Background: Detailed insight into the role of chloroplastic H2O2 in cell signaling is necessary.
Results: A large change in gene expression occurred in response to chloroplastic H2O2, resulting in positive and negative effects on the response to stresses.
Conclusion: Chloroplastic H2O2 regulates abiotic and biotic stress response.
Significance: We provided a new insight into the role of chloroplastic H2O2 in stress response.
Keywords: Arabidopsis, Reactive Oxygen Species (ROS), Signal Transduction, Stress Response, Transcriptomics
Abstract
Recent findings have suggested that reactive oxygen species (ROS) are important signaling molecules for regulating plant responses to abiotic and biotic stress and that there exist source- and kind-specific pathways for ROS signaling. In plant cells, a major source of ROS is chloroplasts, in which thylakoid membrane-bound ascorbate peroxidase (tAPX) plays a role in the regulation of H2O2 levels. Here, to clarify the signaling function of H2O2 derived from the chloroplast, we created a conditional system for producing H2O2 in the organelle by chemical-dependent tAPX silencing using estrogen-inducible RNAi. When the expression of tAPX was silenced in leaves, levels of oxidized protein in chloroplasts increased in the absence of stress. Microarray analysis revealed that tAPX silencing affects the expression of a large set of genes, some of which are involved in the response to chilling and pathogens. In response to tAPX silencing, the transcript levels of C-repeat/DRE binding factor (CBF1), a central regulator for cold acclimation, was suppressed, resulting in a high sensitivity of tAPX-silenced plants to cold. Furthermore, tAPX silencing enhanced the levels of salicylic acid (SA) and the response to SA. Interestingly, we found that tAPX silencing-responsive genes were up- or down-regulated by high light (HL) and that tAPX silencing had a negative effect on expression of ROS-responsive genes under HL, suggesting synergistic and antagonistic roles of chloroplastic H2O2 in HL response. These findings provide a new insight into the role of H2O2-triggered retrograde signaling from chloroplasts in the response to stress in planta.
Introduction
Reactive oxygen species (ROS)3 such as hydrogen peroxide (H2O2), superoxide radical (O2˙̄), singlet oxygen (1O2), and hydroxyl radical act not only as toxic compounds but also as signaling molecules associated with responses to abiotic and biotic stress in living organisms including animals, plants, and microbes (1–4). In plant cells, ROS are produced as byproducts of central metabolism, such as photosynthesis, respiration, and photorespiration, in chloroplasts, mitochondria, and peroxisomes (1, 3, 5). Additionally, plant cells contain ROS-producing enzymes, such as NADPH oxidase, which generates ROS in apoplast (4, 6–8). Recently, to identify ROS-responsive genes and to clarify the function of ROS as signaling molecules, transcriptomic analyses have been performed using ROS-generating agents or mutants lacking antioxidative enzymes (9–15). Interestingly, these analyses have revealed the existence of source- and kind-specific pathways for ROS signaling (15). The synergistic and antagonistic interactions of multiple signaling pathways in plants are thought to be important to the fine-tuning of responses to abiotic and biotic stress.
Chloroplasts are one of the most significant sources of ROS in plant cells (3). The ROS generated in chloroplasts act as a retrograde signal to the nucleus for regulating plant responses to environmental stress. The identification of a conditional fluorescent (flu) mutant of Arabidopsis, which allowed the production of 1O2 within plastids in a controlled manner, provided genetic evidence that the release of 1O2 is involved in the regulation of programmed cell death (9, 16–19). In the flu mutants, 1O2 generated within the first minute of re-illumination had a negative effect on growth and development. After re-illumination, distinct sets of genes were activated that were different from those induced by paraquat, a producer of H2O2 and O2˙̄, suggesting that 1O2 acts as a signal with a high degree of specificity (9). Rizhsky et al. (10) reported that Arabidopsis mutants (KD-CSD2) with suppressed expression of the thylakoid-attached copper/zinc superoxide dismutase (CSD2) showed a dwarf phenotype even under normal growth conditions, but were more tolerant to oxidative stress due to the induction of other defense-related genes. Furthermore, we recently found that although oxidative damage under high light (HL) was enhanced in knock-out mutants lacking stromal and thylakoid membrane-bound ascorbate peroxidase (respective sAPX and tAPX), which are key enzymes for the scavenging of H2O2 in chloroplasts, the HL-induced expression of ROS-responsive genes, such as heat shock transcription factor A2 (HsfA2) and cytosolic APX (APX1 and APX2), was significantly suppressed in these mutants (20), indicating that H2O2 derived from chloroplasts has a negative effect on the expression of ROS-responsive genes under HL. Thus, it seems likely that different kinds of ROS produced in the same organelle are associated with distinct signaling pathways.
To clarify the signaling function of H2O2 derived from chloroplasts, a system for producing H2O2 in a controlled manner is needed. For the system, the chemical-inducible RNAi would be more useful than the knock-out or constitutive knockdown method because plants may acclimate to the knock-out or constitutive knockdown of target gene during growth and development. Furthermore, this system should not require any application of stress to plants because other signaling molecules, including hormones, are produced by stress and may act synergistically or antagonistically. Here we created a novel system for producing H2O2 in Arabidopsis chloroplasts by chemical-dependent tAPX silencing using an estrogen-inducible RNAi method (21, 22). When the expression of tAPX was silenced in the leaves, the levels of oxidized protein in the chloroplasts were increased. Microarray analysis revealed that the expression of a large set of genes changed in response to tAPX silencing. Among them, only a single gene was known to be responsive to ROS, but many genes were associated with cold acclimation and pathogen resistance. These findings suggest that H2O2 derived from chloroplasts acts as a specific signal for regulating plant responses to abiotic and biotic stress. Furthermore, we found that tAPX silencing has negative and positive effects on cold acclimation and salycilic acid (SA) response, respectively, and that there are synergistic and antagonistic roles of chloroplastic H2O2 on HL response. These findings provide a new insight into the role of H2O2-triggered retrograde signaling from chloroplasts in the response to stress in planta.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Transformation of Arabidopsis
The method used for plasmid construction is described in supplemental Fig. S1. To construct the plasmid for the estrogen-inducible silencing of tAPX, a DNA fragment containing the 3′-untranslational region of tAPX (530 bp) was cloned into pGWB80, a vector for constitutive RNAi. An open reading frame of the β-glucuronidase (GUS) gene was also cloned into pGWB80 as control. The regions of the RNAi construct having an inverted repeat of tAPX and GUS were transferred to pMDC7 (pMDC7-tAPX and pMDC7-GUS, respectively), a vector for estrogen-inducible expression (21). Agrobacterium tumefaciens, which was transformed with the obtained constructs by electroporation, was used to infect Arabidopsis via the vacuum infiltration method (23). T1 seedlings were selected on basic Murashige & Skoog medium in Petri dishes containing 3% sucrose and 20 mg liter−1 spectinomycin for 2 weeks and transferred to soil. T3 seeds were harvested and used for the experiments.
Plant Materials and Growth Conditions
Surface-sterilized Arabidopsis thaliana wild-type (Col-0) and transgenic seeds were sown on Murashige & Skoog medium containing 3% sucrose. Plates were stratified in darkness for 2 days at 4 °C and then transferred to a growth chamber. After 7 days, seedlings were potted in soil and grown in the same growth chamber. In this study we use a continuous light (100 μmol of photons m−2 s−1, 25 °C) condition for plant growth to abolish light/dark effect on the production of ROS in chloroplasts. Seventeen-day-old transgenic plants were treated with a 100 μm estrogen solution containing 0.1% (v/v) Tween 20 under light. T-DNA insertion lines for tAPX in the Col-0 background (KO-tAPX; WiscDsLox457–460A17) were previously obtained from the Arabidopsis Biological Resource Center (20).
Preparation of Total RNA and cDNA Synthesis
Total RNA was semi-automatically purified from leaves of Arabidopsis plants using a QuickGene RNA cultured cell kit and QuickGene-Mini80 (FUJIFILM Corp., Tokyo, Japan). The first strand cDNA was synthesized using reverse transcriptase (ReverTra Ace; Toyobo) with an oligo(dT) primer. These analyses were performed according to the manufacturer's instructions.
Semiquantitative RT-PCR Analysis
The semiquantitative RT-PCR analysis was performed according to Ogawa et al. (24). Primer sequences were as follows: APX1-forward (F) (5′-TGGCCGTTGAAGTTACTGGTG-3′), APX1-reverse (R) (5′-CCAACCAAAAACAGCCATGAC-3′), APX3-F (5′-AGGCTTTGATGGACCATGGAC-3′), APX3-R (5′-GCAAAAATGAATCACGGACCC-3′), sAPX-F (5′-ATGGCAGAGCGTGTGTCTCT-3′), sAPX-R (5′-GTACCAGCATCATGCCAACC-3′), tAPX-F (5′-ATTTCACCAAAATGTGCCGC-3′), tAPX-R (5′-TTTTCCCCAACCACTACGGTC-3′), actin8-F (5′-GAGATCCACATCTGCTGG-3′), and actin8-R (5′-GCTGAGAGATTCAGGTGCCC-3′). The semiquantitative RT-PCR experiments were repeated at least three times with cDNA prepared from three batches of plant leaves.
Quantitative Real-time PCR Experiments
Quantitative Real-time PCR (q-PCR) experiments were performed according to Nishizawa et al. (25). Primer sequences were described in supplemental Table S1. The q-PCR experiments were also repeated at least three times with cDNA prepared from three batches of plant leaves.
