Background: E-site tRNA and Shine-Dalgarno interactions have been proposed to increase the fidelity of protein synthesis.
Results: Neither E-site tRNA nor the Shine-Dalgarno interactions impact the fidelity of tRNA selection during protein synthesis.
Conclusion: The allosteric three-site model for the ribosome cannot be confirmed.
Significance: This work will contribute to understanding the molecular mechanisms that dictate fidelity during protein synthesis.
Keywords: Protein Synthesis, Ribosomal RNA (rRNA), Ribosomes, Transfer RNA (tRNA), Translation, E-site, Fidelity, Shine-Dalgarno
Abstract
Ongoing debate in the ribosome field has focused on the role of bound E-site tRNA and the Shine-Dalgarno-anti-Shine-Dalgarno (SD-aSD) interaction on A-site tRNA interactions and the fidelity of tRNA selection. Here we use an in vitro reconstituted Escherichia coli translation system to explore the reported effects of E-site-bound tRNA and SD-aSD interactions on tRNA selection events and find no evidence for allosteric coupling. A large set of experiments exploring the role of the E-site tRNA in miscoding failed to recapitulate the observations of earlier studies (Di Giacco, V., Márquez, V., Qin, Y., Pech, M., Triana-Alonso, F. J., Wilson, D. N., and Nierhaus, K. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10715–10720 and Geigenmüller, U., and Nierhaus, K. H. (1990) EMBO J. 9, 4527–4533); the frequency of miscoding was unaffected by the presence of E-site-bound cognate tRNA. Moreover, our data provide clear evidence that the reported effects of the SD-aSD interaction on fidelity can be attributed to the binding of ribosomes to an unanticipated site on the mRNA (in the absence of the SD sequence) that provides a cognate pairing codon leading naturally to incorporation of the purported “noncognate” amino acid.
Introduction
A series of in vitro studies over several decades has argued that occupation of the E-site by a deacylated cognate tRNA allosterically regulates affinity for A-site substrates, thus increasing the overall fidelity of tRNA selection (2–4). A recent study extended these ideas by arguing that just following initiation, when no E-site tRNA is available to occupy the E-site, the SD-aSD3 pairing interaction functionally replaces the E-site codon-anticodon interaction, similarly increasing the fidelity of tRNA selection (1). These studies were all biochemical in nature, using well defined reconstituted systems with either homopolymeric or defined mRNA sequences and with direct analysis of miscoding events by HPLC. The observed effects in these studies are generally striking, and it has been difficult to identify a conceptual error.
Despite the appeal of such allosteric models, other studies have argued strongly against such a role for the E-site tRNA (5). Most compellingly perhaps is a recent single-molecule study performed under relatively normal translation conditions (that include physiologically relevant concentrations of ternary complex and EFG) that failed to show any coupling between tRNA binding events in the E- and A-sites, arguing against models for allosteric coupling (6). Another more recent single-molecule study under less physiological conditions also argued for uncoupled activity between E- and A-sites during some stages of translation elongation (7). Also, although x-ray structures of the ribosome often contain E-site-bound tRNA, there is little, albeit some, evidence for the existence of defined codon-anticodon interactions in these structures (8–11), consistent with earlier biophysical studies (12, 13). It should be acknowledged, however, that the E-site tRNA in these structures has typically not been cognate, but instead a heterogeneous collection of tRNAs that happened to copurify with the ribosomes. We note that although the role for E-site tRNA during tRNA selection is highly contested, its role in frame maintenance is less controversial and has been documented by multiple groups both in vitro and in vivo (14–16). Moreover, our own studies of post-peptidyl quality control suggested that the quality of codon-anticodon interactions in the E-site might impact interactions with both aminoacyl-tRNA and release factor substrates in the A-site (17).
Here we examined the effects of the SD-aSD interaction on the fidelity of the decoding process and found no evidence supporting a role for this interaction in increasing the fidelity of protein synthesis. Instead, in the absence of the SD-aSD interaction, the ribosome fails to quantitatively initiate on the first AUG of the mRNA, hence leading to multiple initiation events, both of which lead to the incorporation of cognate amino acids corresponding to the codon poised in the A-site. We also examined the role of E-site-bound tRNA on the decoding process and failed to observe the dramatic effects previously reported by Nierhaus and colleagues (1–4). We further provide experimental evidence to suggest that direct competition in the A-site by excess deacylated tRNA may account for some of the previously reported surprising effects on fidelity.
