Abstract
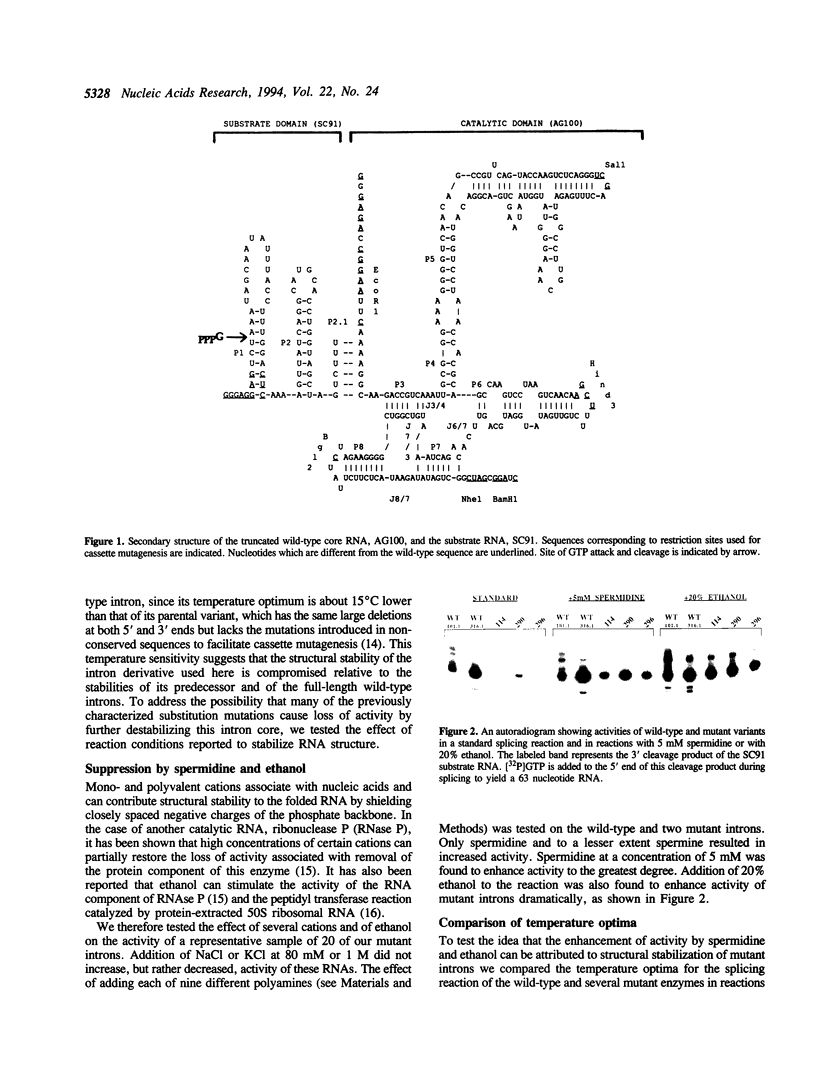
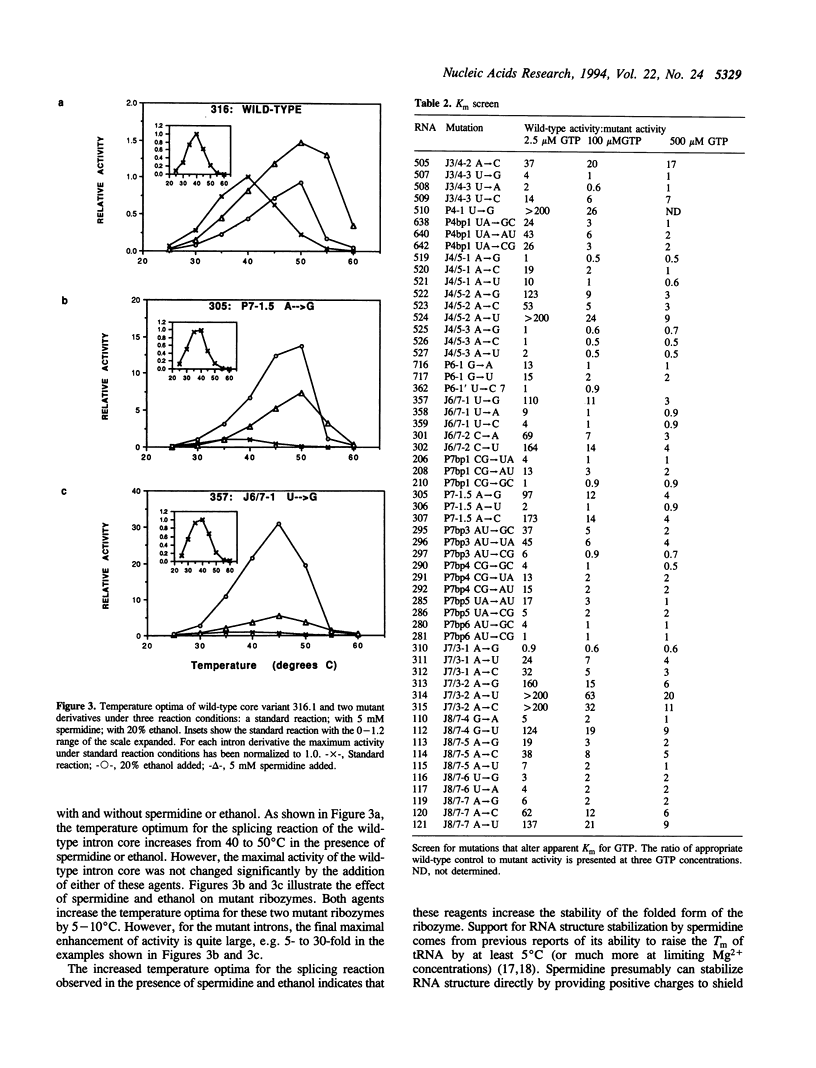
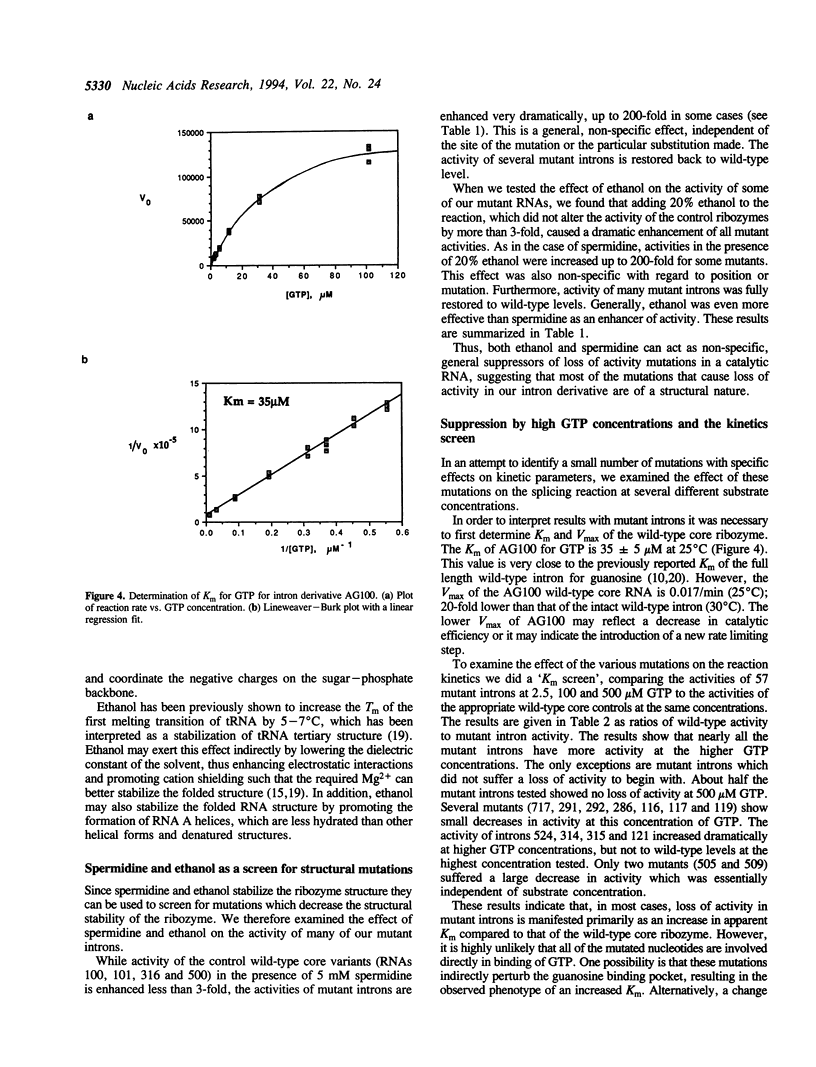
We have previously described a collection of mutations in conserved residues of the core of the Tetrahymena self-splicing intron. Most of these single base substitutions have less than 10% of the activity of their parental intron derivative [Couture, S., et al., (1990) J. Mol. Biol., 215, 345-358]. We examined the effect of two agents known to stabilize RNA structure, spermidine and ethanol, on the activity of many of these mutant RNAs. In the presence of either 5 mM spermidine or 20% ethanol most substitution mutations were partially or completely suppressed. These conditions also increased the temperature optima of both wild-type and mutant ribozymes. In addition, we find that mutations are also suppressed by a high concentration of GTP, a substrate in the reaction which is bound specifically by the intron. Thus we observe a general suppression of mutations in an RNA enzyme (ribozyme) by spermidine, ethanol and by substrate stabilization. These results are consistent with the idea that most mutations destabilize the folded structure of the ribozyme and can be suppressed by any of a variety of stabilizing influences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Cech T. R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. 1984 Apr 26-May 2Nature. 