Western Blotting
The Western blot analysis was carried out as described previously (20). The protein bands were detected using specific antibody (anti-tAPX) as the primary antibody and anti-mouse IgG-HRP conjugate (Bio-Rad, CA) as the secondary antibody. The anti-tAPX cross-reacted not only with tAPX but also with sAPX and APX1 (20).
Enzyme Assay
The activities of APX isoenzymes were measured as the decrease in absorbance at 290 nm (e = 2.8 mm−1 cm−1) due to AsA oxidation according to Yoshimura et al. (26). Leaf tissues (0.5 g) were ground to a fine powder in liquid N2 and then homogenized in 1 ml of 100 mm potassium phosphate buffer (pH 7.6), 20%(w/v) sorbitol, 1 mm EDTA, 5 mm AsA, and 2% (w/v) polyvinylpyrrolidone. The homogenate was centrifuged for 30 min at 12,000 × g. The soluble fraction contained the activities of the sAPX and cytosolic APX isozymes, whereas the membrane fraction had the activities of the tAPX and peroxisomal APX isozymes.
Measurements of Chlorophyll Fluorescence
Chlorophyll fluorescence was measured following Maruta et al. (20). The maximum quantum yield of photosystem II (Fv/Fm), and quantum yield of photosystem II (ΦPSII) in Arabidopsis leaves was determined after dark adaptation for 20 min. Chlorophyll fluorescence in the Arabidopsis leaves was measured at 25 °C with a Closed FluorCam 800MF (Photon Systems Instruments, Brno, Czech Republic).
Detection of H2O2 and Oxidized Protein
H2O2 was detected by diaminobenzidine (DAB) staining performed according to Maruta et al. (20). A detached leaf was vacuum-infiltrated with 1 mg ml−1 of DAB (pH 3.8). The DAB-treated leaf was kept under normal light (NL; 100 μmol photons m−2 s−1) and then decolorized by incubation in 70% ethanol. H2O2 was visualized as a reddish-brown color. Oxidized proteins were detected using a protein gel blot assay with an OxyBlot protein oxidation kit (Chemicon International) according to Davletova et al. (14).
Measurement of Antioxidants
The rosette leaves of Arabidopsis plants (0.5 g wet weight) were frozen in liquid N2 and used for the measurement of reduced and oxidized ascorbate (AsA and dehydroascorbate, respectively) and glutathione (GSH and GSSG, respectively). The levels of AsA and dehydroascorbate were determined spectrophotometrically using AsA oxidase as described previously (20). Levels of GSH and GSSG were determined using a GSH reductase recycling system coupled to 5,5′-dithiobis(2-nitrobenzoic acid) (27). All measurements were repeated at least three times with extracts from three batches of plants.
Isolation of Chloroplasts from Arabidopsis Leaves
The chloroplasts were isolated from Arabidopsis leaves according to Phee et al. (28) with minor modifications. Arabidopsis leaves (20 g) were cut into 1-cm2 pieces and chopped using a blender in 120 ml of extraction buffer (0.33 m sorbitol, 50 mm HEPES/KOH (pH 8.0), 5 mm MgCl2, and 2 mm isoascorbate). The homogenates were then filtered through 0.2-mm mesh and centrifuged at 2200 × g for 30 s. After the washing pellets once with extraction buffer, the pelleted chloroplasts were suspended in a small volume of the rupture buffer (100 mm Tris/HCl (pH 8.0), 16 mm MgCl2, 1 mm EDTA, and 2% Triton X).
Microarray Analysis
The microarray analysis was performed by using an Agilent Arabidopsis 3 Oligo Microarray (Agilent Technologies, Palo Alto, CA) as reported by Morishita et al. (29). Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were treated with estrogen for 48 h as described above. Total RNA was isolated using Sepasol-RNA I (Nakarai Tesque, Kyoto, Japan) from the control and tAPX-silenced plants; mRNA was prepared using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA). The total cDNA probe was synthesized and labeled using a Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies) following the manufacturer's instructions. Feature extraction software (GeneSpring; Agilent Technologies) was used to locate and delineate every spot in the array and to integrate the intensity, filtering, and normalization of each spot. Furthermore, the data from two replicates of IS-GUS-2-17 plants were normalized using GeneSpring to remove unusual data. Similarly, those from two replicates of IS-tAPX-19-23 plants were also normalized. Finally, we selected the genes whose ratio of induction in the tAPX-silenced plants was >2.0 (up-regulated genes) or <0.5 (down-regulated genes) by comparing both normalized data from IS-GUS-2-17and IS-tAPX-19-23 plants. Thus, two biological replicates were analyzed. The microarray data were deposited in the public NCBI Gene Expression Omnibus data base under the GEO accession number GSE35526.
Quantification of Free and Total SA
Free and total SA were extracted and measured according to Malamy et al. (30) with minor modifications. Arabidopsis leaves (0.2 g) were ground in 3 ml of 90% methanol and centrifuged at 7000 × g for 15 min. The pellet was resuspended with 3 ml of 90% methanol and centrifuged. Methanol extracts were combined, centrifuged at 7000 × g for 15 min, and dried at 35 °C under vacuum. The dried methanol extract was resuspended in 5 ml of water at 80 °C, and the solution was divided into two equal portions. An equal volume of 0.2 m sodium acetate buffer (pH 4.5) was added to one portion for assaying free SA, and an equal volume of 0.2 m sodium acetate buffer (pH 4.5) containing 0.24 mg/ml β-glucosidase (16.9 unit/mg; Roche) was added to the other for assaying total SA. Both samples were incubated at 37 °C overnight. After digestion, samples were acidified to pH 1–1.5 with HCI. The fractions were then extracted with two volumes of ethyl acetate:cyclopentane:isopropyl alcohol (50:50:1 [v/v]), dried down, resuspended in 23% (v/v) methanol/sodium acetate (pH 5.0), and filtered through a centrifugal filter (0.2 μm; Millipore). SA was quantified by reverse-phase HPLC on a 5C18 column (COSMOSIL; AR-II, 4.6ID × 150 mm) and detected using a Shimazu RF-10AXL fluorescence detector (excitation energy 313 nm, emission energy 405 nm).
Data Analysis
The significance of differences between data sets was evaluated with a t test. Calculations were carried out with Microsoft Excel software.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for major genes mentioned in this article are as follows: Actin2 (At3g18780), Actin8 (At2g42100), APX1 (At1g07890), APX3 (At4g35000), sAPX (At4g08390), tAPX (At1g77490), AtbZIP65 (At5g06839), AtNUDX6 (At2g04450), CBF1/DREB1B (At4g25490), CBF2/DREB1C (At4g25470), COR6.6 (At5g15970), COR15B (At2g42530), COR414-TM1 (At1g29395), COR414-TM2 (At1g29390), ICS2 (At1g18870), LCR68 (At2g02130), LCR70 (At2g02120), NIMIN-3 (At1g09415), RLP7 (At1g47890), RLP23 (At2g32680), RLP34 (At3g11010), RLP39 (At3g24900), RLP41 (At3g25010), Toll-interleukin resistance (TIR) domain protein (At4g11340), TolB-related protein (At4g01870).
RESULTS
Estrogen-induced Silencing of tAPX in Transgenic Plants
To generate estrogen-inducible tAPX RNAi Arabidopsis plants (inducible silencing (IS)-tAPX), we prepared an RNAi construct having an inverted repeat corresponding to a 530-bp fragment of the 3′-terminal region of tAPX or full-length GUS gene (as a negative control) and placed it under the control of an estrogen-inducible promoter (21) (Supplemental Fig. S1). We introduced these plasmids into Arabidopsis plants and then selected more than 100 independent lines of IS-tAPX and IS-GUS plants for spectinomycin resistance. Among transgenic lines (T2 generation), we screened the lines with high expression of a chimeric transcription factor, XVE, essential for estrogen-inducible RNAi (21) and then isolated two lines, IS-tAPX-19 and IS-tAPX-2, with higher and lower expression of XVE, respectively (supplemental Fig. S2).