EXPERIMENTAL PROCEDURES
Buffers and Reagents
Buffers used were: (i) polymix buffer (95 mm KCl, 5 mm NH4Cl, 5 mm magnesium acetate, 0.5 mm CaCl2, 8 mm putrescine, 1 mm spermidine, 5 mm potassium phosphate, pH 7.5, 1 mm DTT) (18); (ii) polyamine buffer (20 mm HEPES-potassium hydroxide, pH 7.6, 150 mm NH4Cl, 4.5 mm MgCl2, 2 mm spermidine, 0.05 mm spermine, 4 mm β-mercaptoethanol) (19); and (iii) buffer A (50 mm Tris-HCl, pH 7.5, 70 mm NH4Cl, 30 mm KCl, 7 mm MgCl2, 1 mm DTT).
Escherichia coli MRE600 (ATCC29417) tight couple ribosomes were prepared as described previously (20). For 30 S and 50 S subunits, crude 70 S ribosomes were first isolated by pelleting over sucrose cushions and dialyzed 2× in 1 mm Mg2+ buffer (50 mm Tris-HCl, pH 7.5, 150 mm NH4Cl, 1 mm MgCl2, and 6 mm β-mercaptoethanol). The subunits were then separated using a 10–40% sucrose gradient on a Ti-15 rotor. Overexpressed native IF1, IF3 and His-tagged IF2, EFTu, and EFG were purified on a 5-ml His-Trap FF FPLC column (GE Healthcare) as described previously (21).
tRNAfMet, tRNALys, tRNAGlu, and tRNAPhe were purchased from Chemical Block Ltd, tRNA2Leu was from Subriden RNA, and tRNAbulk was from Sigma (all from E. coli). mRNAs were transcribed by T7 RNA polymerase from double-stranded DNA templates. mRNA used for the SD experiments had the following sequence: 5′-GUGUGGGAAGAAAAGGAGGUCACAUAUGGUAUUCAAAGAAAAGAAUGGACUCAGAGCUAC-3′. The sequence of this mRNA is similar to the one used by Di Giacco et al. (1), except for a deletion of a (GA4)5 repeat and a shorter 3′-end. For the minus SD experiments, the underlined nucleotides were deleted. In the case of the mutagenesis of the second AUG, its sequence (in bold) was changed to CCC. Poly(U) template was purchased from Sigma.
tRNA Charging
Charging of tRNAfMet, tRNAVal, tRNAPhe, tRNA2Leu, and tRNALys was performed as described previously (17). In cases where a single tRNA in the complete tRNA mixture was aminoacylated (Asp-tRNAAsp and Leu-tRNALeu), the tRNAbulk mixture was first deacylated by incubating the mixture with 10 mm KOH for 5 s at 37 °C followed by neutralization with HCl and ethanol precipitation. For the subsequent charging reaction, only the desirable synthetase and amino acid were supplied. The acetylation (N-acetylated) of Phe-tRNAPhe was generated by incubating phenylalanine-tRNAPhe (1.6 μm) with 1/100 volume concentrated acetic anhydride in 0.3 m sodium acetate on ice for 1 h followed by the addition of an equivalent volume of acetic anhydride and incubation for an additional 1 h.
Formation of Ribosomal Complexes
The initiator fMet-tRNAfMet (3 μm) was enzymatically loaded into the P-site by incubating with 70 S ribosomes (2 μm), IF1, IF2, and IF3 (3 μm each), GTP (2 mm), and mRNA (6 μm in polymix buffer (or in buffer A)) at 37 °C for 45 min. The initiation complex (IC) was mixed with preincubated ternary complex containing EFTu (15 μm), EFG (6 μm), GTP (2 mm), and charged tRNAs (6 μm each). The mixture was incubated for 5 min at 37 °C, purified from unincorporated tRNAs and factors over a sucrose cushion (1.1 m sucrose, 20 mm Tris-HCl, pH 7.5, 500 mm NH4Cl, 10 mm MgCl2, 0.5 mm EDTA), and spun at 69 krpm in a TLA 100.3 rotor for 2 h.
