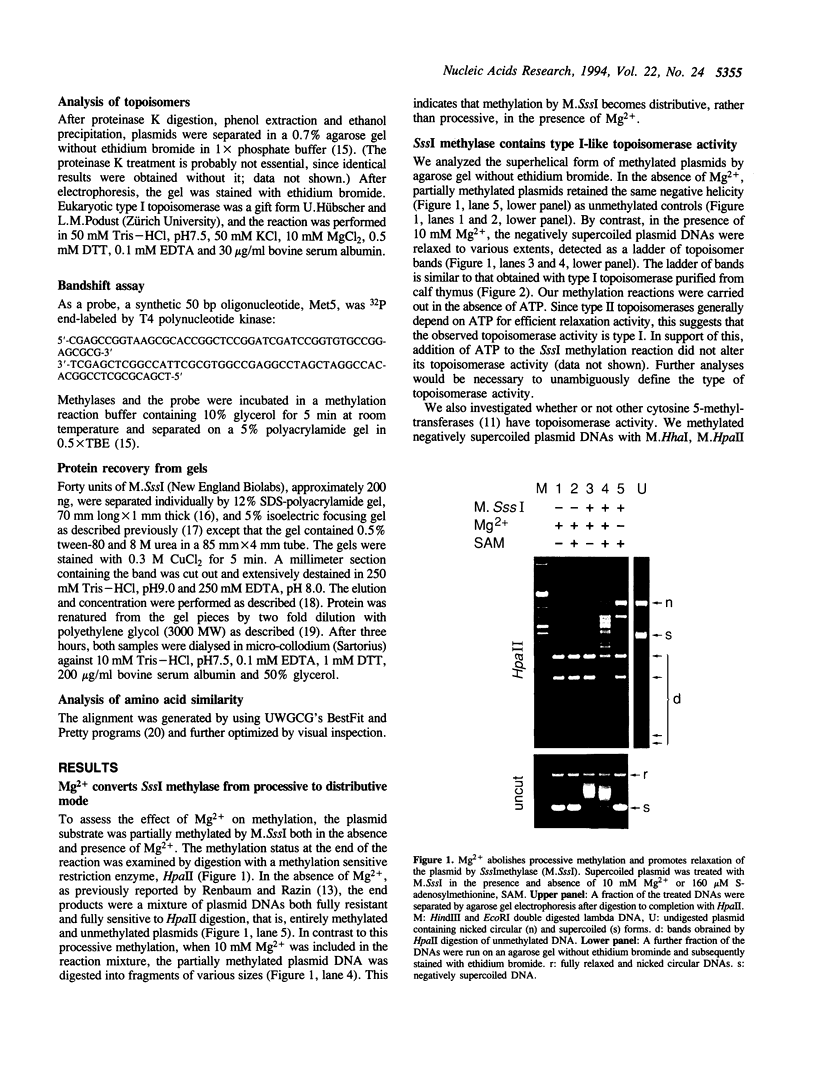
Abstract
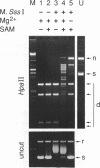
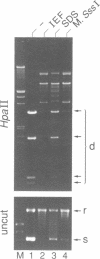
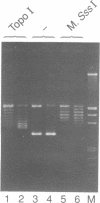
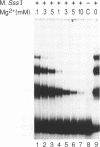
A prokaryotic CpG-specific methylase from Spiroplasma, SssI methylase, is now widely used to study the effect of CpG methylation in mammalian cells, and can processively modify cytosines in CpG dinucleotides in the absence of Mg2+. In the presence of Mg2+, we found (i) that the methylation reaction is distributive rather than processive as a result of the decreased affinity of SssI methylase for DNA, and (ii) that a type I-like topoisomerase activity is present in SssI methylase preparations. This topoisomerase activity was still present in SssI methylase further purified by either SDS-polyacrylamide or isoelectric focusing gel electrophoresis. We show that methylase and topoisomerase activities are not functionally interdependent, since conditions exist where only one or the other enzymatic activity is detectable. The catalytic domains of SssI methylase and prokaryotic topoisomerases show similarity at the amino acid level, further supporting the idea that the topoisomerase activity is a genuine activity of SssI methylase. Mycoplasmas, including Spiroplasma, have the smallest genomes of all living organisms; thus, this condensation of two enzymatic activities into the same protein may be a result of genome economy, and may also have functional implications for the mechanism of methylation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balaghi M., Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993 Jun 30;193(3):1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T. Supercoiling-dependent sequence specificity of mammalian DNA methyltransferase. Nucleic Acids Res. 1987 May 11;15(9):3835–3843. doi: 10.1093/nar/15.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryans M., Kass S., Seivwright C., Adams R. L. Vector methylation inhibits transcription from the SV40 early promoter. FEBS Lett. 1992 Aug 31;309(1):97–102. doi: 10.1016/0014-5793(92)80748-6. [DOI] [PubMed] [Google Scholar]
- Castell S. E., Jordan S. L., Halford S. E. Site-specific recombination and topoisomerization by Tn21 resolvase: role of metal ions. Nucleic Acids Res. 1986 Sep 25;14(18):7213–7226. doi: 10.1093/nar/14.18.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. C., Chae C. B. Demethylation of somatic and testis-specific histone H2A and H2B genes in F9 embryonal carcinoma cells. Mol Cell Biol. 1993 Sep;13(9):5538–5548. doi: 10.1128/mcb.13.9.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland J. L., Builder S. E., Swartz J. R., Winkler M., Chang J. Y., Wang D. I. Polyethylene glycol enhanced protein refolding. Biotechnology (N Y) 1992 Sep;10(9):1013–1019. doi: 10.1038/nbt0992-1013. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Guenette D. K., Ritzenthaler J. D., Foley J., Jackson J. D., Smith B. D. DNA methylation inhibits transcription of procollagen alpha 2(I) promoters. Biochem J. 1992 May 1;283(Pt 3):699–703. doi: 10.1042/bj2830699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hwu W. L., Lee Y. M., Lee S. C., Wang T. R. In vitro DNA methylation inhibits FMR-1 promoter. Biochem Biophys Res Commun. 1993 May 28;193(1):324–329. doi: 10.1006/bbrc.1993.1627. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Kass S. U., Goddard J. P., Adams R. L. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993 Dec;13(12):7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kochanek S., Renz D., Doerfler W. Probing DNA-protein interactions in vitro with the CpG DNA methyltransferase. Nucleic Acids Res. 1993 May 25;21(10):2339–2342. doi: 10.1093/nar/21.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein M., Keini G., Cedar H., Bergman Y. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 1994 Mar 11;76(5):913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Lima C. D., Wang J. C., Mondragón A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994 Jan 13;367(6459):138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- Mizutani M., Ura K., Hirose S. DNA superhelicity affects the formation of transcription preinitiation complex on eukaryotic genes differently. Nucleic Acids Res. 1991 Jun 11;19(11):2907–2911. doi: 10.1093/nar/19.11.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Razin A. Mode of action of the Spiroplasma CpG methylase M.SssI. FEBS Lett. 1992 Nov 30;313(3):243–247. doi: 10.1016/0014-5793(92)81201-v. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Stöver T., Köhler E., Fagin U., Wende W., Wolfes H., Pingoud A. Determination of the DNA bend angle induced by the restriction endonuclease EcoRV in the presence of Mg2+. J Biol Chem. 1993 Apr 25;268(12):8645–8650. [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Laigret F., Whitley J. C., Citti C., Finch L. R., Carle P., Renaudin J., Bové J. M. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res. 1992 Apr 11;20(7):1559–1565. doi: 10.1093/nar/20.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]