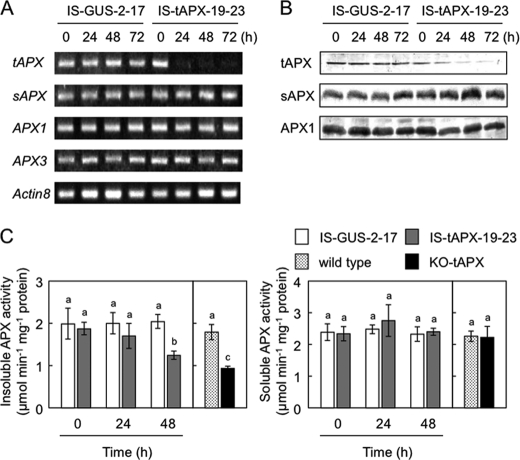
Seventeen-day-old IS-GUS-2, IS-tAPX-2, and IS-tAPX-19 plants (T2 generation) were sprayed with a 100 μm estrogen and incubated under continuous light at 100 μmol photons m−2 s−1. In the leaves of IS-tAPX-19, the transcript levels of tAPX started to decrease at 16 h after the treatment and were suppressed almost completely at 20 h, corresponding to the expression of the RNA triggers (Fig. 1 and supplemental Fig. S2). On the other hand, the treatment with estrogen had no effect on the transcript levels of tAPX in the leaves of not only IS-GUS-2 but also IS-tAPX-2 plants, suggesting that high expression of XVE is required for the induction of RNAi. The following analyses were performed using the T3 generation of IS-tAPX-19-23 and IS-GUS-2-17 plants (homozygous lines selected by spectinomycin). The protein levels of tAPX in the leaves of IS-tAPX-19-23, but not in those of IS-GUS-2-17, were drastically decreased at 48 h after treatment with estrogen (Fig. 1B). At 48 h after the treatment, the activity of APX in the membrane fraction of the IS-tAPX-19-23 plants was ∼61% that in the IS-GUS-2-17 plants (Fig. 1C). We also checked the activity of APX in the membrane fraction of the knock-out mutants (KO-tAPX) and found that the activity of the mutants was ∼52% that in the wild-type plants (Fig. 1C), indicating that an almost complete silencing of tAPX occurs in the IS-tAPX-19-23 plants under these conditions. Although the 530-bp fragment of the RNAi-trigger had partially homology to APX1 and sAPX genes, the treatment with estrogen had no effect on the expression levels of other APX isoenzymes, APX1, APX3, and sAPX, and the activities of soluble APX isoenzymes in either IS-GUS-2-17 or IS-tAPX-19-23 plants (Fig. 1). No responsivity of APX2 to the treatment was confirmed by q-PCR (data not shown). These findings indicated that the treatment with estrogen specifically suppresses the expression of tAPX in the IS-tAPX-19-23 plants.
FIGURE 1.
Estrogen-dependent silencing of tAPX in the transgenic plants. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants, grown under NL, were sprayed with a 100 μm estrogen. A, shown is the transcript levels of APX genes. Semiquantitative RT-PCR was performed using specific primers for APX genes and Actin8 on total RNA from transgenic plants. PCR amplification was performed with 18–28 cycles of 95 °C for 60 s, 55 °C for 60 s, and 72 °C for 60 s. Aliquots of the products were analyzed on 2% agarose gel. B, shown are Western blots of tAPX, sAPX, and APX1 proteins using anti-tAPX antibody. C, activities of soluble and insoluble APXs in crude extracts of Arabidopsis leaves are shown. Seventeen-day-old wild-type and KO-tAPX plants were also used in this study. Error bars indicate S.D. (n = 3). Values without a common letter are significantly different according to t tests (p < 0.05).
We also studied the dose-dependent effect of estrogen treatment on tAPX expression and found that treatment with low concentrations of estrogen (25∼50 μm) effectively induces tAPX silencing in the IS-tAPX-19-23 plants (data not shown). However, the effect of 25∼50 μm estrogen on tAPX expression was often inconsistent. Therefore, we used 100 μm concentrations of estrogen in this study according to rigorous routine safety precautions.
tAPX Silencing Results in Increased Oxidative Damage in Chloroplasts under NL
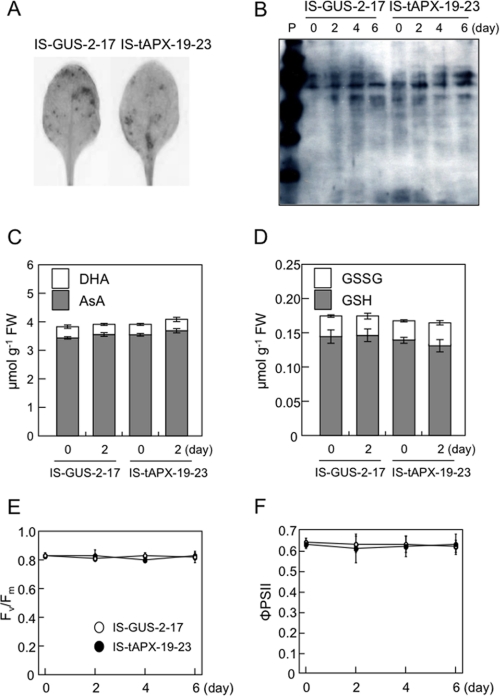
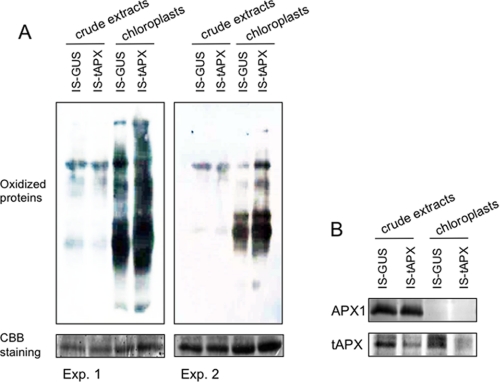
To clarify whether tAPX silencing causes H2O2 to accumulate in chloroplasts under NL, we checked the levels of H2O2, oxidized proteins, AsA, and GSH in the leaves of transgenic plants treated with estrogen. Surprisingly, the levels of H2O2 were almost the same between IS-tAPX-19-23 and IS-GUS-2-17 plants at 48 h after the treatment with estrogen (Fig. 2A). Additionally, as shown in Fig. 2B, the oxidation of total proteins was not affected by tAPX silencing. Furthermore, there were no differences in the levels and redox state of AsA and GSH between the transgenic lines (Fig. 2, C and D). These findings suggest that tAPX silencing does not cause an accumulation of H2O2 at high levels in whole leaves under NL. Next, to investigate the effect of tAPX silencing on the oxidative damage in more detail, we determined the levels of oxidized proteins in chloroplasts isolated from IS-tAPX-19-23 and IS-GUS-2-17 leaves at 48 h after the treatment with estrogen because the almost complete silencing of tAPX was observed (Fig. 1). The levels of oxidized proteins were greater in the chloroplasts of IS-tAPX-19-23 leaves than those of IS-GUS-2-17 leaves (Fig. 3), indicating that tAPX silencing causes an increase in oxidative damage. On the other hand, there was no difference in Fv/Fm and ΦPSII between the IS-tAPX-19-23 and IS-GUS-2-17 plants treated with estrogen (Fig. 2, E and F). Thus, it seems likely that the levels of H2O2 in the chloroplasts were increased by tAPX silencing under NL, although they were not very high, leading to inhibition of the photosynthetic machinery.
FIGURE 2.
Effect of tAPX silencing on cellular redox state and photosynthesis under NL. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL for indicated period. A, H2O2 was detected by DAB staining. A detached leaf at 48 h after the estrogen treatment was vacuum-infiltrated with DAB solution and incubated under NL for 3 h. The leaf was then decolorized by incubation in 70% ethanol. The same results were obtained in five independent experiments. Results of representative leaves were photographed. B, total proteins were extracted from leaves of IS-GUS-2-17 and IS-tAPX-19-23 plants. Oxidized proteins were detected using a protein gel blot assay with an OxyBlot protein oxidation kit as described under “Experimental Procedures.” C and D, levels of AsA, dehydroascorbate (DHA), GSH, and GSSG were measured. White bars (showing dehydroascorbate and GSSG) are started on top of gray bars (showing AsA and GSH). E and F, quantum yields of photosystem II, Fv/Fm and ΦPSII, were also measured using a Closed FluorCam 800MF as described under “Experimental Procedures.”
FIGURE 3.
Protein oxidation in chloroplasts isolated from tAPX-silenced leaves. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL. At 48 h after estrogen treatment, the chloroplasts were isolated from leaves of IS-GUS-2-17 and IS-tAPX-19-23 plants. A, oxidized proteins in the crude extracts and chloroplasts were detected by Western blotting. The same results were obtained in five independent experiments. Two representative results are shown as Exp. 1 and 2. CBB, Coomassie Brilliant Blue. B, shown are Western blots of tAPX, sAPX, and APX1 for Exp. 1. The blot of APX1 indicated that there was no contamination of the cytosolic fraction in the chloroplasts. In the figure, IS-GUS-2-17 and IS-tAPX-19-23 plants are shown as IS-GUS and IS-tAPX, respectively.
Identification of Genes Responsive to tAPX Silencing
To identify the genes responsive to H2O2 derived from chloroplasts, a microarray analysis was performed using the leaves of IS-tAPX-19-23 and IS-GUS-2-17 plants treated with estrogen for 48 h. The transcription of 365 genes was up-regulated in response to tAPX silencing in the IS-tAPX-19-23 plants, and that of 409 genes was down-regulated (supplemental Table S2). These genes were named here responsive to tAPX silencing (RTS). In the microarray data, one of the most down-regulated genes was tAPX (supplemental Table S2). Some RTS genes are known to be associated with abiotic and biotic stress responses, hormonal responses, growth, and development. Approximately 25% of all RTS genes encoded signal transduction-related proteins, such as transcription factors and protein kinases.
To confirm whether the induction of RTS genes by tAPX silencing was due to an increase in H2O2 levels, we studied the effect of treatment with AsA, a scavenger of ROS, or dark on the transcript levels of five RTS genes, gibberellin-regulated family protein (GRFP; At5g59845), jacalin lectin family protein (JLFP: At1g60110), ULTRAPETALA 2 (ULT2; At2g20825), cytochrome P450 family protein (CYP72A14: At3g14680), and protein kinase (PK; At4g13190), which were highly induced by tAPX silencing (supplemental Table S2). The induction of all RTS genes by tAPX silencing was significantly inhibited by the treatment with AsA for 3 h (Fig. 4A). Furthermore, an almost complete inhibition of the induction by tAPX silencing was observed in the dark (Fig. 4A). These findings suggest that RTS genes are responsive to the H2O2 produced by photosynthesis in chloroplasts.
