Abstract
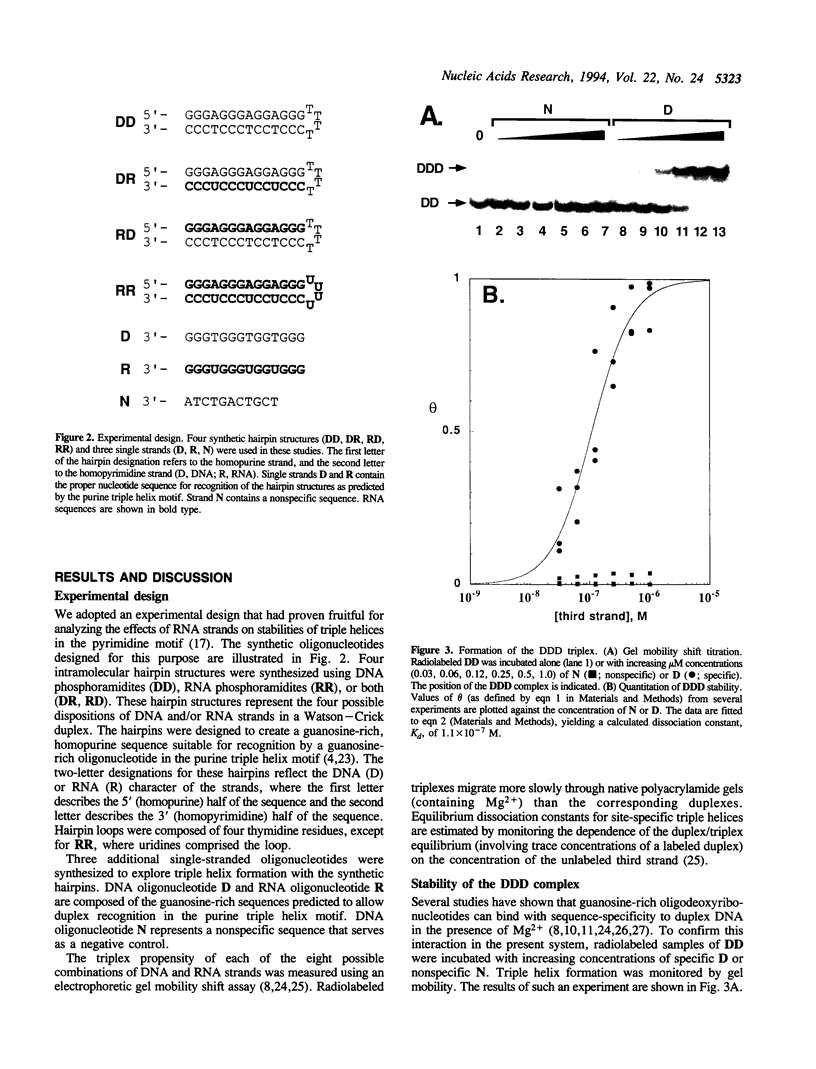
Research concerning oligonucleotide-directed triple helix formation has mainly focused on the binding of DNA oligonucleotides to duplex DNA. The participation of RNA strands in triple helices is also of interest. For the pyrimidine motif (pyrimidine.purine.pyrimidine triplets), systematic substitution of RNA for DNA in one, two, or all three triplex strands has previously been reported. For the purine motif (purine.purine.pyrimidine triplets), studies have shown only that RNA cannot bind to duplex DNA. To extend this result, we created a DNA triple helix in the purine motif and systematically replaced one, two, or all three strands with RNA. In dramatic contrast to the general accommodation of RNA strands in the pyrimidine triple helix motif, a stable triplex forms in the purine motif only when all three of the substituent strands are DNA. The lack of triplex formation among any of the other seven possible strand combinations involving RNA suggests that: (i) duplex structures containing RNA cannot be targeted by DNA oligonucleotides in the purine motif; (ii) RNA strands cannot be employed to recognize duplex DNA in the purine motif; and (iii) RNA tertiary structures are likely to contain only isolated base triplets in the purine motif.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beru N., Smith D., Goldwasser E. Evidence suggesting negative regulation of the erythropoietin gene by ribonucleoprotein. J Biol Chem. 1990 Aug 25;265(24):14100–14104. [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Poly(rA) binds poly(rG).poly(rC) to form a triple helix. Nucleic Acids Res. 1992 Jan 25;20(2):315–318. doi: 10.1093/nar/20.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. J., Van Dyke M. W. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993 Dec 11;21(24):5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Davis T. L., Firulli A. B., Kinniburgh A. J. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Kessler D. J., Gunnell S., Duvic M., Pettitt B. M., Hogan M. E. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991 Sep 24;30(38):9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- Escudé C., François J. C., Sun J. S., Ott G., Sprinzl M., Garestier T., Hélène C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993 Dec 11;21(24):5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J. P. The structures and functions of transfer RNA. Prog Biophys Mol Biol. 1977;32(3):233–308. [PubMed] [Google Scholar]
- Guschlbauer W., Chantot J. F., Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J Biomol Struct Dyn. 1990 Dec;8(3):491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- Han H., Dervan P. B. Sequence-specific recognition of double helical RNA and RNA.DNA by triple helix formation. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hélène C. The anti-gene strategy: control of gene expression by triplex-forming-oligonucleotides. Anticancer Drug Des. 1991 Dec;6(6):569–584. [PubMed] [Google Scholar]