For the poly(U) experiments, IC without E-site tRNA was prepared by incubating 70 S ribosomes (2 μm), N-Ac-[14C]Phe-tRNAPhe (4 μm), and mRNA (6 μm) for 30 min at 37 °C in polyamine buffer. IC with E-site tRNA was prepared using a three-step procedure. First, we mixed 70 S ribosomes (2 μm) with mRNA (6 μm) and tRNAPhe (4 μm) for 15 min at 37 °C. Next, we added N-Ac-[14C]Phe-tRNAPhe (4 μm) for 30 min. Finally, we added EFG (6 μm) and GTP (0.12 mm) for 10 min at 37 °C. In both cases, with or without the E-site tRNA, the ICs were pelleted over a sucrose cushion. The experiments were performed by mixing the preformed ribosomal complexes (ICs with and without E-site tRNA) with preformed ternary complexes as described previously.
For the HPLC experiments, ICs were prepared by mixing 30 S and 50 S subunits (2 μm each) with N-Ac-[14C]Phe-tRNA (4 μm) and mRNA (6 μm) with or without the deacylated E-site tRNA (tRNAfMet or tRNATyr; 4 μm) for 30 min at 37 °C in polyamine buffer. The ICs were then pelleted over sucrose cushion and resuspended in the appropriate concentration (2 μm). The ICs were then incubated with an equivalent volume of ternary complex containing 40 μm of each aa-tRNA.
Identification of Peptides
Samples were treated with KOH (0.3 m) at 50 °C for 30 min to hydrolyze the peptidyl-tRNAs. Peptides were then resolved using electrophoretic cellulose TLC in pyridine acetate buffer, pH 2.7 (submerged in Stoddard solvent) and applying a voltage of 1200 V over a distance of 8 cm for ∼30 min (22).
For the HPLC analysis, we used a Zorbax Eclipse XDB-C18 (4.6 × 150 mm) column equilibrated in 0.1% TFA, 2% acetonitrile. After incubating the reaction time points in 0.3 m KOH, HCl was added to neutralize. Samples were then injected onto the HPLC and resolved with a 2–60% acetonitrile gradient over 90 min, flow rate 0.5 ml/min, temperature 60 °C. Dipeptide products were readily resolved from the individual amino acids. 1-ml fractions were collected and quantified by liquid scintillation counting to account for amounts of [14C]Phe, [3H]Leu, and [3H]Lys residue.
Toeprinting Assay
Initiated and elongated peptidyl-tRNA complexes were prepared as described above using nonradiolabeled fMet-tRNAfMet. The mRNA used had additional sequence at the 3′-end to allow the radiolabeled oligonucleotide primer to anneal and be extended by reverse transcriptase (23).
RESULTS
The Shine-Dalgarno-Anti-Shine-Dalgarno Interaction Is Not Critical in Specifying Fidelity of First Round of Elongation
A previous study argued that the SD-aSD interaction between mRNA and the 16 S rRNA functionally compensates for the lack of codon-anticodon interaction at the E-site during the first round of elongation (1). Using a defined mRNA species, the authors argue that the incorporation of noncognate Asp-tRNAAsp on a valine codon (GUA) in the A-site is reduced in the presence of a functional SD sequence in the mRNA.
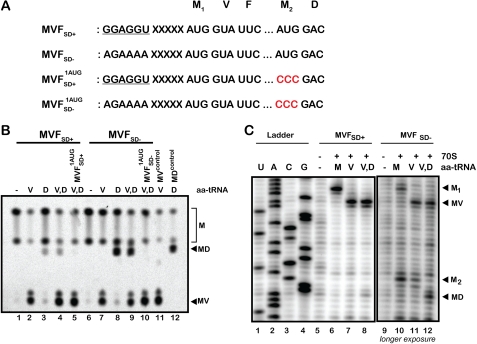
We began to explore this phenomenon in our own E. coli in vitro reconstituted translation system, utilizing mRNAs and tRNA isoacceptors similar to those used in the earlier study. In these experiments, ribosomes were programmed with heteropolymeric mRNAs either with or without a Shine-Dalgarno sequence (GGAGGU) five nucleotides upstream of the coding sequence AUG-GUA-UUC (MVF) (Fig. 1A), and the P-site was loaded with f-[35S]Met-tRNAfMet through a standard initiation process (24). These ICs were subsequently reacted with two different ternary complexes, EFTu·GTP·Val-tRNAVal or EFTu·GTP·Asp-tRNAAsp, or both together, and the products were resolved by electrophoretic TLC (22). As reported previously (1), when mRNAs encoding MVF, with and without a Shine-Dalgarno sequence, were compared, there was a greater extent of incorporation of Asp (into MD product), a noncognate species for the GUA codon, in the absence of the SD sequence (Fig. 1B, lanes 8 and 9). Indeed, when both Val-tRNAVal and Asp-tRNAAsp are provided, in the presence of the SD sequence, primarily MV product is generated, whereas in the absence of the SD sequence, a near equal mixture of MD and MV product is generated (Fig. 1B, lane 9). These data might suggest that there is considerable promiscuity of the ribosome in decoding the second amino acid of a protein sequence when there is no SD sequence found in the mRNA. However, we noted that in the absence of the SD sequence, the reactions with either Val-tRNAVal or Asp-tRNAAsp failed to go to completion, and there appears to be no competition between the two aa-tRNAs when both are added together in the same reaction. In contrast, in the presence of the SD sequence, the reaction with Val-tRNAVal proceeded to near completion where most of the fMet-tRNAfMet was consumed (Fig. 1B, lane 2). These observations suggest that in the absence of the SD sequence, there exists a population of ribosomes that preferentially react with a distinct aa-tRNA (Asp-tRNAAsp), thus explaining the observed end-point defects.
