Background: It is unclear how YAP gene expression is controlled in colorectal cancer (CRC) cells.
Results: β-Catenin/TCF4 complexes directly regulate YAP gene expression through a novel DNA enhancer element.
Conclusion: YAP is a direct Wnt/β-catenin target gene, and its expression is required for CRC cell growth.
Significance: Aberrant Wnt/β-catenin signaling may account for oncogenic YAP expression in intestinal cells.
Keywords: β-Catenin, Cancer Biology, Cell Proliferation, Gene Expression, Transcription Enhancers, Wnt Signaling
Abstract
Mutations in the Wnt/β-catenin pathway occur in most colorectal cancers (CRCs), and these mutations lead to increased nuclear accumulation of the β-catenin transcriptional co-activator. In the nucleus, β-catenin associates with TCF/LEF sequence specific transcription factors to activate target gene expression. The Hippo pathway restricts cellular growth by preventing nuclear accumulation of the Yes-associated protein (YAP) transcriptional co-activator. YAP expression is elevated in CRCs suggesting that, like Wnt/β-catenin signaling, the Hippo pathway may contribute to colorectal carcinogenesis. Regulation of YAP at the post-translational level has been well studied but the transcription factors that control YAP gene expression are unknown. Here we demonstrate that β-catenin/TCF4 complexes bind a DNA enhancer element within the first intron of the YAP gene to drive YAP expression in CRC cells. As such, reducing β-catenin expression in CRC cells using shRNAs leads to decreased YAP mRNA and protein levels. YAP is abundantly expressed in the cytoplasm and nuclei of several established human colon cancer cell lines and this localization pattern is insensitive to plating density. Finally, we show that YAP expression is elevated in the majority of a panel of primary human colorectal tumors compared with its expression in uninvolved colonic mucosa, and that YAP and β-catenin localize to the nuclear compartment of tumor cells. Together, these results implicate YAP as an oncogene whose expression is driven by aberrant Wnt/β-catenin signaling in human CRC cells.
Introduction
The Wnt/β-catenin signaling pathway is critical for homeostasis of the intestinal tract (1–4). The key effector protein in the canonical Wnt pathway is the transcriptional co-activator, β-catenin (5). A multiprotein destruction complex that contains the adenomatous polyposis coli protein (APC)3 tightly regulates β-catenin levels. Mutations in components of the Wnt pathway, most often in APC, are found in ∼90% of colon cancers. These mutations lead to heightened β-catenin levels in the nucleus where it associates with members of the TCF/LEF (T-cell factor/lymphoid enhancer factor) family of sequence specific transcription factors. TCF4 is the predominant TCF family member expressed in colonocytes, and β-catenin/TCF4 complexes activate target gene expression by recruiting multiprotein complexes that remodel or modify chromatin into a transcriptionally permissive state (5, 6). To identify Wnt/β-catenin target genes in human colorectal cancer (CRC) cells, we previously conducted a ChIP-Seq screen using β-catenin-specific antibodies (7). One of the targets identified in that screen was the gene encoding the Yes-associated protein, YAP. YAP is a transcriptional co-activator and functions in the Hippo signaling pathway.
The Hippo signaling pathway controls organ size in Drosophila melanogaster and higher eukaryotes (8). The pathway was discovered in D. melanogaster using screens designed to identify genes that negatively regulated tissue growth (9–11). Mutations in a Ste-20 family protein kinase resulted in cell overgrowth and decreased cell death, and this protein was named Hippo because of the phenotype observed in mosaic flies. YAP is a key effector protein in the Hippo pathway, and its subcellular localization is controlled by phosphorylation. When the Hippo pathway is engaged, activation of a core kinase complex containing Hippo (Mst1/2 in mammals), Lats1/2 kinases, Mob, and Salvador, leads to YAP phosphorylation, and its retention in the cytoplasm through interactions with 14-3-3 proteins. When the Hippo pathway is disengaged, YAP is translocated into the nucleus where it associates with members of the TEA domain containing transcription factors (TEADs). YAP/TEAD complexes activate expression of growth promoting genes including connective tissue growth factor (CTGF) and the EGF family member amphiregulin (AREG) (12, 13).
The Hippo/YAP pathway is highly conserved in mammals and as in D. melanogaster, it is required for proper control of organ size. In the mouse, YAP overexpression or conditional deletion of Mst1/2 leads to hepatomegaly (14–16). Removal of Mst kinases in the mouse gastrointestinal tract leads to increased numbers of stem cells and proliferative transit amplifying cells and an impairment of epithelial cell differentiation (17). Moreover, the Hippo pathway is critical for establishment and maintenance of the intestinal crypt microenvironment and it is required for repair of the intestinal epithelium following injury (14, 17, 18).
Deregulation of Hippo/YAP signaling has been implicated in intestinal tumorigenesis (14, 17, 18). YAP expression is elevated in human primary colonic tumors compared with its expression in uninvolved tissue and YAP expression is increased in the intestinal tumors that arise in the adenomatous polyposis coli multiple intestinal neoplasia (APCmin) mouse (14, 18, 19). Whether YAP is required for increased tumorigenesis in the APCmin mouse model has not been tested; however, YAP is required for growth of human colon cancer cells in colonies (17). While these studies indicate that increased YAP expression correlates with tumorigenesis, mutations in components of the Hippo pathway have not been identified in the colon cancers examined to date. Thus, the molecular mechanisms that explain YAP overexpression in intestinal tumors are incompletely understood.
Here, we demonstrate that β-catenin/TCF4 complexes directly associate with the YAP gene locus and that β-catenin is required for YAP expression in CRC cells. β-Catenin/TCF4 complexes bind a site within the first intron of YAP and a DNA element that harbors this site increased expression of luciferase reporter plasmids in transient assays. We show that YAP promotes CRC cell metabolism, anchorage independent growth in soft agar and tumor growth in a xenograft mouse model. We also find that YAP expression is elevated in primary and metastatic human colorectal tumors. Together, our findings suggest that Wnt/β-catenin and Hippo/YAP signaling pathways converge to promote colon cancer. These results shed light on the potential to target the Hippo/YAP signaling pathway for the development of therapeutics to treat individuals affected by this disease.