FIGURE 4.
Effects of treatment with AsA or dark on the induction of RTS genes by tAPX-silencing. A, seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL or dark. At 45 h after the treatment with estrogen, the leaves of IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with 10 mm AsA or water and kept under NL. The transcript levels of five RTS genes (GRFP, JLFP, ULT2, CYP72A14, and PK) were measured by q-PCR as described under “Experimental Procedures.” Error bars indicate S.D. (n = 3). Values without a common letter are significantly different (p < 0.05). B, transcript levels of five RTS genes (described in Fig. 4A) in the wild-type and KO-tAPX plants, grown under light for 17 days, were measured by q-PCR. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for wild-type plants.
As described in the introduction section, we thought the estrogen-inducible RNAi method has a benefit for identifying the source-specific ROS signaling than the knock-out method. To clarify this, the expression of the above five RTS genes in KO-tAPX was investigated. Surprisingly, the transcript levels of the genes were similar to those in the wild-type plants (Fig. 4B). Therefore, a further 7 RTS genes, bZIP family transcription factor (AtbZIP65; At5g06839), 1-aminocyclopropane-1-carboxylate synthase (ACS7; At4g26200), unknown protein (UP; At3g20340), F-box family protein (F-box; At5g15660), hydrolase (At1g49640), protein disulfide isomerase 1-like 1-1 (PDIL1-1; At1g21750), and Arabidopsis NAC domain-containing protein 74 (ANAC074; At4g28530) selected at random were analyzed. As shown in supplemental Fig. S3, the transcription of these genes was significantly up-regulated by the treatment with estrogen in the IS-tAPX-19-23 plants. These results reflected the microarray data (Table 1 and supplemental Table S2). Only one gene (hydrolase) was highly expressed in the KO-tAPX plants. However, the transcript levels of the other RTS genes were lower in the KO-tAPX than wild-type plants (supplemental Fig. S3). These findings indicated that little transcriptomic change in response to tAPX silencing was observed in the KO-tAPX plants and thus that estrogen-inducible RNAi is useful for identifying source-specific H2O2 signaling.
TABLE 1.
The microarray and q-PCR data of RTS genes expression
| AGI code | Annotation | -Fold change IS-tAPX/IS-GUS |
Figure | |
|---|---|---|---|---|
| Microarray | q-PCR | |||
| At5g59845 | GRFP | 163.5 | 64.3 | 4 |
| At1g60110 | JLFP | 83.6 | 41.7 | 4 |
| At2g20825 | ULT2 | 51.0 | 20.7 | 4 |
| At3g14680 | CYP72A14 | 45.1 | 16.1 | 4 |
| At4g13190 | PK | 16.5 | 16.1 | 4 |
| At4g25490 | CBF1/DREB1B | 0.3 | 0.3 | 5 |
| At4g25470 | CBF2/DREB1C | 0.5 | 0.5 | 5 |
| At5g15970 | COR6.6 | 0.5 | 0.5 | 5 |
| At2g42530 | COR15B | 0.3 | 0.6 | 5 |
| At1g29395 | COR414-TM1 | 0.3 | 0.6 | 5 |
| At1g29390 | COR414-TM2 | 0.5 | 0.6 | 5 |
| At1g18870 | ICS2 | 25.2 | 7.3 | 6 |
| At4g11340 | TIR domain protein | 4.1 | 13.9 | 6 |
| At4g01870 | TolB-related protein | 4.4 | 6.4 | 6, Supplemental Fig. 4 |
| At1g47890 | RLP7 | 2.1 | 4.3 | 6 |
| At2g32680 | RLP23 | 4.7 | 4.3 | 6 |
| At3g11010 | RLP34 | 2.1 | 3.2 | 6 |
| At3g24900 | RLP39 | 9.4 | 3.1 | 6 |
| At3g25010 | RLP41 | 4.3 | 3.3 | 6 |
| At1g09415 | NIMIN-3 | 2.7 | 3.5 | 6 |
| At2g04450 | AtNUDX6 | 2.1 | 2.8 | 6 |
| At2g02130 | LCR68 | 5.0 | 5.2 | 6 |
| At2g02120 | LCR70 | 2.0 | 5.9 | 6 |
| At5g06839 | AtbZIP65 | 2.7 | 4.4 | Supplemental Fig. 3 |
| At4g26200 | ACS7 | 33.5 | 5.5 | Supplemental Fig. 3 |
| At3g20340 | UP | 16.5 | 7.7 | Supplemental Fig. 3 |
| At5g15660 | F-box | 2.1 | 2.2 | Supplemental Fig. 3 |
| At1g49640 | Hydrolase | 14.6 | 10.7 | Supplemental Fig. 3 |
| At1g21750 | PDIL1-1 | 2.4 | 2.2 | Supplemental Fig. 3 |
| At4g28530 | ANAC074 | 2.3 | 2.0 | Supplemental Fig. 3 |
Gadjev et al. (15) conducted a comparative transcriptome analysis using microarray data sets obtained by the application of oxidative stress-causing agents and from a mutant (flu) and transgenic plants lacking APX1, catalase, and copper/zinc superoxide dismutase and identified more than 30 genes as “general oxidative stress response markers” (see supplemental Fig. S4) whose steady-state levels of transcription were elevated at least 5-fold in most experiments. However, our microarray data showed that among RTS genes, there were only a few of these marker genes (At2g41380 and At4g01870). Therefore, we checked the effect of tAPX silencing on the transcript levels of all marker genes by q-PCR analysis. Corresponding to the results of the microarray assay, the transcript levels of At4g01870 were increased in response to tAPX silencing (supplemental Fig. S4). Additionally, the transcript levels of At2g41380 were also higher in the IS-tAPX-19-23 plants (supplemental Fig. S4), although not so high as compared with the microarray data (supplemental Table S2). However, among the other markers, we could not detect any whose transcript level was increased more than 2-fold by the tAPX silencing (supplemental Fig. S4). These findings suggest that H2O2 derived from chloroplasts acts as a signal that has a different role from that derived from other sources.
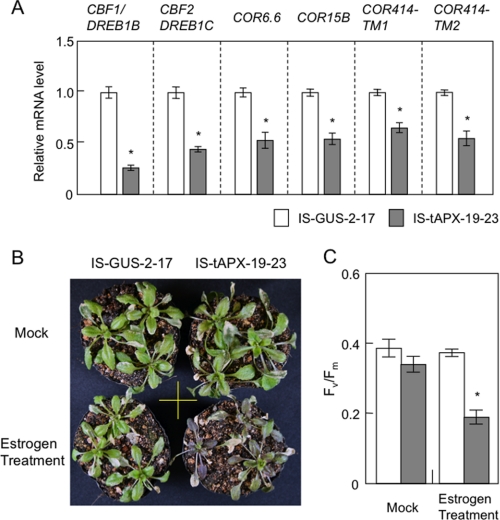
Suppression of Cold-responsive Genes by tAPX Silencing Causes Plant Sensitivity to Cold Stress
The microarray data indicated that the expression of cold-responsive genes was suppressed by tAPX silencing under NL, which was consistent with the results of the q-PCR analysis (Fig. 5A). These genes included C-repeat/DRE binding factor/dehydration-responsive element binding factors (CBF1/DREB1B and CBF2/DREB1C), and cold-regulated genes (COR6.6, COR15B, COR414-TM1, and COR414-TM2). To investigate the involvement of the H2O2 signaling derived from chloroplasts in the regulation of cold response, we checked the effect of tAPX silencing on plant sensitivity to cold. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were incubated under light for 48 h after the treatment with or without estrogen and then subjected to cold stress (continuous light of 100 μmol of photons m−2 s−1, 4 °C) for 2 weeks. Both transgenic lines showed visible symptoms under cold stress, but only the estrogen-treated IS-tAPX-19-23 plants exhibited brown leaves (Fig. 5B). In addition, Fv/Fm decreased significantly in the IS-tAPX-19-23 plants compared with the IS-GUS-2-17 plants under cold stress (Fig. 5C). The cold sensitivity of IS-tAPX-19-23 plants depended on the treatment with estrogen (Fig. 5, B and C). On the other hand, the cold-sensitive phenotypes of the tAPX-silenced plants were not observed under low light (LL; 10 μmol photons m−2 s−1) (supplemental Fig. S5), because the production rate of chloroplastic H2O2 appears to be limited under LL. These findings demonstrated that tAPX silencing negatively affects cold acclimation.
FIGURE 5.
Effect of tAPX silencing on cold acclimation. A, 17-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL. At 48 h after the estrogen treatment, the transcript levels of RTS genes (CBF1/DREB1B, CBF2/DREB1C, COR6.6, COR15B, COR414-TM1, and COR414-TM2), known to be involved in cold acclimation, were measured by q-PCR. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for IS-GUS-2-17 plants. B and C, 17-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen solution or water (Mock) and transferred to cold stress conditions (100 μmol photons m−2 s−1, 4 °C) for 2 weeks. The treatment with estrogen was performed every 3 days to maintain the tAPX silencing. B, 14 days after cold stress, the IS-GUS-2-17 and IS-tAPX-19-23 plants were photographed. The same results were obtained in four independent experiments. C, Fv/Fm values in the leaves of IS-GUS-2-17 and IS-tAPX-19-23 10 days after cold stress were measured using a Closed FluorCam 800MF. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value of IS-GUS-2-17 plants.