FIGURE 1.
Shine-Dalgarno sequence is responsible for initiation-codon choice. A, schematic of the mRNA sequences used to program ribosome complexes. SD+ indicates the presence of a Shine-Dalgarno sequence upstream of the first AUG codon, SD− indicates its absence, and 1AUG indicates that the second AUG codon in the mRNA was altered to CCC (proline). B, autoradiograph of an electrophoretic TLC used to follow the reactivity of the depicted initiation complexes with the indicated aa-tRNA. C, toeprinting analysis reveals the importance of Shine-Dalgarno sequence in specifying the initiation codon. In the presence of a Shine-Dalgarno sequence, the ribosome occupies only the first codon, whereas in its absence, the ribosome occupies both AUG codons equivalently. U, A, C, G indicate sequencing lanes, and − indicates no addition. M1 indicates ribosome positioned at the first AUG, and MV and MD indicate formation of dipeptide that accompanies three-nucleotide movement. M2 indicates ribosome positioned at the second AUG.
Toeprinting Analysis Identifies Two AUG Codons in the mRNA Sequence
To interrogate ribosome positioning on the mRNA during this unusual decoding event (Asp-tRNAAsp efficiently decoding a noncognate valine codon, GUA), a “toeprinting” assay was performed in which primer extension by reverse transcriptase from the extended mRNA 3′-tail identifies the 3′-proximal (A-site) side of bound ribosomes (23). As before, ribosomal complexes were assembled with mRNAs that either did or did not carry an SD sequence upstream of the “initiating” AUG; however, in this case, the mRNAs carried 3′-terminal extensions to allow priming for the primer extension reaction (see “Experimental Procedures”). The ICs were then reacted (or not) with EFTu·GTP ternary complex carrying either Val-tRNAVal or a mixture of Val-tRNAVal and Asp-tRNAAsp; these complexes were then purified over a sucrose cushion. As anticipated, the primer extension reaction for ribosomal complexes with an SD sequence containing mRNA and fMet-tRNAfMet at the P-site revealed a single prominent “toeprint” at the appropriate position (Fig. 1C, lane 6). By contrast, when no SD sequence is present in the mRNA, initiation complexes (with only fMet-tRNAfMet at the P-site) reveal two nearly equivalent ribosome footprints on the mRNA (Fig. 1C, lane 10). Indeed, inspection of the mRNA sequence utilized here (and previously (1)) reveals the presence of two distinct AUG codons, one upstream and typically associated with the SD sequence and another downstream and not positioned next to an SD sequence (this latter AUG simply represents an internal methionine in the coding sequence). The presence of two equivalent toeprints is thus not surprising in the context of a mutated upstream SD sequence. Strikingly, the downstream AUG codon sits just upstream of an intact cognate aspartate codon (GAC).