EXPERIMENTAL PROCEDURES
Cell Lines
The human CRC cell lines HCT116, SW620, SW480, RKO, LS174, and HT29 were obtained from the American Type Culture Collection (ATCC). HEK293FT cells were obtained from Invitrogen. Cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, 2 mm Glutamax, and 0.1 mg/ml streptomycin at 37 °C in 5% CO2. HEK293FT cells were grown in media supplemented with 500 μg/ml neomycin.
ChIP
ChIP analysis was performed as described (7) using 3 μg of anti-β-catenin (BD Transduction, 610154), anti-TCF4 (Millipore, 05–511), and anti-RNAP (Covance, 8WG16/MMS-126R) antibodies. Immunoprecipitated DNA was analyzed by real-time PCR as described (7) using primer sequences listed in supplemental Table S1. Data are presented as relative levels obtained using a standard curve generated from formaldehyde fixed, sonicated, and purified input DNA.
Reverse Transcription-PCR (RT-PCR)
RNA isolation, cDNA synthesis, and quantitative PCR (qPCR) were performed as described (7) except that cDNA was diluted 1:100 for qPCR. Primer sequences are listed in supplemental Table S1. Relative expression was measured using the 2ΔCT method, and data are represented as fold levels over control.
Plasmids and Luciferase Reporter Assays
HCT116 genomic DNA was isolated using a DNeasy kit (Qiagen, 69504). A 624 bp element within the first intron of the YAP gene was amplified in a standard PCR reaction containing 300 ng of genomic DNA, and 0.6 μm of the forward and reverse primers listed in supplemental Table S1. The PCR products were subcloned into the pGL3 promoter vector as KpnI- NheI fragments (Promega E1761) using standard molecular biology techniques. The pGL3-YAP mutant (Mut.) construct was derived from pGL3-YAP wild-type (WT) by mutating the TCF motif from CTTTGAT to CGCTGAT using the Stratagene Lightning Site-directed Mutagenesis Kit. For luciferase assays, HCT116 cells were seeded in a 12-well dish in triplicate, and were then transfected the following day using Lipofectamine 2000 (Invitrogen). Each transfection contained 250 ng of the firefly luciferase reporter plasmid, 12.5 ng of pRL-SV40 Renilla luciferase, and the total DNA was adjusted to 2 μg with pBlueScript. Where indicated, 250 ng of pcDNA3-mycDNTCF4 (Addgene, kindly deposited by B. Vogelstein) was included in the transfection mix. Where indicated, 2 μm BIO (6-bromoindirubin-3′-oxime, Calbiochem 361550) was added for 12 h prior to measuring luciferase levels. The next day, cells were lysed in 200 μl of 1× passive lysis buffer (Promega), and luciferase levels were measured using a Dual-Glo Luciferase kit (Promega, 2920) on a GloMax 20/20 luminometer (Promega).
DNA Binding Assay
A set of 28 bp oligonucleotides that centered on the TCF4 motif within the YAP DNA enhancer element were synthesized with the 5′ nucleotide biotinylated. A second set of oligonucleotides was synthesized and contained a mutated TCF4 site and the oligonucleotide sequences are available in supplemental Table S1. The oligonucleotides were diluted to 1.5 μm in annealing buffer (50 mm NaCl, 10 mm Tris pH 8.0, 1 mm EDTA pH 8.0), heated to 95° for 5 min and then slowly cooled to room temperature. Double-stranded DNA (500 ng) was bound to 50 μl streptavidin-coated magnetic beads (Promega, Z5481) in 1× binding buffer (1 m NaCl, 10 mm Tris, 1 mm EDTA, pH 8.0) at room temperature. HCT116 nuclear lysates were prepared as described (20), and 100 μg were preincubated in buffer B (20 mm Tris pH 7.9, 15% glycerol, 150 mm KCl, 1 mm EDTA, 0.05% Nonidet P-40) supplemented with 50 μg of salmon sperm DNA. The proteins were incubated with the DNA/Bead mixture for 1 h at room temperature in buffer B, and the complexes were captured on a magnetic stand. Following three washes in buffer B, the proteins were eluted in SDS-PAGE loading buffer and analyzed by Western blotting using anti-TCF4 or anti-β-catenin antibodies.
β-Catenin and YAP shRNA-mediated Knockdown
Lentiviral plasmids (pLKO.1) encoding the β-catenin shRNA and a scrambled shRNA control were obtained from Open Biosystems (21). Four independent sequences that target the YAP gene were identified using guidelines provided in the pLKO.1 product insert and oligonucleotides corresponding to these sequences were annealed and subcloned into pLKO.1 (supplemental Table S1). To generate lentiviruses, ∼5 × 106 HEK293FT cells seeded in a 10 cm dish were transfected with 3.0 μg of the pLKO.1 shRNA plasmid, 3 μg each of the pLP1 and pLP2 packaging plasmids, and 3 μg of the pLP/VSVG envelope plasmid using Lipofectamine 2000 (Invitrogen). Media containing virus particles was harvested at 24 and 48 h following transfection and centrifuged at 1500 × g for 5 min at room temperature. The media was supplemented with 6 μg/ml hexadimethrine bromide and was added to HCT116 and SW620 cells. The next day, 1 μg/ml puromycin was added. The efficiency of knockdown was assessed by RT-PCR, and Western blot analysis at 3 and 5 days following infection.
Western Blot Analysis
Preparation of protein extracts and Western blot analysis were conducted as described (22) using the following antibodies and dilutions: anti-β-catenin (BD Transduction, 610154, 1:1000), anti-YAP1 (Santa Cruz SC15407X, 1:1000), anti-α-tubulin (Sigma T9026, 1:1000), and anti-histone H3 (Millipore 07–690, 1:25,000).