We also checked the sensitivity of KO-tAPX plants to cold stress. Although the transcript levels of CBF2/DREB1C and COR414-TM1 were significantly lower in the KO-tAPX plants than wild-type plants, those of other cold-responsive genes were similar in the two lines (Supplemental Fig. S6). In addition, there was no visible difference in sensitivity to cold stress between the wild-type and KO-tAPX plants. Furthermore, the Fv/Fm in the leaves of wild-type plants was almost the same as that in the KO-tAPX plants under cold stress (Supplemental Fig. S6). As shown in Fig. 1C, the activity of APX in the membrane fraction was much lower in the KO-tAPX plants than in the estrogen-treated (48 h) IS-tAPX-19-23 plants. Thus, it seems likely that the cold-sensitive phenotypes of tAPX-silenced plants are due to the down-regulation of cold-responsive genes by H2O2 signaling and not due to a decrease in ROS-scavenging capacity. Thus, the H2O2 signaling derived from chloroplasts seems to be involved in the suppression of cold acclimation.
tAPX Silencing Activates Expression of Disease-resistance Genes and SA Response
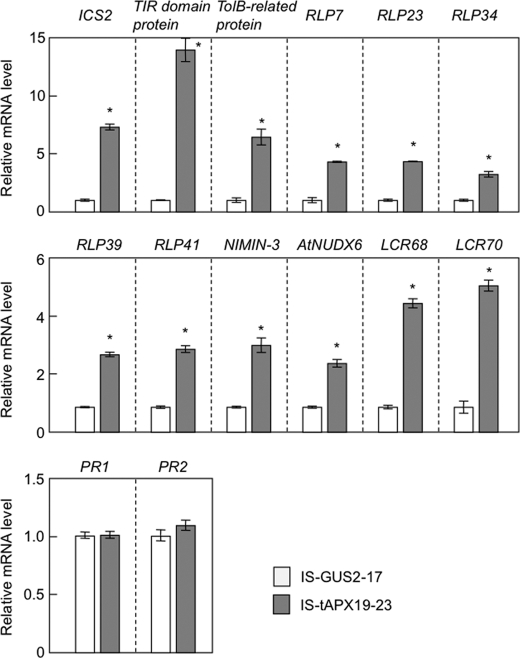
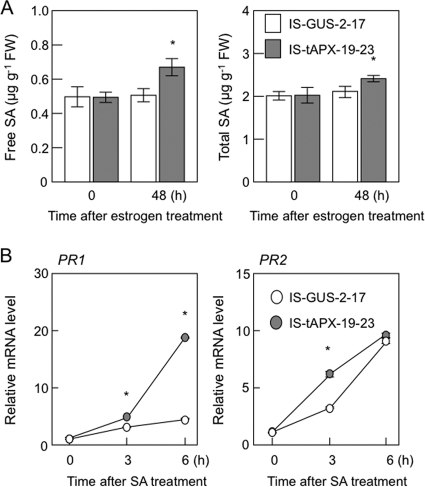
As shown in supplemental Table S2, some RTS genes are known to be involved in pathogen response/resistance, suggesting that H2O2 derived from chloroplasts plays a role in plant responses to biotic stress. For example, low molecular weight cysteine-rich proteins (LCR68 and LCR70) are defensin-type pathogenesis-related (PR) proteins (31). Receptor-like proteins (RLP7, -23, -34, -39, and -41) as well as Toll-interleukin resistance (TIR) domain protein (At4g11340) and a related protein (At4g01870, annotated as TolB-related protein) are generally thought to be associated with plant responses to pathogen attack (32, 33). Cytosolic Nudix hydrolase (AtNUDX6) and NIM1-interacting 3 (NIMIN-3) were recently identified as regulators of the SA response in Arabidopsis (24, 34–36). Furthermore, an isochorismate synthase (ICS2) is involved in the biosynthesis of SA (37). The q-PCR analysis confirmed that the transcription of these genes was up-regulated by the tAPX silencing (Fig. 6). In addition to the induction of ICS2, the levels of free and total SA were slightly but significantly increased in the IS-tAPX-19-23 plants at 48 h after estrogen treatment (Fig. 7A). Thus, it seems likely that the tAPX silencing enhances SA biosynthesis.
FIGURE 6.
Effect of tAPX silencing on the transcript levels of disease-resistance genes. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL. At 48 h after the estrogen treatment, the transcript levels of RTS genes (ICS2, TolB, TIR, RLP7, RLP23, RLP34, RLP39, RLP41, NIMIN3, NUDX6, LCR68, LCR70), PR1, and PR2, known to be involved in disease resistance, were measured by q-PCR. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for IS-GUS-2-17 plants. TIR, Toll-interleukin resistance.
FIGURE 7.
Effect of tAPX silencing on the response to SA. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL for 48 h. A, levels of free and total SA in the IS-GUS-2-17 and IS-tAPX-19-23 plants before and after estrogen treatment were measured as described under “Experimental Procedures.” B, 48 h after treatment with estrogen, IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm SA. The transcript levels of PR1 and PR2, SA-responsive genes, were measured by q-PCR. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for IS-GUS-2-17 plants.
As shown in Fig. 6, however, the transcript levels of pathogenesis-related genes (PR1 and PR2), known to be SA-responsive genes (38), were not affected by the tAPX silencing in the IS-tAPX-19-23 plants. Therefore, we checked the effect of tAPX silencing on the SA response. At 48 h after the treatment with estrogen, the IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with 50 μm SA. The transcript levels of PR1 increased in both plants under the SA treatment but were higher in the IS-tAPX-19-23 plants (Fig. 7B). Furthermore, the transcript levels of PR2 were also higher in the IS-tAPX-19-23 plants at 3 h after SA treatment. These findings suggest that the H2O2 signaling derived from chloroplasts activates SA biosynthesis and SA-inducible gene expression.
In contrast, the levels of ICS2 transcripts and SA in the KO-tAPX plants were almost the same as those in the wild-type plants (supplemental Fig. S7 and S8). Furthermore, the transcript levels of the other RTS genes, except for LCR70, were not higher in the KO-tAPX plants but, rather, lower in the mutants than in the wild-type plants (supplemental Fig. S7). Thus, as was the case for the cold acclimation response, the plant responses to SA and pathogen were not enhanced in the KO-tAPX plants.
Interplay of Chloroplastic H2O2 and HL in Gene Regulation
HL is one of the most characterized stresses to enhance ROS production in chloroplasts. Therefore, we tried to investigate the interplay between chloroplastic H2O2 and HL in gene regulation. First, to check the effect of tAPX silencing on HL sensitivity, at 48 h after estrogen treatment, the IS-GUS-2-17 and IS-tAPX-19-23 plants were exposed to HL at 1000 μmol photons m−2 s−1. As shown in supplemental Fig. S9, there was no visible effect of tAPX silencing on the HL sensitivity. This result was consistent with our previous work using the KO-tAPX plants (20). Fv/Fm was similarly decreased by HL in both IS-GUS-2-17 and IS-tAPX-19-23 plants (Supplemental Fig. S9). Furthermore, the sensitivity of the tAPX-silenced plants to various concentrations of methylviologen treatment was almost the same as that of the control plants (data not shown). These findings suggest that the tAPX silencing in the IS-tAPX-19-23 plants had no effect on tolerance to HL- and methylviologen treatment-induced photooxidative stress.
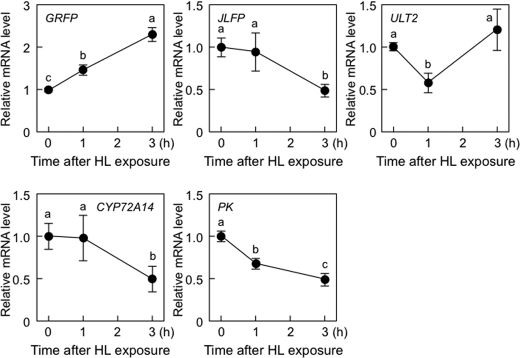
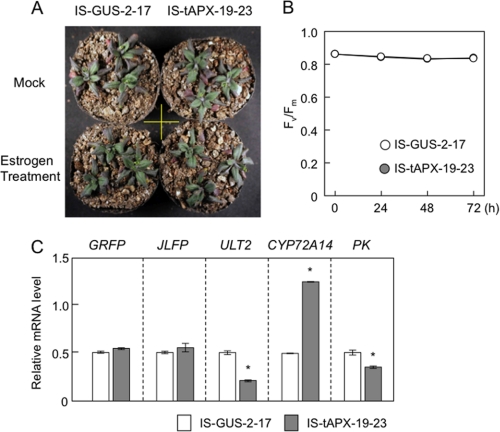
Next, to determine the effect of HL on the expression of five RTS genes (GRFP, JLFP, ULT2, CYP72A14, and PK), 19-day-old wild-type plants were exposed to HL. The transcript levels of GRFP were significantly increased by HL (Fig. 8), suggesting that chloroplastic H2O2 acts as a positive signal for HL response. However, the transcript levels of the other RTS genes were decreased or not affected by HL exposure (Fig. 8). We also checked the effect of tAPX silencing on expression of these genes under moderate light (ML; 400 μmol photons m−2 s−1). One-week-old IS-GUS-2-17 and IS-tAPX-19-23 plants, grown under NL, were further grown for 10 days under ML and then treated with estrogen. There was no difference in phenotype and Fv/Fm between IS-GUS-2-17 and IS-tAPX-19-23 plants (Fig. 9, A and B). At 48 h after estrogen treatment, the transcript levels of GRFP, ULT2, and PK were significantly suppressed by tAPX silencing. Furthermore, although the transcript levels of JLFP and CYP72A14 were increased by tAPX silencing, the induction levels of these genes under ML were largely lower than those under NL (Fig. 9C). In the IS-GUS-2-17 and IS-tAPX-19-23 plants, the expression of these genes was similar under both ML and NL (data not shown). Thus, it seems likely that there are not only synergistic but also antagonistic effects of chloroplastic H2O2 on HL response.