Toeprinting experiments were also performed on initiation complexes that had been elongated with either Val-tRNAVal or a mix of Val-tRNAVal and Asp-tRNAAsp. The addition of Val-tRNAVal to the complexes results in a characteristic three-nucleotide shift of the upstream ribosome on complexes with or without the SD sequence (and MV dipeptide is produced by this elongation step). By contrast, the downstream toeprinting pattern was shifted three nucleotides only in the presence of Asp-tRNAAsp. These results are strikingly consistent with the observations from the peptidyl transfer reactions in Fig. 1B. The miscoding by Asp-tRNAAsp (to form MD dipeptide) apparently resulted from ribosomal initiation on the downstream AUG codon (with no SD sequence), whereas normal decoding (to form MV dipeptide) resulted from ribosomal initiation on the upstream AUG codon. We subsequently confirmed that the MD dipeptide is produced from the second AUG codon by mutating the second AUG codon to CCC and showing that no MD dipeptide product is then formed (Fig. 1B, lanes 5 and 10).
Although the earlier results of Di Giacco et al. (1) were confirmed in our own reconstituted system, the explanation that the quality of the SD-aSD interaction affects the fidelity of tRNA selection is flawed. The miscoding effects that were previously documented can be understood in terms of a second, downstream AUG codon with a proximal aspartate (GAC) codon and a normal cognate tRNA selection event.
Bound E-site tRNA Fails to Impact Fidelity of tRNA Selection in A-Site
Previous experiments that argued for allostery between the E- and A-sites were performed using ribosomes programmed either with poly(U) (2, 3) or with specific heteropolymeric mRNAs (1). Here we extended these studies using a number of distinct homo- and heteropolymeric mRNAs to examine the impact of E-site tRNA binding on the fidelity of tRNA selection in the A-site.
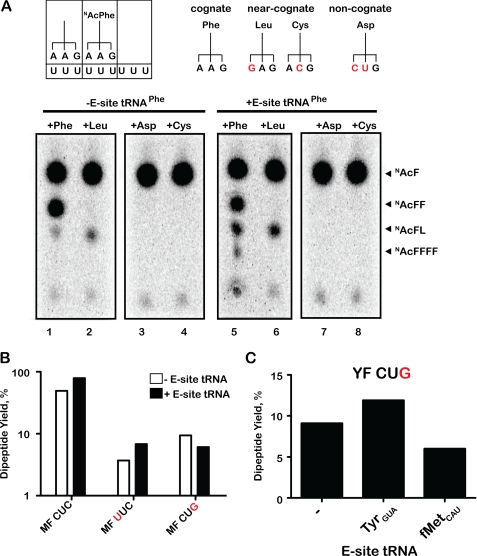
We began by programming ribosomes with poly(U) mRNA, N-acetyl-[14C]Phe-tRNAPhe in the P-site, and deacylated tRNAPhe in the E-site (or not). Ribosomal complexes without an E-site tRNA were prepared by simply adding N-Ac-Phe-tRNAPhe to the poly(U)-programmed ribosomes; binding of the acylated tRNA should be to the classical P-site. Ribosomal complexes loaded with E-site tRNA were prepared as described previously (25, 26) by first binding deacylated tRNAPhe (which binds in either the P/P- or the P/E-state) and then by subsequently adding N-Ac-Phe-tRNAPhe (which binds in the A/P-state, forcing the deacylated tRNA into the P/E-state). Following this so-called nonenzymatic loading, EFG was added to translocate the complex from the pre- to post-translocation state. Ribosomal complexes with or without E-site tRNA were next purified over a sucrose cushion and were subsequently reacted with various ternary complexes (EFTu·GTP with Phe-tRNAPhe, Leu-tRNALeu, Asp-tRNAAsp, or Cys-tRNACys). We note that these experiments were performed in the presence of polyamines, which have been previously shown to stabilize any E-site-bound tRNA approaching 100% occupancy (5, 27). First, when Phe-tRNAPhe is added, we see the production of dipeptide (N-AcFF) (Fig. 2A, lane 1) using our previously described electrophoretic TLC system (22) and even subsequent additions of Phe in complexes that are likely contaminated with EFG (e.g. N-AcFFF in Fig. 2A, lane 5). Contrary to earlier studies (1), we observe that the presence of E-site tRNA modestly increases the amount of miscoding as reported on by the reactivity of Leu-tRNALeu to form N-AcFL dipeptide product (Fig. 2A, lane 6) (Leu on Phe has a codon-anticodon interaction with a mismatch in the first position in this case). We see that both the absolute amount of product and the fraction of N-Ac-Phe converted to dipeptide are increased by the presence of the bound E-site tRNA (from 4.7 to 9.8%). In the case of reactions with other tRNAs including Asp-tRNAAsp (noncognate) or Cys-tRNACys (near cognate), we see no evidence of dipeptide formation in the electrophoretic system, suggesting that if miscoding occurs in this case, the reaction is very inefficient.