MTT Assays
In each well of a 24-well plate, 10,000 cells were seeded in 0.5 ml of DMEM containing 0.5% fetal bovine serum. Cells were seeded into quadruplicate wells for each experimental condition. At one and 5 days after seeding, 50 μl of MTT salt solution (5 mg/ml, Sigma M5655) was added and incubated for 2 h at 37 °C. An equal volume of solubilization buffer (3% Triton X, 0.008% HCl in isopropanol) was added, and the samples were incubated on a rocker for 10 min. The solubilized crystals were collected and measured using a spectrophotometer at an absorbance of 570 nm.
Anchorage-independent Growth Assays
In each well of a 6-well dish, 5000 HCT116 or SW620 cells were seeded in 0.3% agar atop a 0.5% agar underlay. Agar layers contained DMEM supplemented with 5% fetal bovine serum. The cells were incubated for 21 days and were then fixed with 10% methanol/10% acetic acid and stained with 0.01% crystal violet. Images were taken with an EC3 Imaging system (Ultra Violet Products) and colonies were counted using the particle analyzing function of ImageJ software. Minimum colony sizes of 50 pixels2 and 20 pixels2 were used to score HCT116 and SW620 colonies, respectively.
Xenograft Assays
SW620 cells were infected with control shRNA or YAP shRNA2 lentivirus and transduced cells were selected by supplementing growth media with 1 μg/ml puromycin. Two days after infection, 5 × 106 cells were suspended in 200 μl of 1× PBS and injected subcutaneously into both flanks of athymic nude mice. Tumor growth was monitored every 3 days and volumes were measured using standard calipers. Upon completion of the experiment (d21), tumors were isolated, photographed, and weighed.
Immunofluorescence
HCT116 and SW480 cells were seeded on glass coverslips, fixed with 3.7% paraformaldehyde for 10 min and then permeabilized with 0.2% Triton X-100 in 1× PBS for 2 min at room temperature. Cells were blocked in 5% goat serum for 30 min at room temperature and then incubated with primary antibody for 1 h at room temperature. Primary antibodies and dilutions used were β-catenin (BD transduction, BD610154, 1:1000) and YAP (Santa Cruz Biotechnology SC15407X, 1:100). The slides were rinsed in 1X PBS and incubated with secondary antibody for 1 h at room temperature. The secondary antibodies and dilutions used were goat anti-mouse Alexa Fluor 488 (Invitrogen, A11001, 1:2500) and donkey anti-rabbit Cy3 (Jackson ImmunoResearch, 711–166-152, 1:1000). Nuclei were stained with DAPI. Slides were mounted using Fluoro-Gel (Electron Microscopy Sciences, 17983–20) and visualized with a Nikon microscope.
Immunohistochemistry
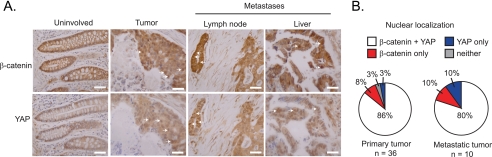
Colon cancer tumor arrays were purchased from Imgenex (IMH-359). Immunohistochemistry was performed as described (19, 23) using anti-β-catenin (BD transduction, BD610154, 1:50) and anti-YAP (Cell Signaling Technology, 4912, 1:200), antibodies. Briefly, slides were dewaxed, and antigen retrieval was performed as described (23). The samples were blocked in 5% goat serum for YAP or 5% horse serum for β-catenin and treated with the Avidin/Biotin blocking kit (Vector Laboratories, SP-2001). Primary antibodies in 5% serum were added and following an overnight incubation at 4 °C, the slides were washed in 1× TBST and then incubated with a 1:400 dilution of biotinylated anti-rabbit (Vector Laboratories, BA-1000) or anti-mouse (Vector Laboratories, BA-2000) secondary antibodies for 1 h. Slides were washed with 1× TBST and then treated with Vectastain ABC Elite reagents (Vector Laboratories, PK-6100). Color was developed using a DAB peroxidase kit (Vector Laboratories, SK-4100), and the samples were counterstained with hematoxylin (Fisher Scientific, CS401-1D). Slides were dehydrated and mounted with cytoseal 60 (Thermo Scientific, 8310-16). Images were taken on a Nikon microscope and two independent investigators scored cells within tumor sections for nuclear β-catenin or nuclear YAP staining. Three fields per section were evaluated and each section was assigned a score of 1–3. Sections with scores of 1–3 were considered positive and the results were used to generate the pie charts in Fig. 6B. For additional details see supplemental Table 1.
FIGURE 6.
β-catenin and YAP are expressed in the nuclear compartment of cells comprising primary and metastatic colorectal tumors. A, immunohistochemical analysis of tissue arrays prepared from primary colorectal tumors, normal tissue, and colorectal tumors that metastasized to the lymph node and liver. Shown are representative images of the 9 uninvolved colonic mucosa samples, the 36 primary tumors, and the 10 metastatic tumors examined. Hematoxylin staining was used to evaluate tissue architecture and β-catenin, and YAP localization was detected using specific antibodies. White arrows identify representative cells with nuclear β-catenin or nuclear YAP staining. The scale bar is 20 μm. B, tumor sections were scored for cells with nuclear β-catenin only (red), nuclear YAP only (blue), nuclear YAP and nuclear β-catenin (white), or no detectable staining (gray).
