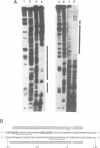
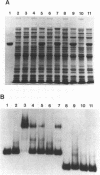
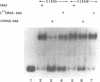
Abstract
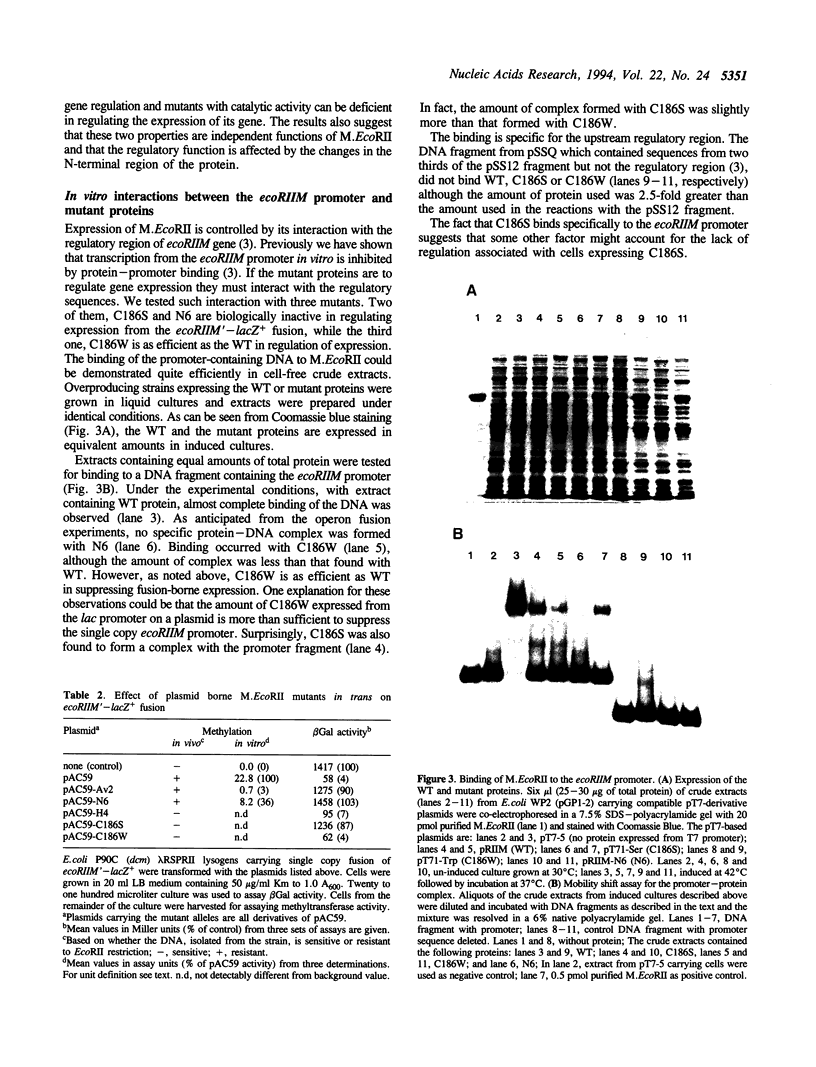
EcoRII Methyltransferase (M.EcoRII) which methylates the second C in the sequence CCWGG (W = A/T) is autogenously regulated by binding to the 5' regulatory region of its gene. DNase I footprinting experiments demonstrated that purified M.EcoRII protected a 47-49 bp region of DNA immediately upstream of the ecoRIIM coding region. We have studied this interaction with mutants of the enzyme, in vitro by DNA binding and in vivo by investigating the repression in trans of expression of beta-galactosidase from an ecoRIIM-lacZ operon fusion. Two catalytically active mutants failed to repress expression of the fusion whereas catalytically inactive mutants had repressor activity. However, with one of the catalytically inactive mutants, C186S, in which the catalytic Cys was replaced with Ser, and which bound unmethylated CCWGG sequences, repression could only be demonstrated when those sequences in cellular DNA were methylated by supplying a cloned dcm gene in trans. In vitro binding of the DNA fragment containing the ecoRIIM regulatory region was detected only with the mutants that showed repressor activity, including C186S. Results indicate that down-regulation of the gene in vivo and binding to the promoter in vitro are not dependent on the catalytic properties of M.EcoRII. Mobility shift experiments with C186S also revealed that it could bind either the promoter or unmethylated CCWGG sites, but not both. We conclude that the concentration of unmethylated CCWGG sites controls expression from the ecoRIIM promoter.