FIGURE 2.
In vitro translation reactions fail to support allosteric model. A, schematic of the poly(U)-programmed complex and the A-site aa-tRNA substrates (showing their anticodons). We used four different aa-tRNAs to test A-site reactivity with or without E-site deacylated tRNA. Mismatched nucleotides are indicated in red. The bottom panel shows an autoradiograph of an electrophoretic TLC used to follow the reactivity of a complex carrying N-Ac-[14C]Phe-tRNAPhe in the P-site with the indicated aa-tRNA (+Phe, +Leu, +Asp, +Cys) in the absence and presence of E-site tRNA. Products of the reaction are indicated as N-Ac-Phe (NAcF), N-Ac-Phe-Phe (NAcFF), N-Ac-Phe-Phe-Phe-Phe (NAcFFFF), and N-Ac-Phe-Leu (NAcFL). B, bar graph depicting the yield of dipeptide product (N-AcFL) for complexes programmed with the indicated heteropolymeric mRNA and reacted with Leu-tRNA2Leu. In all cases, the E-site codon is AUG (or M for methionine), the P-site is filled with N-Ac-[14C]Phe-tRNAPhe on a Phe (F) codon, whereas the A-site codon is denoted by its full nucleotide sequence. The reaction yield was determined by HPLC separation of the reaction products followed by scintillation counting of the fractions. C, bar graph depicting yield of dipeptide product (N-AcFL) for the YF CUG complex reacting with Leu-tRNA2Leu in the presence of the indicated tRNAs. In this complex, the E-site codon is UAC (or Y for tyrosine), the P-site is filled with N-Ac-[14C]Phe-tRNAPhe on a Phe (F) codon, whereas the A-site codon is CUG.
Although initial experiments by Nierhaus and colleagues (2) used homopolymeric mRNAs, the group switched to heteropolymeric mRNAs and again reported that bound E-site tRNA increases the fidelity of tRNA selection in the A-site (1). As a result, we used the heteropolymeric message MFK to prepare complexes with and without E-site-bound tRNA, as described previously (1). As with the homopolymeric mRNA, for no E-site tRNA complexes, N-Ac-Phe-tRNAPhe was simply added to the MFK mRNA-programmed ribosomes; for E-site tRNA containing complexes, deacylated tRNAfMet was first added, then N-Ac-Phe-tRNAPhe, and then EFG for translocation from the pre- to the post-translocation state. Again, these ribosome complexes were purified over a sucrose cushion and used for subsequent elongation reactions. Ternary complexes (EFTu·GTP carrying Lys-tRNALys or Leu-tRNALeu) were next added as indicated, and the products were resolved by electrophoretic TLC. Although the cognate dipeptide N-Ac-Phe-Lys was readily formed with and without E-site-bound tRNA, we were unable to detect the formation of noncognate dipeptide N-Ac-[14C]Phe-Leu in this system (<7% relative to N-Ac-Phe-Lys) (data not shown). In equivalent experiments performed by Di Giacco et al. (1), the levels of miscoding detected were on the order of 4%.
Resolution of Products by HPLC Analysis Yields Same Results
We further explored the incorporation of near cognate or noncognate aminoacyl-tRNAs during tRNA selection using the HPLC system to increase the sensitivity of the experiment. Moreover, we increased the overall concentration of ternary complex in the experiments to favor the production of dipeptide product (concentration of 20 μm, well above the Kd of the interaction between the ribosome and ternary complex). In an extensive set of experiments, we used a related group of mRNA sequences encoding MFLCUC, MFLUUC, MFLCUG, MFLUUA, MFKAAA, and MFMAUG to evaluate potential tRNA selection of [3H]Leu-tRNA on cognate, near cognate (first or third position mismatch between the tRNA-mRNA codon anticodon interaction), and noncognate (more than one mismatch) codons at the A-site (Fig. 2B). The dipeptide (N-Ac-[14C]Phe-[3H]Leu) formed was resolved from reactants via HPLC and quantified based on the amount of 14C and 3H in the product species. These complexes were prepared as described previously with and without E-site tRNA on the described heteropolymeric mRNA sequences.