RESULTS
Identifying YAP as a Direct Wnt/β-Catenin Target Gene
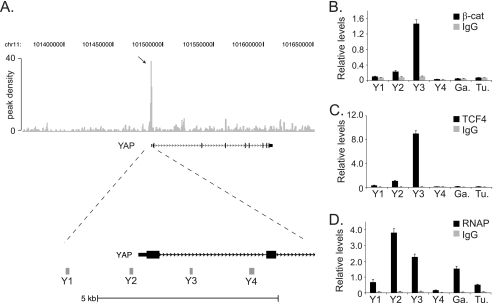
To identify direct Wnt/β-catenin-regulated genes, our laboratory previously conducted a β-catenin ChIP-Seq screen in the HCT116 human CRC cell line (7). A peak of 40 read sequences aligned near the transcript start site of YAP suggesting that this region harbored a β-catenin binding site (Fig. 1A). To confirm the ChIP-seq results, we conducted ChIP assays using β-catenin specific antibodies and an irrelevant immunoglobulin as a control. Oligonucleotides that spanned the YAP proximal promoter and putative binding site were designed (see gray bars Y1-Y4, Fig. 1A), and these primer sets were used to detect β-catenin-precipitated DNA using quantitative and real-time PCR. Strong β-catenin binding was detected at region Y3 with little or no binding at regions Y1 and Y4 (Fig. 1B). Intermediate binding was seen at region Y2, and background levels of β-catenin binding were observed at control regions in the GAPDH or Tubulin genes. Because β-catenin is recruited to chromatin through interactions with TCF4, we next tested whether TCF4 bound to the YAP gene using ChIP assays. As was the case for β-catenin, TCF4 binding was detected at region Y3 with little or no binding detected at regions Y1, Y4, or GAPDH or Tubulin controls (Fig. 1C). To determine whether β-catenin/TCF4 bound a transcriptionally active YAP gene locus, we precipitated RNA polymerase II (RNAP) in ChIP assays and found that RNAP bound YAP (Fig. 1D). These results suggest that β-catenin/TCF4 complexes associate with a transcriptionally active YAP gene locus in CRC cells.
FIGURE 1.
β-Catenin, TCF4, and RNAP bind the first intron of the YAP gene in vivo. A, diagram of the YAP gene locus and the β-catenin-enriched regions identified in a ChIP-Seq screen. Top, gray vertical lines represent peak density plot from the β-catenin ChIP-Seq screen. The arrow indicates the accumulated ChIP-Seq reads that mapped near the YAP transcription start site. Bottom, enlargement of the 5′-end of the YAP1 gene with Y1, Y2, Y3, and Y4 indicating the position of the PCR primer sets used in B, C, and D. B, quantitative real-time PCR analysis of DNA fragments isolated in ChIP assays conducted in HCT116 cells using β-catenin specific antibodies or an irrelevant IgG as a control. The primer sets Y1-Y4 were used to detect β-catenin binding and primers designed against regions in GAPDH (Ga.) and Tubulin (Tu.) were used as negative controls. C and D, as in B except anti-TCF4 antibodies were used in C and anti-RNAP antibodies were used in D to detect TCF4 and RNAP binding in ChIP assays, respectively. In B, C, and D, error is S.E.
The β-Catenin/TCF4 Binding Region of YAP Demarcates an Enhancer Element
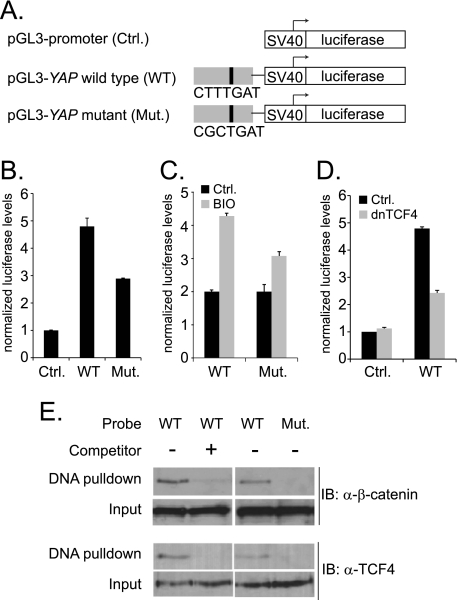
In CRC cells, constitutive β-catenin/TCF4 complexes drive target gene expression. Regulatory DNA elements that assemble β-catenin/TCF complexes and recruit RNAP have been described as Wnt responsive elements (WREs) (24). WREs often, but not always, contain a consensus motif known to bind TCF4 (7, 25, 26). To determine whether the β-catenin/TCF4 binding site in the YAP gene identified a putative enhancer element, we first searched a 600 base pair sequence region centered under the apex of the β-catenin ChIP-Seq peak for the core consensus TCF binding motif CTTTG A/T A/T or A/T A/T CAAAG (7, 25). The 600 bp segment was chosen because it was twice the size of the largest DNA fragments included in construction of the β-catenin ChIP-Seq library (7). This search found the sequence CTTTGAT. To test whether a DNA fragment that harbored this motif functioned as an enhancer, we first performed luciferase reporter assays. A 624 base pair fragment of the YAP gene containing the consensus TCF binding site was inserted into the pGL3 promoter-firefly luciferase plasmid (Fig. 2A). When transiently expressed in human CRC cells, the pGL3-YAP wild-type (WT) plasmid yielded 5-fold higher luciferase levels compared with levels obtained from cells transfected with vector control (Fig. 2B).
FIGURE 2.
The β-catenin/TCF4 binding region of YAP enhances SV40-driven luciferase gene expression. A, diagram of the pGL3-promoter plasmids used in luciferase assays with the SV40 minimal promoter and firefly luciferase gene shown as white rectangles and the 624 bp YAP enhancer in gray. A black vertical line represents the position of the consensus TCF motif within the YAP enhancer. The TCF sequence was changed by site-directed mutagenesis to generate the pGL3-YAP mutant plasmid (Mut.). B, HCT116 cells were transfected with vector control, pGL3-YAP wild-type (WT) or pGL3-YAP mutant (Mut.) firefly luciferase plasmids and a plasmid expressing Renilla luciferase. Twenty-four hours later, luciferase levels were measured and firefly luciferase levels were normalized to Renilla luciferase levels to control for differences in transfection efficiency between samples. C, cells transfected with the indicated reporters were untreated (black bars) or treated (gray bars) with the GSK3β inhibitor BIO for 12 h prior to measuring luciferase levels. D, cells were transfected with vector control or the pGL3-YAP (WT) luciferase reporter and, where indicated, the cells were co-transfected with a dominant negative TCF4 plasmid. Luciferase levels were measured as in B. E, nuclear proteins prepared from HCT116 cells were incubated with a 28 bp biotinylated and double-stranded DNA probe that contained either the wild-type or mutant TCF consensus sequence. Where indicated, unlabeled YAP duplexes were included in the binding reactions. Western blot analysis was used to detect bound β-catenin or TCF4. In B, C, and D, error is standard deviation.