Full text
PDF

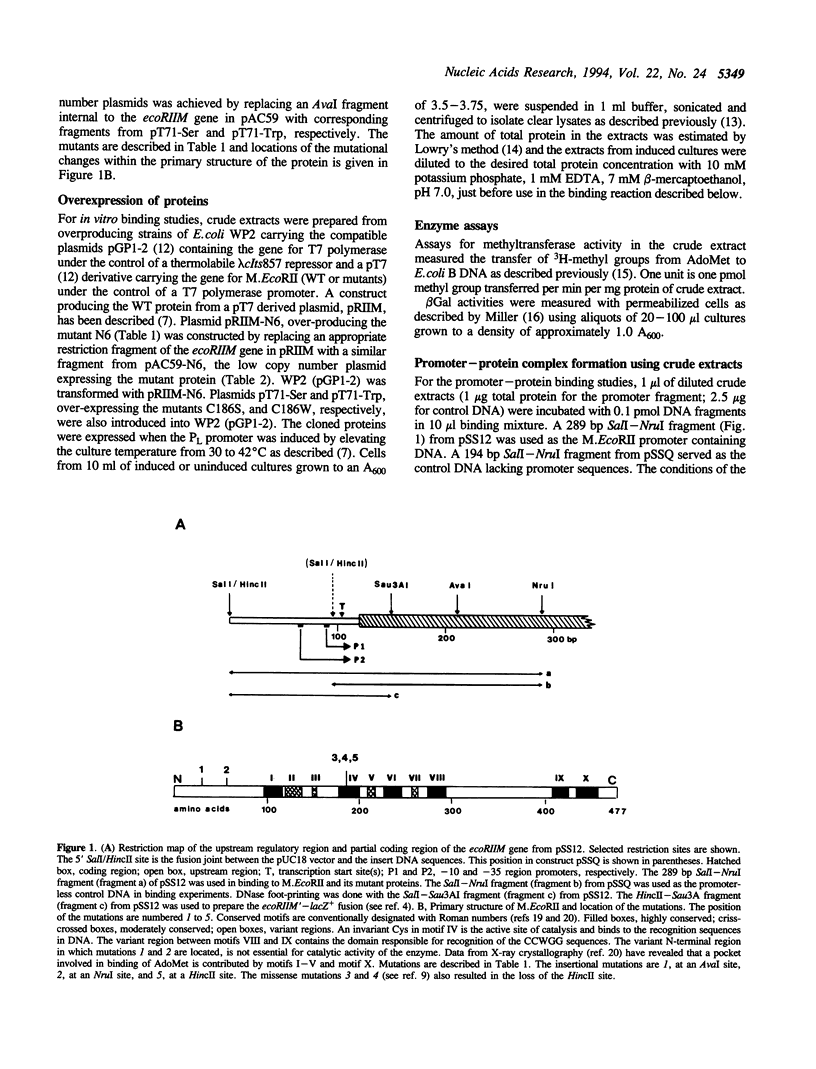



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhagwat A. S., Johnson B., Weule K., Roberts R. J. Primary sequence of the EcoRII endonuclease and properties of its fusions with beta-galactosidase. J Biol Chem. 1990 Jan 15;265(2):767–773. [PubMed] [Google Scholar]
- Bhagwat A. S., Sohail A., Roberts R. J. Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol. 1986 Jun;166(3):751–755. doi: 10.1128/jb.166.3.751-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J. E., Jr The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989 Aug 11;58(3):427–429. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]
- Friedman S., Som S. Induction of EcoRII methyltransferase: evidence for autogenous control. J Bacteriol. 1993 Oct;175(19):6293–6298. doi: 10.1128/jb.175.19.6293-6298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S., Som S., Yang L. F. The core element of the EcoRII methylase as defined by protease digestion and deletion analysis. Nucleic Acids Res. 1991 Oct 11;19(19):5403–5408. doi: 10.1093/nar/19.19.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Friedman S. The irreversible binding of azacytosine-containing DNA fragments to bacterial DNA(cytosine-5)methyltransferases. J Biol Chem. 1985 May 10;260(9):5698–5705. [PubMed] [Google Scholar]
- Hattman S., Schlagman S., Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973 Sep;115(3):1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives C. L., Nathan P. D., Brooks J. E. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J Bacteriol. 1992 Nov;174(22):7194–7201. doi: 10.1128/jb.174.22.7194-7201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Maloy S., Stewart V. Autogenous regulation of gene expression. J Bacteriol. 1993 Jan;175(2):307–316. doi: 10.1128/jb.175.2.307-316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringquist S., Smith C. L. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sohail A., Lieb M., Dar M., Bhagwat A. S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990 Aug;172(8):4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Friedman S. Autogenous regulation of the EcoRII methylase gene at the transcriptional level: effect of 5-azacytidine. EMBO J. 1993 Nov;12(11):4297–4303. doi: 10.1002/j.1460-2075.1993.tb06114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Friedman S. Direct photolabeling of the EcoRII methyltransferase with S-adenosyl-L-methionine. J Biol Chem. 1990 Mar 15;265(8):4278–4283. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T., Blumenthal R. M. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J Bacteriol. 1992 May;174(10):3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Bhagwat A. S. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992 Jan 25;20(2):319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Kubareva E. A., Romanova E. A., Oretskaya T. S., Gromova E. S., Shabarova Z. A., Bhagwat A. S. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993 Jan 25;21(2):295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]