi: 10.1016/j.bios.2010.11.046. http://dx.doi.org/10.1016/j.bios.2010.11.046. [DOI] [PubMed] [Google Scholar]
- 6.Kirchhoff JR, Shulga O. An acetylcholinesterase enzyme electrode stabilized by an electrodeposited gold nanoparticle layer. Electrochemistry Communications. 2007;9(5):935–940. http://dx.doi.org/10.1016/j.elecom.2006.11.021. [Google Scholar]
- 7.Jia CP, et al. Highly sensitive protein detection using enzyme-labeled gold nanoparticle probes. Analyst. 2010;135(2):327–331. doi: 10.1039/b916629g. http://dx.doi.org/10.1039/b916629g. [DOI] [PubMed] [Google Scholar]
- 8.Nam JM, et al. Restriction-Enzyme-Coded Gold-Nanoparticle Probes for Multiplexed DNA Detection. Small. 2009;5(23):2665–2668. doi: 10.1002/smll.200901105. http://dx.doi.org/10.1002/smll.200901105. [DOI] [PubMed] [Google Scholar]
- 9.Moskovitz Y, Srebnik S. Mean-field model of immobilized enzymes embedded in a grafted polymer layer. Biophysical Journal. 2005;89(1):22–31. doi: 10.1529/biophysj.104.053686. http://dx.doi.org/10.1529/biophysj.104.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergstrom K, et al. Reduction of fibrinogen adsorption on PEG-coated polystyrene surfaces. J Biomed Mater Res. 1992;26(6):779–90. doi: 10.1002/jbm.820260607. http://dx.doi.org/10.1002/jbm.820260607. [DOI] [PubMed] [Google Scholar]
- 11.Okamura Y, et al. Hemostatic effects of phospholipid vesicles carrying fibrinogen gamma chain dodecapeptide in vitro and in vivo. Bioconjug Chem. 2005;16(6):1589–96. doi: 10.1021/bc050178g. http://dx.doi.org/10.1021/bc050178g. [DOI] [PubMed] [Google Scholar]
- 12.Sebra RP, et al. Surface grafted antibodies: controlled architecture permits enhanced antigen detection. Langmuir. 2005;21(24):10907–11. doi: 10.1021/la052101m. http://dx.doi.org/10.1021/la052101m. [DOI] [PubMed] [Google Scholar]
- 13.Bronfman FC, et al. Ligand-induced internalization of the p75 neurotrophin receptor: a slow route to the signaling endosome. J Neurosci. 2003;23(8):3209–20. doi: 10.1523/JNEUROSCI.23-08-03209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara T, Matsuyama S, Tokuda H. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J Biol Chem. 2003;278(41):40408–14. doi: 10.1074/jbc.M307836200. http://dx.doi.org/10.1074/jbc.M307836200. [DOI] [PubMed] [Google Scholar]
- 15.Pihlajamaa T, et al. Characterization of recombinant amino-terminal NC4 domain of human collagen IX: interaction with glycosaminoglycans and cartilage oligomeric matrix protein. J Biol Chem. 2004;279(23):24265–73. doi: 10.1074/jbc.M402865200. http://dx.doi.org/10.1074/jbc.M402865200. [DOI] [PubMed] [Google Scholar]