To test whether β-catenin/TCF4 complexes were responsible for enhancer activity, we mutated the consensus TCF motif to CGCTGAT. In transfected cells, levels of luciferase expressed from the YAP (Mut.) reporter were ∼60% of the levels observed with the YAP (WT) reporter (Fig. 2B). To test whether the residual activity of the YAP (Mut.) reporter was due to a second β-catenin binding site, cells were treated with the GSK3β inhibitor BIO. BIO treatment increased both pGL3-YAP (WT) and pGL3-YAP (Mut.) reporter activities suggesting that the YAP enhancer element contains a second, unidentified β-catenin binding site (Fig. 2C). To extend these findings, we co-transfected dominant negative TCF4 (dnTCF4) along with pGL3-YAP (WT) or control luciferase plasmids into HCT116 cells. Dominant negative TCF4 lacks the amino-terminal β-catenin interaction domain and competes with the assembly of functional β-catenin/TCF4 complexes at DNA binding sites (27). Dominant negative TCF4 reduced YAP (WT) enhancer-driven luciferase levels (Fig. 2D).
We next tested whether β-catenin/TCF4 complexes assembled on the consensus TCF site within the YAP element using a DNA pull-down assay. Nuclear extracts were prepared from HCT116 cells, and these proteins were incubated with a biotinylated and double-stranded DNA fragment containing the wild-type YAP sequences or YAP sequences with a mutated TCF binding site. DNA/protein complexes were isolated using streptavidin-coated magnetic beads, and β-catenin and TCF4 binding was assessed by Western blot analysis. β-Catenin and TCF4 bound the biotinylated wild-type probe, and this binding was competed when an excess of unlabeled, wild-type YAP DNA was added to the binding reactions (Fig. 2E). β-Catenin and TCF4 did not bind the mutant probe (Fig. 2E). Together, results from these experiments indicate that the DNA element within the YAP intron functions as an enhancer element that assembles β-catenin/TCF4 complexes to activate transcription.
β-Catenin Is Required for YAP Expression in CRC Cells
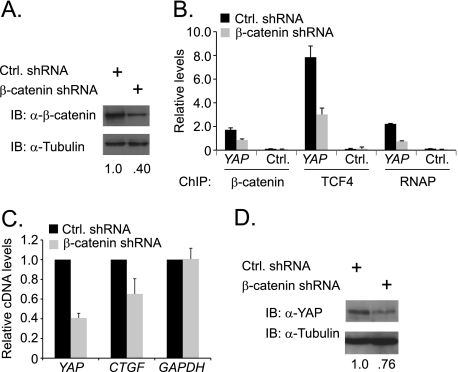
To test whether β-catenin was required for YAP expression in CRC cells, we used a lentiviral system to deliver short hairpin RNAs (shRNAs) designed to reduce CTNNB1 mRNA levels. CTNNB1 is the gene that codes for β-catenin, and for simplicity we will refer to mRNA and protein as β-catenin. We used this system previously and found that β-catenin shRNAs significantly reduced β-catenin protein expression in HCT116 cells (26). As a control, we packaged lentivirus that contained a scrambled shRNA sequence. Five days after infection with β-catenin or control shRNAs, whole cell lysates were prepared and β-catenin protein levels were assessed by Western blot analysis. Infection with the β-catenin shRNA virus resulted in a 60% reduction of β-catenin protein levels compared with levels seen in cells that were infected with the control shRNA (Fig. 3A). To test whether chromatin associated β-catenin was also reduced we conducted ChIP assays and found that cells infected with the β-catenin shRNA virus contained 50% less β-catenin bound to the YAP gene (Fig. 3B). shRNA-mediated reduction of β-catenin also impaired TCF4 binding and RNAP recruitment to YAP (Fig. 3B). These findings suggest that β-catenin/TCF4 complexes recruit RNAP to transcribe YAP. In support of this model, we find that β-catenin depletion resulted in a 60% decrease in YAP mRNA levels (Fig. 3C). Moreover, expression of CTGF, which is a well-established and direct YAP-regulated target gene, was also decreased in β-catenin shRNA-expressing cells (Fig. 3C) (12). To determine whether reduced mRNA expression was reflected at the protein level, we conducted Western blot analysis using YAP specific antibodies and found that β-catenin knockdown decreased YAP expression 24% (Fig. 3D). Thus, β-catenin/TCF4 complexes assembled at the YAP gene recruit RNAP to activate YAP gene expression and this leads to increased YAP protein levels.
FIGURE 3.
β-Catenin controls YAP expression in HCT116 colon cancer cells. A, Western blot analysis of β-catenin protein levels in HCT116 cells expressing a control shRNA or an shRNA targeting β-catenin. HCT116 cells were infected with lentiviruses and Western blots were conducted 5 days after infection. B, quantitative real-time PCR analysis of DNA precipitated using β-catenin, TCF4, or RNAP specific antibodies in ChIP assays conducted in HCT116 cells expressing the indicated shRNAs. Primer set Y3 listed in Fig. 1A was used to detect β-catenin, TCF4, and RNAP binding to the YAP gene. C, quantitative real-time PCR analysis of cDNAs synthesized from RNA isolated from HCT116 cells expressing control (black bars) or β-catenin shRNA (gray bars). Specific primers were used to detect expression of YAP, CTGF, and GAPDH as a control. D, Western blot analysis of YAP protein levels in cells expressing the indicated shRNAs. Whole cell lysates were prepared and analyzed 5 days following lentiviral infection. In B and C error is S.E.