FIGURE 8.
Effect of HL on the expression of RTS genes. Nineteen-day-old wild-type plants were exposed to HL (1000 μmol of photons m−2 s−1). The transcript levels of RTS genes (GRFP, JLFP, ULT2, CYP72A14, and PK) were measured by q-PCR. Error bars indicate S.D. (n = 3). Values without a common letter are significantly different according to t tests (p < 0.05).
FIGURE 9.
Effect of tAPX silencing on the expression of RTS genes under ML. One-week-old IS-GUS-2-17 and IS-tAPX-19-23 plants grown under NL were further grown for 10 days under ML (400 μmol of photons m−2 s−1). The plants were then sprayed with estrogen and kept under ML for 72 h. A, 48 h after estrogen treatment the IS-GUS-2-17 and IS-tAPX-19-23 plants were photographed. B, Fv/Fm values in the leaves of IS-GUS-2-17 and IS-tAPX-19-23 after estrogen treatment were measured using a Closed FluorCam 800MF. Error bars indicate S.D. (n = 3). C, 48 h after estrogen treatment, the transcript levels of RTS genes (GRFP, JLFP, ULT2, CYP72A14, and PK) were measured by q-PCR. Error bars indicate S.D. (n = 3). Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for IS-GUS-2-17 plants.
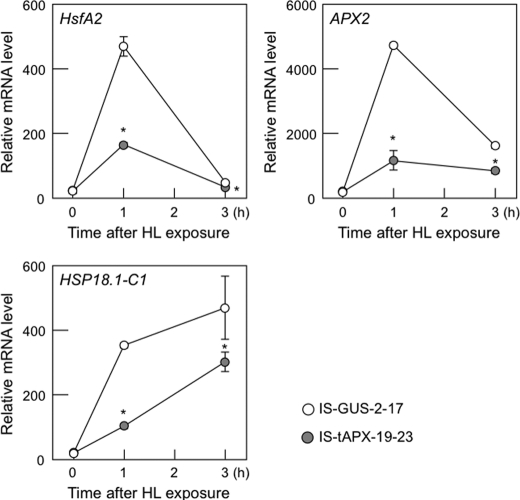
Finally, we checked the effect of tAPX silencing on the expression of known ROS-responsive genes, HsfA2, APX2, heat shock protein 18.1-C1 (HSP18.1-C1) (20, 25), under HL. In both IS-GUS-2-17 and IS-tAPX-19-23 plants, the transcript levels of ROS-responsive genes were drastically increased by HL (Fig. 10). However, the induction of ROS-responsive genes by HL was significantly inhibited by tAPX silencing (Fig. 10), supporting the antagonistic effects of chloroplastic H2O2 on HL response.
FIGURE 10.
Effect of tAPX silencing on expression of ROS-responsive genes under HL. Seventeen-day-old IS-GUS-2-17 and IS-tAPX-19-23 plants were sprayed with a 100 μm estrogen and kept under NL. At 48 h after treatment with estrogen, IS-GUS-2-17 and IS-tAPX-19-23 plants were exposed to HL (1000 μmol photons m−2 s−1). The transcript levels of ROS-responsive genes (HsfA2, APX2, and HSP18.1-C1) were measured by q-PCR. Error bars indicate S.D. (n = 3). Significant differences: *, p < 0.05 versus the value for IS-GUS-2-17 plants.
DISCUSSION
Specificity of Signaling Function of Chloroplast-derived H2O2
In this study we attempted to identify the H2O2-triggered retrograde signaling from chloroplasts to the nucleus without application of stress. To achieve this objective, we created a novel system for producing H2O2 in chloroplasts by chemical-dependent tAPX silencing using estrogen-inducible RNAi. In the IS-tAPX-19-23 plants, levels of the tAPX transcript were suppressed almost completely at 20 h after the treatment with estrogen, and levels of the tAPX protein were decreased at 48 h (Fig. 1, supplemental Fig. S1). Concomitant with the tAPX silencing, the levels of chloroplastic oxidized proteins were increased (Fig. 3), suggesting that the generation and accumulation of H2O2 are enhanced in the chloroplasts during tAPX silencing under NL. We identified 774 RTS genes as candidate genes responsive to H2O2 derived from chloroplasts (supplemental Table S2). Although the 530-bp fragment of the RNAi-trigger contained 18 bp sequences matching At5g38350, At2g24200, and At1g59610 genes, those genes were not included in the down-regulated genes in the tAPX-silenced plants. The induction of RTS genes by tAPX silencing was inhibited by the application of AsA and dark, suggesting the induction to be due to an increase in the H2O2 levels (Fig. 4). Interestingly, among the RTS genes, there were only two genes known to be responsive to ROS (supplemental Fig. S4). Furthermore, comparison of our microarray data with data from the various mutants lacking CSD2, catalase 2 (CAT2), and APX1 (10, 11, 15, 39), revealed that only three (At3g08770, At4g29030, and At2g33850), five (At1g23730, At3g25020, At1g17870, At3g24982, and At2g23150), and four (At4g23150, At4g10500, At2g29120, and At2g32680) RTS genes, whose expressions were up-regulated in response to tAPX silencing, were genes specifically up-regulated in the KD-CSD2, KD-CAT2, and KO-APX1 plants, respectively (data not shown). Consequently, the majority of RTS genes are novel ROS-responsive genes, indicating that H2O2 derived from chloroplasts may act as signaling molecules through a specific pathway. As shown in Fig. 2, the levels and/or redox states of H2O2, oxidized proteins, AsA, and GSH as well as the quantum yields of photosystem II in whole leaves were not affected by the tAPX silencing under NL. As the oxidation of chloroplastic proteins occurred even in the IS-GUS-2-17 plants (Fig. 3), the levels of oxidized proteins in chloroplasts do not directly reflect the degree of photosynthetic activity. Thus, it seems likely that the increased levels of H2O2 and oxidized protein in chloroplasts of IS-tAPX-19-23 plants are not so high, as they have negative effect on photosynthesis. This might be important to identify chloroplast-specific signaling events, because H2O2 can easily permeate biological membranes.
Recently, it has been reported that overexpression of tAPX in flu mutants enhanced 1O2-dependent programmed cell death, suggesting that H2O2 generated in chloroplasts is an antagonist of oxidative signaling through 1O2 (40). In fact, a comparison of the present microarray data with the results of Gadjev et al. (15) revealed the expression of 11 genes (At1g27130, At3g09870, At3g05640, At3g59350, At3g44560, At4g29700, At5g47240, At1g76790, At1g74930, At5g51190, and At5g47230) to be induced by the release of 1O2 in the flu mutants but suppressed by the tAPX silencing (supplemental Fig. S2). These genes may be involved in the antagonistic cross-talk between 1O2- and H2O2-triggered signaling. However, the majority of RTS genes were not known to be responsive to 1O2. Moreover, we have reported that the lack of tAPX and sAPX has no effect on the expression of 1O2-responsive genes during HL irradiation (20). Therefore, further analysis is required for understanding the antagonistic cross-talk between 1O2- and H2O2-triggered signaling.
The sensing of H2O2 within the chloroplasts seems to be essential for its specific action as a signal. This is not surprising, as in plastids there are a number of signals, including plastid gene expression, the redox state of the photosynthetic electron transport chain, and/or the precursor of chlorophyll biosynthesis to regulate nuclear gene expression (41–43). Møller and Sweetlove (44) proposed that peptides derived from the proteolytic breakdown of oxidatively damaged proteins function as secondary ROS messengers and regulate mitochondrion- or chloroplast-specific ROS signaling. This is an interesting possibility to investigate because levels of oxidized proteins in chloroplasts are increased in response to tAPX silencing (Fig. 3).
Some RTS genes encoded signal transduction-related proteins, such as transcription factors and kinases (supplemental Table S2), indicating that these genes are involved in the H2O2 signaling derived from chloroplasts and that there may be diverse pathways downstream the chloroplastic H2O2. Moreover, some of these genes encoded plant-specific transcription factors, such as NAC (NAM, ATAF, CUC2) transcription factors. It would be worth determining whether the H2O2 signaling derived from plant-specific chloroplasts is regulated by these transcription factors.