308(5962):820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- Beneventi S., Onori G. Effect of ethanol on the thermal stability of tRNA molecules. Biophys Chem. 1986 Dec 15;25(2):181–190. doi: 10.1016/0301-4622(86)87009-0. [DOI] [PubMed] [Google Scholar]
- Bolton P. H., Kearns D. R. Effect of magnesium and polyamines on the structure of yeast tRNAPhe. Biochim Biophys Acta. 1977 Jul 5;477(1):10–19. doi: 10.1016/0005-2787(77)90156-3. [DOI] [PubMed] [Google Scholar]
- Couture S., Ellington A. D., Gerber A. S., Cherry J. M., Doudna J. A., Green R., Hanna M., Pace U., Rajagopal J., Szostak J. W. Mutational analysis of conserved nucleotides in a self-splicing group I intron. J Mol Biol. 1990 Oct 5;215(3):345–358. doi: 10.1016/s0022-2836(05)80356-0. [DOI] [PubMed] [Google Scholar]
- Gardiner K. J., Marsh T. L., Pace N. R. Ion dependence of the Bacillus subtilis RNase P reaction. J Biol Chem. 1985 May 10;260(9):5415–5419. [PubMed] [Google Scholar]
- Heerschap A., Walters J. A., Hilbers C. W. Interactions of some naturally occurring cations with phenylalanine and initiator tRNA from yeast as reflected by their thermal stability. Biophys Chem. 1985 Aug;22(3):205–217. doi: 10.1016/0301-4622(85)80044-2. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Krenitsky T. A., Tuttle J. V. Correlation of substrate-stabilization patterns with proposed mechanisms for three nucleoside phosphorylases. Biochim Biophys Acta. 1982 May 3;703(2):247–249. doi: 10.1016/0167-4838(82)90055-3. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Finch J. T., Klug A., Clark B. F. High-resolution x-ray diffraction studies on a pure species of transfer RNA. J Mol Biol. 1972 Dec 14;72(1):99–101. doi: 10.1016/0022-2836(72)90071-x. [DOI] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D. P., Szostak J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989 Nov 23;342(6248):391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992 Jun 5;256(5062):1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Pace C. N., McGrath T. Substrate stabilization of lysozyme to thermal and guanidine hydrochloride denaturation. J Biol Chem. 1980 May 10;255(9):3862–3865. [PubMed] [Google Scholar]
- Prinz H., Furgac N., Cramer F. Spermine stabilizes the conformation of tRNAPhe in crystals. Biochim Biophys Acta. 1976 Sep 20;447(1):110–115. doi: 10.1016/0005-2787(76)90101-5. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Lin B. Genetic analysis of staphylococcal nuclease: identification of three intragenic "global" suppressors of nuclease-minus mutations. Genetics. 1985 Aug;110(4):539–555. doi: 10.1093/genetics/110.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- Tropp J. S., Redfield A. G. Proton exchange rates in transfer RNA as a function of spermidine and magnesium. Nucleic Acids Res. 1983 Apr 11;11(7):2121–2134. doi: 10.1093/nar/11.7.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]