YAP Enhances CRC Cell Metabolism and Anchorage-independent Growth in Soft Agar
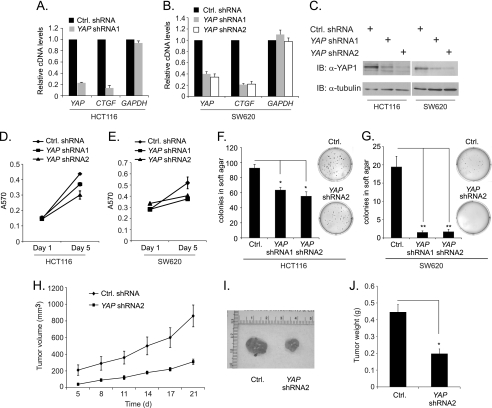
YAP expression is elevated in several epithelial cancers and its expression is required for proliferation of ovarian cancer cells and immortalized breast epithelial cells (19, 28–30). Furthermore, YAP has recently been shown to promote colony growth of CRC cells (17). To examine the role for YAP in these CRC cell properties, we knocked down YAP gene expression in HCT116 cells using lentiviral delivery of an shRNA that targeted the YAP transcript. Five days following infection with the lentivirus containing YAP shRNA1, YAP mRNA levels were measured by quantitative RT-PCR. HCT116 cells expressing YAP shRNA1 resulted in an 80% decrease in YAP mRNA relative to control cells (Fig. 4A). Expression of CTGF was also decreased significantly by YAP knockdown (Fig. 4A). Similar results were seen in the highly metastatic SW620 CRC cell line (Fig. 4B). As an additional control, we designed a second shRNA, YAP shRNA2, that targeted a different site of the YAP mRNA sequence and found that both shRNAs significantly decreased YAP and CTGF mRNA levels when compared with control (Fig. 4B). Furthermore, expression of YAP shRNA1 or shRNA2 in HCT116 and SW620 cells caused a significant decrease in YAP protein levels (Fig. 4C). Therefore the YAP shRNAs specifically and effectively targeted YAP mRNA in both cell lines.
FIGURE 4.
YAP enhances CRC cell metabolism, anchorage-independent growth in soft agar and tumorigenesis. A, quantitative real time PCR analysis of cDNAs prepared from mRNAs isolated from HCT116 cells 5 days after they were infected with a lentivirus expressing a control (black bars) or YAP specific (gray bars) shRNAs. Specific primers were used to measure relative expression of YAP, CTGF, and GAPDH. B, as in A, except that SW620 CRC cells were used and a second YAP specific shRNA (YAP shRNA2) was included as an additional control. C, Western blot analysis of YAP protein levels in HCT116 and SW620 cells expressing the indicated shRNAs. Blots were reprobed with tubulin antibodies to demonstrate that equivalent amounts of proteins were loaded. D and E, MTT assays were conducted in HCT116 and SW620 cells expressing the indicated shRNAs. Assays were performed 1 and 5 days after lentiviral infection of the cells. F and G, anchorage independent growth assays were conducted in HCT116 and SW620 cells expressing the indicated shRNAs. The day following infection with the indicated lentiviruses, cells were plated in soft agar and colony formation was assessed and quantified after 21 days. H, growth curves of tumors that formed after injection of control or YAP shRNA2 expressing SW620 cells into the flanks of athymic mice. I, explanted tumors on day 21 following injection. J, tumor weights at day 21. Error denotes S.E. and *, p < 0.005 and **, p < 0.0005.
To examine the effect of YAP knockdown on CRC cell metabolism, we performed MTT assays using HCT116 and SW620 cells expressing YAP shRNA1, YAP shRNA2, or a scrambled shRNA control. The MTT assay measures mitochondrial activity and is routinely used to as an indirect measure of cellular metabolism and viability. In both CRC cell lines, YAP knockdown resulted in a significant decrease in cell metabolism after 5 days in culture (Fig. 4, D and E). To determine whether YAP impacts CRC colony formation in an anchorage-independent context, HCT116 and SW620 cells expressing YAP shRNA1, YAP shRNA2, or a scrambled shRNA control were plated in soft agar and grown for 21 days. YAP knockdown using either shRNA resulted in a considerable decrease in colony number in both cell lines (Fig. 4, F and G). This decrease was especially dramatic in metastatic SW620 cells, which virtually lost the ability to form detectable colonies when YAP protein levels were reduced (Fig. 4G). These data indicate that YAP is a positive regulator of metabolism and anchorage-independent growth in CRC cells, thereby suggesting a role for YAP in promoting colorectal cancer.
To determine the role of YAP in vivo, control or YAP shRNA2-expressing SW620 cells were injected subcutaneously into the flanks of athymic nude mice. The tumors were measured every 3 days and we found that YAP depletion inhibited tumor growth in this xenograft model (Fig. 4H). At the end of the protocol (d21), explanted tumors derived from YAP knock-down cells were visually smaller and weighed significantly less than control tumors (Fig. 4, I and J). These results indicate that YAP promotes tumorigenesis of implanted CRC cells.
Nuclear YAP Expression Is Retained in Densely Grown CRC Cells
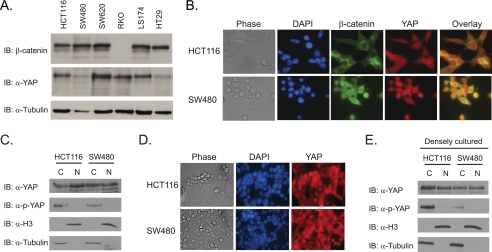
To further examine YAP protein expression in colon cancer, we screened six CRC cell lines by Western blot analysis. YAP protein was readily detectable in all cell lines tested although protein levels were lower in SW480 and HT29 cells (Fig. 5A). As a control, the blots were probed with β-catenin specific antibodies and as expected, β-catenin was expressed at high levels in CRC cells that contained mutations in the Wnt/β-catenin signaling pathway; however β-catenin was undetectable in RKO cells, which contain a wild-type Wnt signaling pathway (Fig. 5A) (31). We then sought to confirm the Western blot results using indirect immunofluorescence. In HCT116 cells, β-catenin staining was strongest at the membrane with diffuse staining throughout the cytoplasm and nucleus, whereas YAP staining was uniform in both compartments (Fig. 5B). In SW480 cells, β-catenin localized predominantly to the nucleus with weak cytoplasmic staining whereas YAP staining was largely confined to the nucleus (Fig. 5B). Moreover, several cells displayed co-localization of YAP and β-catenin in the nucleus which is especially evident in SW480 cells.