Possible Role of H2O2 Signal from Chloroplasts in Cold Acclimation and SA-mediated Biotic Stress Response
Microarray and q-PCR analyses indicate that the H2O2 signaling derived from chloroplasts may be involved in the down-regulation of the cold response. The transcript levels of CBF1/DREB1B and CORs were decreased by the tAPX silencing under NL (Fig. 5). CBF1/DREB1B is a central transcription factor for cold acclimation and positively regulates the expression of CORs (45, 46). The transgenic plants with suppressed CBF1/DREB1B expression showed a high degree of sensitivity to cold stress (45). Like the CBF1/DREB1B transgenic plants, the estrogen-treated IS-tAPX-19-23 plants were highly sensitive to stress, although the high sensitivity was diminished under LL (Fig. 5 and supplemental Fig. S5). Our findings and previous reports suggest that the H2O2 signaling derived from chloroplasts down-regulates CBF1/DREB1B expression, resulting in the suppression of COR expression and cold tolerance. On the other hand, down-regulation of the expression of CBF1/DREB1B and some CORs was not observed in the KO-tAPX plants (supplemental Fig. S6). In addition, there was no difference in sensitivity to cold stress between wild-type and KO-tAPX plants. Furthermore, the estrogen-treated IS-tAPX-19-23 plants as well as the KO-tAPX plants did not show high sensitivity to HL- and methylviologen-induced photooxidative stress (supplemental Fig. S9) (20). Thus, it seems likely that the down-regulation of cold-responsive genes by the H2O2 signaling, but not a decrease in ROS-scavenging capacity, resulted in the cold-sensitive phenotypes of tAPX-silenced plants. Interestingly, the transcript levels of CBF2/DREB1C known to be a negative regulator of CBF1/DREB1B expression were also suppressed by tAPX silencing (47). This may indicate a feedback regulation of cold acclimation by CBF1/DREB1B under control of H2O2 signaling.
Cold stress inhibits activities of some enzymes involved in the Calvin cycle, leading to overreduction of the photosynthetic electron transport chain due to the depletion of NADP+, a final electron acceptor. Thus, the production of H2O2 in chloroplasts is activated by cold stress. Accordingly, it seems unlikely that the suppression of cold-responsive genes by H2O2 signaling from chloroplasts is responsible for plant acclimation to cold stress. Another signaling pathway, possibly 1O2 signaling, may act as a positive regulator for cold acclimation and as an antagonist against H2O2 signaling.
We also found that RTS genes include those involved in the response to pathogens and SA biosynthesis (Fig. 6). Actually, the levels of SA were increased in response to tAPX silencing (Fig. 7A). Although the transcription of PR1 and PR2 was not induced just by the silencing because the increased level of SA was not so high, the expression of PR1 and PR2 was much higher in the tAPX-silenced IS-tAPX-19-23 plants than in the control plants on treatment with SA (Fig. 7). These findings suggest that the H2O2 derived from chloroplasts activates not only SA biosynthesis but also the response to SA. Recently, it was found that SA is synthesized in chloroplasts (48). Furthermore, it is clear that SA can bind to APX isoenzymes and inhibit their activity, leading to an increase in H2O2 (49). Thus, it seems likely that the chloroplastic H2O2 and SA activate each other and that this positive feedback loop is involved in the plant response to biotic stress.
There is growing evidence that ROS are produced in chloroplasts during infections and are involved in the resistance to pathogens (50, 51). It was found that under illumination, the mitogen-activated protein kinase cascade, which is involved in the resistance, rapidly inhibits CO2 fixation in tobacco chloroplasts, leading to the generation of ROS in chloroplasts due to excess excitation energy in the photosynthetic electron transport chain under illumination (50). In fact, ROS accumulated in tobacco chloroplasts in a light-dependent manner after infection with tobacco mosaic virus, which is known to activate the mitogen-activated protein kinase cascade (50), suggesting that the source of ROS is photosynthesis. Furthermore, the overexpression of tAPX inhibited the hypersensitive response in Arabidopsis plants (51). These findings and data presented previously strongly suggest that chloroplast-generated ROS are substantially involved in plant defense and response to pathogens.
Synergistic and Antagonistic Roles of Chloroplastic H2O2 for HL Response
As described above, we previously found that lack of tAPX and sAPX suppresses the expression of ROS-responsive genes, such as APX2, HsfA2, and HSP18.1-C1, under HL, although H2O2 is highly accumulated in the KO-tAPX and KO-sAPX plants (20). This negative regulation of ROS-responsive gene expression under HL occurred in the estrogen-treated IS-tAPX-19-23 plants (Fig. 10), suggesting an antagonistic role of chloroplastic H2O2 for HL response. In fact, the induction of RTS genes by the tAPX silencing was lower under ML than NL (Fig. 9). Furthermore, four RTS genes, all of which were highly expressed in response to tAPX silencing, were not induced by HL (Fig. 8). On the other hand, the transcript levels of GRFP were significantly increased in response to HL as well as tAPX silencing (Fig. 8). Based on these findings, we concluded that there are synergistic and antagonistic roles of chloroplastic H2O2 for HL response.
Expression analyses of APX2 have revealed that several signals including ROS are involved in the HL response. Identification of the regulator of APX2 1-1 (rax1-1) mutant, a GSH-deficient mutant, revealed that GSH acts as a signaling molecule and regulates HL response (52). Moreover, the redox state of photosynthetic electron transport is characterized a signal for regulating the HL response, including APX2 induction (53, 54). Ball et al. (52) identified specifically up- or down-regulated genes in rax1-1, and Adamiec et al. (55) identified HL-responsive genes, whose responses were inhibited by treatment with 3-(3′,4′-dichlorphenyl)-1,1′-dimethylurea, an inhibitor of PET (photosynthetic electron transport). However, we could detect hardly those genes in the RTS genes, suggesting that chloroplastic H2O2 is not involved in the GSH and the redox state of photosynthetic electron transport-dependent HL response. The altered expression of APX2 8 (alx8) mutants, which lacked SAL1 protein, were isolated in a screen for elevated expression of APX2 under LL (56, 57). SAL1, an enzyme that dephosphorylate 3′-phosphoadenosine 5′-phosphate, is a negative regulator of abscisic acid (ABA)-dependent stress response (57). Recently, 3′-phosphoadenosine 5′-phosphate was found to act as a retrograde signal from chloroplasts to nucleus (58). In the alx8 mutants, the expression of APX2 and levels of ABA were elevated even in LL and were more enhanced in HL, suggesting an involvement of ABA in HL response. In fact, Galvez-Valdivieso et al. (59) found that ABA is essential for APX2 induction under HL and concluded that ABA is necessary for HL response. They also identified 816 genes as representing a significant overlap between HL and ABA responses. Comparison of RTS genes with both ABA- and HL-responsive genes revealed that 13 up-regulated and 23 down-regulated genes by the tAPX silencing are up- and down-regulated by both ABA and HL (supplemental Table S3). Furthermore, 20 up-regulated and 55 down-regulated genes by the tAPX silencing were up- and down-regulated in alx8 mutants, respectively (supplemental Table S3). These findings suggest that chloroplastic H2O2 is involved in ABA-mediated HL signaling. In contrast, 17 up-regulated 11 down-regulated genes by the tAPX silencing were conversely down- and up-regulated in the alx8 mutants, respectively (supplemental Table S3). Based on these findings, we concluded that chloroplastic H2O2 is synergistically and antagonistically involved in the signaling pathways for HL response mediated by ABA. However, RTS genes did not encode any enzymes and signaling factors, known to be involved in the ABA biosynthesis and response, respectively. At present, we cannot explain the physiological significance of the antagonistic function of chloroplastic H2O2 in HL response. However, as described above, chloroplastic H2O2 appears to be involved in the cold acclimation and the SA-mediated biotic stress response. Thus, it was indicated that the synergistic and antagonistic interaction among source- and kind-specific ROS signaling pathways and other signals-derived pathways seems to be important for fine-tuning of response to abiotic and biotic stresses, although it needs further consideration regarding the physiological role of chloroplastic H2O2 for stress responses through analysis of gene expression under various types of stress using the tAPX silencing system and functional analysis of RTS genes.
The Estrogen-inducible RNAi Method Is Useful for Identification of Source- and Kind-specific ROS Signaling
The levels of RTS genes expression, SA, and stress sensitivity were changed little in the KO-tAPX plants compared with wild-type plants. In many cases, signaling events are transient. Thus, acclimation to the lack of tAPX might occur during the developmental process in the KO-tAPX plants, and therefore, the effects of tAPX expression on whole gene expression differed between KO-tAPX plants and estrogen-treated tAPX-silenced plants. Accordingly, these findings demonstrated that chemical-inducible RNAi is more useful than knock-out or constitutive knockdown for identifying source-specific oxidative signaling. There is no doubt that reverse genetic analyses of antioxidative enzymes using knock-out or constitutive knockdown plants have provided powerful evidence that ROS act as signaling molecules in plant cells (9–11, 14, 15). However, here we emphasize that the information obtained from such plants does not completely reflect the response to ROS themselves, and thus the plant response should be analyzed by multiple methods, including chemical-inducible RNAi. The application of chemical-inducible RNAi to other antioxidative enzymes, including superoxide dismutase, peroxiredoxin, and other APX, will help to identify source- and kind-specific ROS signaling events and clarify their physiological function in the stress response.
Moreover, in plants, the signaling functions of ROS have been characterized mainly using leaves and/or roots, not stem, flowers, and fruits. The chemical-inducible RNAi method allows tissue-specific gene silencing by limiting the tissue of treatment with the agent and, therefore, may also be useful for elucidating the tissue-specific function of ROS as signals.
Supplementary Material
Acknowledgments
We are grateful to Drs. Nam-Hai Chua (Rockefeller University) and Tsuyoshi Nakagawa (Shimane University) for donating the pMDC7 vector and pGWB80 vector, respectively, and to Drs. Mikio Nishimura, Makoto Hayashi, and Mitsue Fukazawa (National Institute for Basic Biology) for technical support using the microarray system. We also thank Satoru Nakagami and Hiroki Yamada for technical assistance.