As a positive control, we used the mRNA sequence encoding MF(CUC), CUC being a cognate leucine codon that should be readily decoded by Leu-tRNA2Leu ternary complex. The products of the reactions (on ribosome complexes with and without E-site tRNA) were resolved by HPLC (see “Experimental Procedures”) and quantified by measuring the incorporated radiolabeled amino acids N-Ac-[14C]-Phe and [3H]Leu. We see the formation of substantial amounts (50 and 80% fractional amount) of dipeptide (N-Ac-[14C]Phe-[3H]Leu) in the absence and presence of E-site tRNA, respectively (Fig. 2B). We next used an mRNA sequence encoding MF(AAA), AAA being a noncognate codon for leucine. Consistent with the electrophoretic TLC studies described above, we fail to observe the production of detectable amounts of N-Ac-[14C]Phe-[3H]Leu in the HPLC system (data not shown). These results again fail to provide experimental support for the earlier experiments by Nierhaus and colleagues (1). We note that Di Giacco et al. (1) reported a yield for the formation of the N-Ac-Phe-Leu product of 1.5% (i.e. ratio of N-Ac-Phe-Leu to the total N-Ac-Phe added), which would result in a formation of 0.08 pmol of N-Ac-Phe-Leu product in our reaction. This amount of product should have been detected above the background of 0.03 pmol.
To further explore the fidelity of selection in this system, we followed the misincorporation of Leu from Leu-tRNA2Leu on mRNAs programmed with several near cognate codons, UUC (first position mismatch) and CUG (third position mismatch). As anticipated, we see modest amounts of product formed both with and without the E-site tRNA (Fig. 2B); in the case of the UUC codon, the E-site tRNA modestly stimulates misincorporation, whereas for the CUG codon, the E-site tRNA modestly inhibits misincorporation. In fact, in our hands, the presence of the E-site tRNA generally tends to modestly stimulate incorporation of aminoacyl-tRNA in the elongation reaction, contrary to earlier studies (1–4). In the rare case where the E-site tRNA modestly inhibits the elongation reaction (with MFLCUG), we noticed that there was significant complementarity between the deacylated tRNAfMet being used to fill the P-site and the A-site codon CUG.
We speculated that the effects of the E-site on miscoding might be explained simply by direct competition in the A-site from the deacylated tRNAfMet in the reaction. This idea was tested by altering the identity of the codon in the E-site to UAC (Tyr) and showing that the addition of deacylated tRNATyr again modestly stimulated the misincorporation of Leu on YFLCUG, whereas deacylated tRNAfMet (no longer having its cognate codon in the E-site) now more substantially inhibits the misincorporation event (Fig. 2C). These data are consistent with the hypothesis that previously documented increases in selectivity (and the allosteric model for E-site function) may in some cases be rationalized by effects on tRNA selection resulting simply from direct competition of the free deacylated tRNA in the reaction.
DISCUSSION
In this study, we investigated the influence of the Shine-Dalgarno sequence and of the E-site tRNA on the fidelity of tRNA selection during translation elongation. In the case of the SD sequence, the earlier studies are readily rationalized by the presence of a downstream previously unnoticed AUG codon that led to the surprising results. In the case of the E-site tRNA, we are broadly unable to recapitulate the unusual miscoding results reported in several earlier studies despite the fact that the system we used was designed to mimic the particular features of those studies. Our data thus fail to support earlier models arguing for allosteric interactions between the E- and the A-site that modulate the fidelity of tRNA selection. We continue to try to understand at a mechanistic level how interactions in the E-site can impact retrospective editing during translation elongation as characterized in the E. coli system (17, 28).
Acknowledgments
We thank Hani Zaher for critical reading of the manuscript and members of the Green laboratory for useful discussions.
Footnotes
- SD
- Shine-Dalgarno
- SD-aSD
- Shine-Dalgarno-anti-Shine-Dalgarno
- aa-tRNA
- aminoacyl-tRNA
- IC
- initiation complex
- EFG
- elongation factor G
- EFTu
- elongation factor Tu
- IF
- initiation factor.