FIGURE 5.
Nuclear YAP expression is retained in densely grown CRC cells. A, Western blot analysis of YAP and β-catenin expression in six CRC cell lines. Tubulin served as a loading control. B, immunocytochemical analysis of YAP and β-catenin expression in HCT116 and SW480 cells. C, Western blot analysis of YAP protein levels in cytoplasmic and nuclear fractions from HCT116 and SW480 cells. Blots were reprobed with anti-histone H3 antibodies and anti-tubulin antibodies to mark the nuclear and cytoplasmic fractions, respectively. D, immunocytochemical analysis of YAP and β-catenin expression in HCT116 and SW480 cells that were densely cultured on glass coverslips. E, Western blot analysis of YAP protein levels in cytoplasmic and nuclear proteins prepared from densely cultured HCT116 and SW480.
YAP function as a transcriptional co-activator is restricted by its subcellular localization. When the Hippo pathway is engaged, YAP is phosphorylated on serine 127 (S127) which sequesters it in the cytoplasm through interactions with 14-3-3 proteins (8). The fact that YAP protein is detected in the cytoplasm of CRC cells suggests that the Hippo pathway is intact in these cells at least to some degree (Fig. 5B). To determine whether cytoplasmic YAP was phosphorylated, we separated HCT116 and SW480 cells into nuclear and cytoplasmic fractions and conducted Western blot assays using anti-phospho-S127 YAP antibodies. While YAP protein was detected in nuclear and cytoplasmic fractions in both cells, phospho-YAP was found primarily in the cytoplasmic fraction (Fig. 5C). Re-probing the blots with anti-histone H3 antibodies and anti-tubulin antibodies, to mark nuclear and cytoplasmic compartments, respectively, attests to the purity of our fractionation protocols. Recent reports have demonstrated that YAP nuclear localization is sensitive to cell density (28, 32). When breast epithelial cell lines reach confluence on the tissue culture dish, YAP is redistributed from the nucleus to the cytoplasm (28, 32). To determine whether density affects YAP localization in CRC cells, HCT116 and SW480 cells were heavily seeded, and YAP was analyzed by indirect immunofluorescence. Interestingly, YAP nuclear localization was clearly retained in several cells (Fig. 5D). Western blot analysis on nuclear and cytoplasmic fractions isolated from these cells confirmed that cell density does not affect nuclear YAP localization outright (Fig. 5E). These findings indicate a defect in the ability of CRC cells to properly regulate the subcellular localization of YAP.
β-Catenin and YAP Are Expressed in the Nuclear Compartment of Cells Comprising Primary and Metastatic Colorectal Tumors
β-Catenin is stabilized in the majority of colon cancers and previous studies have shown that YAP expression is increased in colorectal tumors compared with normal tissue (17, 19, 33). To determine whether β-catenin and YAP localize to the nucleus in cells within the same tumor, we performed an immunohistochemical analysis on primary colonic tumors and uninvolved colonic mucosa using antibodies against β-catenin and YAP. In uninvolved tissue, β-catenin and YAP localize primarily to the cell membrane and cytoplasm (Fig. 6A). In line with previous reports, we find that β-catenin and YAP expression is increased in tumor cell nuclei (17, 19, 33). Importantly, several tumors contain cells with elevated levels of β-catenin and YAP in the nucleus (Fig. 6A, white arrows). Of the 36 primary tumors examined, 86% scored positive for nuclear localization of β-catenin and YAP, while only 3% lacked nuclear expression of either protein (Fig. 6B). We also examined ten CRC derived metastatic tumors for expression of β-catenin and YAP and found that both proteins were highly expressed in the cell nucleus in metastatic tumors (Fig. 6A). Of the 10 metastatic tumor samples examined, 80% contained regions with nuclear β-catenin and YAP localization (Fig. 6B). This analysis indicates that elevated and nuclear expression of β-catenin and YAP is found in cells comprising the majority of primary and metastatic colorectal carcinomas examined.
DISCUSSION
Wnt/β-catenin signaling is required for proper establishment and maintenance of the colonic crypt microenvironment (3). Mutations in components of the Wnt/β-catenin signaling pathway are found in the vast majority of colon cancers, and these mutations lead to the stabilization and nuclear import of the β-catenin transcriptional co-activator (34). Because chromatin-associated β-catenin complexes drive expression of underlying target genes, misexpression of these targets contributes to the pathogenesis of colorectal cancers (5). Therefore, identification of these targets is paramount to understanding the mechanisms that initiate cellular transformation and the progression of these neoplastic cells to adenomas and carcinomas. To identify direct β-catenin target genes in human colon cancer cells, our lab conducted a ChIP-Seq screen using β-catenin specific antibodies (7). One target identified in that screen was the gene encoding the YAP transcriptional co-activator. In the present study, we demonstrate that β-catenin/TCF4 complexes directly regulate YAP gene expression in human CRC cells. These findings suggest that aberrant Wnt/β-catenin signaling directly deregulates the Hippo/YAP pathway in CRC cells.