This work was supported by Grants-in-aid for Scientific Research (A) 22248042 (to S. S.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and by Scientific Research for Plant Graduate Students from the Nara Institute of Science and Technology (to T. M.).

This article contains supplemental Tables S1–S3 and Figs. S1–S9.
- ROS
- reactive oxygen species
- APX
- ascorbate peroxidase
- AsA
- ascorbate
- RTS
- responsive to tAPX silencing
- sAPX
- stromal APX
- tAPX
- thylakoid membrane-bound APX
- HL
- high light
- q-PCR
- quantitative real-time PCR
- DAB
- diaminobenzidine
- IS-
- inducible silencing
- GRFP
- gibberellin-regulated family protein
- JLFP
- jacalin lectin family protein
- CYP72A14
- cytochrome P450 family protein
- LL
- low light
- NL
- normal light
- ML
- moderate light
- PR
- pathogenesis-related
- HSP18.1-C1
- heat shock protein 18.1-C1
- ABA
- abscisic acid
- SA
- salicylic acid.
REFERENCES
- 1. Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 2. Apel K., Hirt H. (2004) Reactive oxygen species. Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 [DOI] [PubMed] [Google Scholar]
- 3. Foyer C. H., Shigeoka S. (2011) Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011) ROS signaling. The new wave? Trends Plant Sci. 16, 300–309 [DOI] [PubMed] [Google Scholar]
- 5. Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., Yoshimura K. (2002) Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319 [PubMed] [Google Scholar]
- 6. Torres M. A., Dangl J. L., Jones J. D. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sagi M., Fluhr R. (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maruta T., Inoue T., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. (2011) Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 180, 655–660 [DOI] [PubMed] [Google Scholar]
- 9. op den Camp R. G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Göbel C., Feussner I., Nater M., Apel K. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rizhsky L., Liang H., Mittler R. (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J. Biol. Chem. 278, 38921–38925 [DOI] [PubMed] [Google Scholar]
- 11. Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H. K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58 [DOI] [PubMed] [Google Scholar]
- 12. Gechev T. S., Gadjev I. Z., Hille J. (2004) An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell. Mol. Life Sci. 61, 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gechev T. S., Minkov I. N., Hille J. (2005) Hydrogen peroxide-induced cell death in Arabidopsis. Transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57, 181–188 [DOI] [PubMed] [Google Scholar]
- 14. Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17, 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gadjev I., Vanderauwera S., Gechev T. S., Laloi C., Minkov I. N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001) FLU. A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K. P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185 [DOI] [PubMed] [Google Scholar]
- 18. Lee K. P., Kim C., Landgraf F., Apel K. (2007) Executer1- and Executer2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim C., Meskauskiene R., Apel K., Laloi C. (2008) No single way to understand singlet oxygen signaling in plants. EMBO Rep. 9, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maruta T., Tanouchi A., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. (2010) Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol. 51, 190–200 [DOI] [PubMed] [Google Scholar]
- 21. Zuo J., Niu Q. W., Chua N. H. (2000) Technical advance. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 [DOI] [PubMed] [Google Scholar]
- 22. Guo H. S., Fei J. F., Xie Q., Chua N. H. (2003) A chemical-regulated inducible RNAi system in plants. Plant J. 34, 383–392 [DOI] [PubMed] [Google Scholar]
- 23. Bechtold N., Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266 [DOI] [PubMed] [Google Scholar]
- 24. Ogawa T., Ueda Y., Yoshimura K., Shigeoka S. (2005) Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana. J. Biol. Chem. 280, 25277–25283 [DOI] [PubMed] [Google Scholar]
- 25. Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547 [DOI] [PubMed] [Google Scholar]
- 26. Yoshimura K., Yabuta Y., Ishikawa T., Shigeoka S. (2000) Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stresses. Plant Physiol. 123, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shigeoka S., Onishi T., Nakano Y., Kitaoka S. (1987) Characterization and physiological function of glutathione reductase in Euglena gracilis z. Biochem. J. 242, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phee B. K., Cho J. H., Park S., Jung J. H., Lee Y. H., Jeon J. S., Bhoo S. H., Hahn T. R. (2004) Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics 4, 3560–3568 [DOI] [PubMed] [Google Scholar]
- 29. Morishita T., Kojima Y., Maruta T., Nishizawa-Yokoi A., Yabuta Y., Shigeoka S. (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50, 2210–2222 [DOI] [PubMed] [Google Scholar]
- 30. Malamy J., Hennig J., Klessig D. F. (1992) Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus Infection. Plant Cell 4, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sels J., Mathys J., De Coninck B. M., Cammue B. P., De Bolle M. F. (2008) Plant pathogenesis-related (PR) proteins. A focus on PR peptides. Plant Physiol. Biochem. 46, 941–950 [DOI] [PubMed] [Google Scholar]
- 32. Kruijt M., DE Kock M. J., de Wit P. J. (2005) Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97 [DOI] [PubMed] [Google Scholar]
- 33. Burch-Smith T. M., Dinesh-Kumar S. P. (2007) The functions of plant TIR domains. Sci. STKE. 2007, pe46. [DOI] [PubMed] [Google Scholar]
- 34. Ishikawa K., Yoshimura K., Harada K., Fukusaki E., Ogawa T., Tamoi M., Shigeoka S. (2010) AtNUDX6, an ADP-ribose/NADH pyrophosphohydrolase in Arabidopsis, positively regulates NPR1-dependent salicylic acid signaling. Plant Physiol. 152, 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ishikawa K., Yoshimura K., Ogawa T., Shigeoka S. (2010) Distinct regulation of Arabidopsis ADP-ribose/NADH pyrophosphohydrolases, AtNUDX6 and -7, in biotic and abiotic stress responses. Plant Signal. Behav. 5, 839–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weigel R. R., Bäuscher C., Pfitzner A. J., Pfitzner U. M. (2001) NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46, 143–160 [DOI] [PubMed] [Google Scholar]
- 37. Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J. P. (2008) Characterization and biological function of the Isochorismate Synthase2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reuber T. L., Plotnikova J. M., Dewdney J., Rogers E. E., Wood W., Ausubel F. M. (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485 [DOI] [PubMed] [Google Scholar]
- 39. Pnueli L., Liang H., Rozenberg M., Mittler R. (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J. 34, 187–203 [DOI] [PubMed] [Google Scholar]
- 40. Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nott A., Jung H.S., Koussevitzky S., Chory J. (2006) Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759 [DOI] [PubMed] [Google Scholar]
- 42. Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 [PubMed] [Google Scholar]
- 43. Kleine T., Voigt C., Leister D. (2009) Plastid signalling to the nucleus. Messengers still lost in the mists? Trends Genet. 25, 185–192 [DOI] [PubMed] [Google Scholar]
- 44. Møller I. M., Sweetlove L. J. (2010) ROS signalling-specificity is required. Trends Plant Sci. 15, 370–374 [DOI] [PubMed] [Google Scholar]
- 45. Novillo F., Medina J., Salinas J. (2007) Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. U.S.A. 104, 21002–21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Medina J., Catalá R., Salinas J. (2011) The CBFs. Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 180, 3–11 [DOI] [PubMed] [Google Scholar]
- 47. Novillo F., Alonso J. M., Ecker J. R., Salinas J. (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fragnière C., Serrano M., Abou-Mansour E., Métraux J. P., L'Haridon F. (2011) Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 585, 1847–1852 [DOI] [PubMed] [Google Scholar]
- 49. Dempsey D. A., Shah J., Klessig D. F. (1999) Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575 [Google Scholar]
- 50. Liu Y., Ren D., Pike S., Pallardy S., Gassmann W., Zhang S. (2007) Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51, 941–954 [DOI] [PubMed] [Google Scholar]
- 51. Yao N., Greenberg J. T. (2006) Arabidopsis Accelerated Cell Death2 modulates programmed cell death. Plant Cell 18, 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ball L., Accotto G. P., Bechtold U., Creissen G., Funck D., Jimenez A., Kular B., Leyland N., Mejia-Carranza J., Reynolds H., Karpinski S., Mullineaux P. M. (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16, 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karpinski S., Escobar C., Karpinska B., Creissen G., Mullineaux P. M. (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9, 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yabuta Y., Maruta T., Yoshimura K., Ishikawa T., Shigeoka S. (2004) Two distinct redox signaling pathways for cytosolic APX induction under photooxidative stress. Plant Cell Physiol. 45, 1586–1594 [DOI] [PubMed] [Google Scholar]
- 55. Adamiec M., Drath M., Jackowski G. (2008) Redox state of plastoquinone pool regulates expression of Arabidopsis thaliana genes in response to elevated irradiance. Acta Biochim. Pol. 55, 161–173 [PubMed] [Google Scholar]
- 56. Rossel J. B., Walter P. B., Hendrickson L., Chow W. S., Poole A., Mullineaux P. M., Pogson B. J. (2006) A mutation affecting Ascorbate Peroxidase 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 29, 269–281 [DOI] [PubMed] [Google Scholar]
- 57. Wilson P. B., Estavillo G. M., Field K. J., Pornsiriwong W., Carroll A. J., Howell K. A., Woo N. S., Lake J. A., Smith S. M., Harvey Millar A., von Caemmerer S., Pogson B. J. (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58, 299–317 [DOI] [PubMed] [Google Scholar]
- 58. Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., Brearley C., Hell R., Marin E., Pogson B. J. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Galvez-Valdivieso G., Fryer M. J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W. J., Jones A. M., Baker N. R., Mullineaux P. M. (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21, 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.