- Ing N. H., Beekman J. M., Kessler D. J., Murphy M., Jayaraman K., Zendegui J. G., Hogan M. E., O'Malley B. W., Tsai M. J. In vivo transcription of a progesterone-responsive gene is specifically inhibited by a triplex-forming oligonucleotide. Nucleic Acids Res. 1993 Jun 25;21(12):2789–2796. doi: 10.1093/nar/21.12.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Malkhosyan S. R., Kohwi-Shigematsu T. Intramolecular dG.dG.dC triplex detected in Escherichia coli cells. J Mol Biol. 1992 Feb 20;223(4):817–822. doi: 10.1016/0022-2836(92)90242-c. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher L. J., 3rd DNA triple-helix formation: an approach to artificial gene repressors? Bioessays. 1992 Dec;14(12):807–815. doi: 10.1002/bies.950141204. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. J. Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry. 1990 Sep 18;29(37):8820–8826. doi: 10.1021/bi00489a045. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd Inhibition of T7 RNA polymerase initiation by triple-helical DNA complexes: a model for artificial gene repression. Biochemistry. 1992 Aug 25;31(33):7587–7594. doi: 10.1021/bi00148a021. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Sobell H. M. A molecular model for gene repression. Proc Natl Acad Sci U S A. 1966 May;55(5):1201–1205. doi: 10.1073/pnas.55.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Krawczyk S. H., Wadwani S., Matteucci M. D. An anti-parallel triple helix motif with oligodeoxynucleotides containing 2'-deoxyguanosine and 7-deaza-2'-deoxyxanthosine. Nucleic Acids Res. 1993 Jan 25;21(2):327–333. doi: 10.1093/nar/21.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. W. The triple helix: a potential mechanism for gene regulation. J Exp Pathol. 1985 Fall;2(3):135–148. [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Olivas W. M., Maher L. J., 3rd DNA recognition by alternate strand triple helix formation: affinities of oligonucleotides for a site in the human p53 gene. Biochemistry. 1994 Feb 1;33(4):983–991. doi: 10.1021/bi00170a017. [DOI] [PubMed] [Google Scholar]
- Pieles U., Zürcher W., Schär M., Moser H. E. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a powerful tool for the mass and sequence analysis of natural and modified oligonucleotides. Nucleic Acids Res. 1993 Jul 11;21(14):3191–3196. doi: 10.1093/nar/21.14.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. W., Crothers D. M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992 Nov 27;258(5087):1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990 Mar 29;344(6265):410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
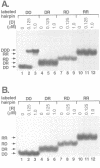
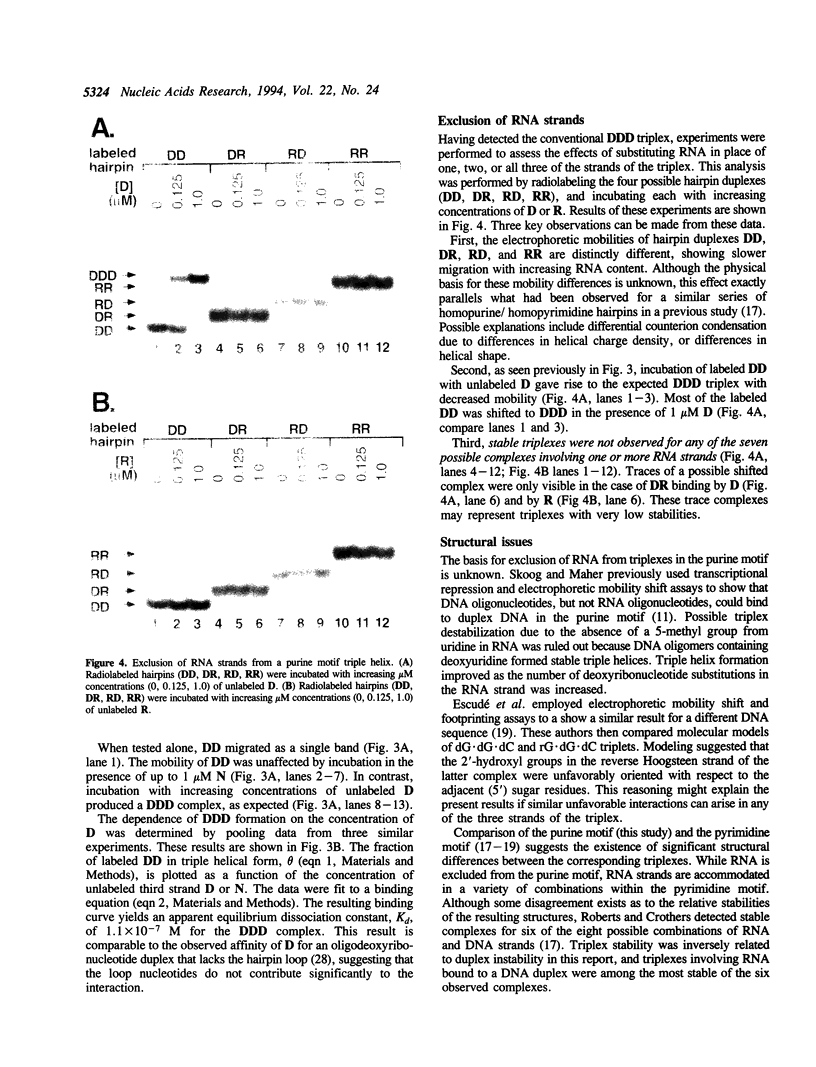
- Skoog J. U., Maher L. J., 3rd Repression of bacteriophage promoters by DNA and RNA oligonucleotides. Nucleic Acids Res. 1993 May 11;21(9):2131–2138. doi: 10.1093/nar/21.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]