REFERENCES
- 1. Di Giacco V., Márquez V., Qin Y., Pech M., Triana-Alonso F. J., Wilson D. N., Nierhaus K. H. (2008) Shine-Dalgarno interaction prevents incorporation of noncognate amino acids at the codon following the AUG. Proc. Natl. Acad. Sci. U.S.A. 105, 10715–10720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geigenmüller U., Nierhaus K. H. (1990) Significance of the third tRNA binding site, the E-site, on E. coli ribosomes for the accuracy of translation: an occupied E-site prevents the binding of non-cognate aminoacyl-tRNA to the A-site. EMBO J. 9, 4527–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schilling-Bartetzko S., Bartetzko A., Nierhaus K. H. (1992) Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J. Biol. Chem. 267, 4703–4712 [PubMed] [Google Scholar]
- 4. Nierhaus K. H. (2006) Decoding errors and the involvement of the E-site. Biochimie 88, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 5. Semenkov Y. P., Rodnina M. V., Wintermeyer W. (1996) The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 12183–12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uemura S., Aitken C. E., Korlach J., Flusberg B. A., Turner S. W., Puglisi J. D. (2010) Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Stevens B., Kaur J., Smilansky Z., Cooperman B. S., Goldman Y. E. (2011) Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 16980–16985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jenner L., Rees B., Yusupov M., Yusupova G. (2007) Messenger RNA conformations in the ribosomal E-site revealed by X-ray crystallography. EMBO Rep. 8, 846–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korostelev A., Trakhanov S., Laurberg M., Noller H. F. (2006) Crystal structure of a 70 S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 10. Selmer M., Dunham C. M., Murphy F. V., 4th, Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., Ramakrishnan V. (2006) Structure of the 70 S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 11. Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 [DOI] [PubMed] [Google Scholar]
- 12. Lill R., Wintermeyer W. (1987) Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 196, 137–148 [DOI] [PubMed] [Google Scholar]
- 13. Robertson J. M., Wintermeyer W. (1987) Mechanism of ribosomal translocation: tRNA binds transiently to an exit site before leaving the ribosome during translocation. J. Mol. Biol. 196, 525–540 [DOI] [PubMed] [Google Scholar]
- 14. Devaraj A., Shoji S., Holbrook E. D., Fredrick K. (2009) A role for the 30 S subunit E-site in maintenance of the translational reading frame. RNA 15, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Márquez V., Wilson D. N., Tate W. P., Triana-Alonso F., Nierhaus K. H. (2004) Maintaining the ribosomal reading frame: the influence of the E-site during translational regulation of release factor 2. Cell 118, 45–55 [DOI] [PubMed] [Google Scholar]
- 16. Sanders C. L., Curran J. F. (2007) Genetic analysis of the E-site during RF2 programmed frameshifting. RNA 13, 1483–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaher H. S., Green R. (2009) Quality control by the ribosome following peptide bond formation. Nature 457, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jelenc P. C., Kurland C. G. (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. U.S.A. 76, 3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartetzko A., Nierhaus K. H. (1988) Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 164, 650–658 [DOI] [PubMed] [Google Scholar]
- 20. Moazed D., Noller H. F. (1986) Transfer RNA shields specific nucleotides in 16 S ribosomal RNA from attack by chemical probes. Cell 47, 985–994 [DOI] [PubMed] [Google Scholar]
- 21. Brunelle J. L., Youngman E. M., Sharma D., Green R. (2006) The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. Rna 12, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Youngman E. M., Brunelle J. L., Kochaniak A. B., Green R. (2004) The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 [DOI] [PubMed] [Google Scholar]
- 23. Hartz D., McPheeters D. S., Traut R., Gold L. (1988) Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164, 419–425 [DOI] [PubMed] [Google Scholar]
- 24. Zaher H. S., Green R. (2010) Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell 39, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moazed D., Noller H. F. (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 [DOI] [PubMed] [Google Scholar]
- 26. Rheinberger H. J., Schilling S., Nierhaus K. H. (1983) The ribosomal elongation cycle: tRNA binding, translocation, and tRNA release. Eur. J. Biochem. 134, 421–428 [DOI] [PubMed] [Google Scholar]
- 27. Gnirke A., Geigenmüller U., Rheinberger H. J., Nierhaus L. H. (1989) The allosteric three-site model for the ribosomal elongation cycle: analysis with a heteropolymeric mRNA. J. Biol. Chem. 264, 7291–7301 [PubMed] [Google Scholar]
- 28. Zaher H. S., Green R. (2011) A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell 147, 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]