Regulation of YAP function at the post-translational level has been well studied with Hippo signaling serving to retain YAP in the cytoplasm through direct phosphorylation-dependent mechanisms (8). At the post-transcriptional level, YAP mRNA is negatively regulated by microRNA-375 in hepatocellular carcinoma cells (35). Our report is the first to address how YAP expression is controlled at the transcriptional level. Results from our previous ChIP-Seq screen strongly suggested that the primary β-catenin binding site localized to the first intron of YAP; however, we noticed that several minor peaks were also found within the YAP gene body (Fig. 1A). Repeat ChIP assays with β-catenin specific antibodies failed to detect significant binding to these sites suggesting that these peaks represent nonspecific associations in the ChIP-Seq screen. However, we demonstrate that the binding site identified by the cluster of 40 peak reads within the first intron of YAP demarcates an enhancer element that binds β-catenin/TCF4 complexes to control YAP expression in CRC cells. While our data support the role of β-catenin/TCF4 complexes in recruiting RNAP, it is also possible that β-catenin may play a role in RNAP promoter clearance and facilitation of an elongation RNAP complex. Regardless of the precise mechanism, β-catenin/TCF4 binding likely enhances production of full-length YAP mRNA as we observe that β-catenin knockdown reduces YAP protein levels (Fig. 3). Ongoing studies in our laboratory are aimed at fully defining the proximal promoter region of YAP to identify additional cis regulatory DNA elements. These efforts will be critical to determine whether additional transcription factors that cooperate with β-catenin/TCF4 complexes are required to deregulate YAP gene expression in CRC cells.
The Hippo pathway serves to restrict YAP nuclear localization when cells are contact inhibited. Recent evidence indicates that signaling from the adherens junction complexes results in YAP phosphorylation and cytoplasmic retention (8, 32, 36, 37). Interestingly, our analysis in densely plated colon cancer cells demonstrated that YAP protein remained in the nucleus under these conditions (Fig. 5). As phosphorylated YAP on serine 127 was retained, we conclude that the Hippo pathway itself is at least partially intact. This cancer cell characteristic was also recently reported in studies of MDA-MB-231 breast adenocarcinoma cells (32). When E-cadherin expression was restored in MDA-MB-231 cells, YAP was redistributed to the cytoplasm in densely plated cells (32). It is possible that levels of E-cadherin and/or α-catenin are insufficient to fully stimulate the Hippo pathway in CRC cells. Because cancer cells in general are refractory to contact growth inhibition, nuclear accumulation of YAP may be a general mechanism that allow for cancers to escape this growth suppression pathway (39). Understanding precisely how YAP escapes cellular redistribution in densely grown CRC cells requires further investigation.
An intersection between Wnt/β-catenin and Hippo signaling pathways has recently been described (40, 41). In an elegant study, Varelas et al. found that TAZ, the YAP paralog, restricted Wnt/β-catenin signaling (40). They showed that TAZ interacted with Dishevelled and that this interaction impaired Wnt3A activation of the Wnt pathway (40). These results demonstrated that Hippo signaling negatively impacted the Wnt/β-catenin pathway, whereas our results suggest that Wnt/β-catenin pathway promotes YAP expression. There are important differences between the cell systems used in our studies that could offer an explanation for this apparent disparity. Varelas et al. primarily studied the role for TAZ in HEK293 cells stimulated with Wnt3A ligand. These cells contain an intact Wnt/β-catenin signaling pathway whereas the CRC cells used in our studies contain elevated β-catenin levels due to mutations in the pathway downstream of Dishevelled. In addition, their study focused on TAZ, while the potential role for YAP in impairing Wnt signaling was not addressed. Together, our results indicate that a complex interplay between Wnt/β-catenin and Hippo pathways exists to properly control cellular growth.
Recently, β-catenin and YAP were shown to interact in nuclear extracts prepared from day E14.5 murine hearts (42). Composite enhancer elements that bind YAP/TEAD and β-catenin/TCF complexes were required for expression of at least some target genes. It was proposed that an antagonistic relationship between Hippo and Wnt signaling pathways controls cardiomyocyte growth by regulating expression of a common set of targets. Our initial efforts using co-immunoprecipitation/Western blot analysis have failed to detect an interaction between β-catenin and YAP in HCT116 cells. However, our analysis of DNA elements identified in our β-catenin ChIP-Seq library has found numerous targets that contain coupled TEAD and TCF consensus motifs. We are currently investigating whether Wnt/β-catenin and Hippo/YAP pathways directly regulate expression of these targets in CRC cells.
Our histochemical analysis of YAP and β-catenin expression in human tissues indicate that expression of both proteins is elevated in primary tumors compared with their expression in uninvolved tissue (Fig. 6A). We also find robust nuclear expression of both factors in metastatic disease. In addition, our results, and those reported by Zhou et al., clearly demonstrate that YAP activity is required for growth of established human CRC cell lines and we find that this feature may be due to increased cellular metabolism (17). While it is widely accepted that deregulated Wnt/β-catenin signaling leads to colon cancer, therapeutics designed to mitigate persistent nuclear β-catenin/TCF complexes have achieved limited success (43). Recent reports and findings presented here strongly implicate YAP functioning as an oncogene in colon cancers (17, 19). We propose that compounds designed to either retain YAP in the cytoplasm or reduce nuclear YAP activity would be beneficial for the treatment of colon cancer.
Supplementary Material
Acknowledgments
We thank Drs. Omar Quintero and Chris Yengo (Pennsylvania State University College of Medicine) for assistance with immunofluorescence protocols and Daniel Bottomly (Oregon Health & Science University) for bioinformatic support. We also thank members of the Yochum laboratory for helpful discussions pertaining to this work.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK080805 (to G. S. Y.) and institutional funds provided by the Pennsylvania State University College of Medicine.

This article contains supplemental Table S1.
- APC
- adenomatous polyposis coli
- CRC
- colorectal cancer
- YAP
- Yes-associated protein
- TCF
- T-cell factor
- LEF
- lymphoid enhancer factor
- WRE
- Wnt responsive element
- TAZ
- transcriptional coactivator with PDZ binding motif
- TEAD
- TEA domain-containing transcription factor
- dnTCF4
- dominant negative TCF4
- CTGF
- connective tissue growth factor.
REFERENCES
- 1. MacDonald B. T., Tamai K., He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rizvi A. Z., Wong M. H. (2005) Epithelial stem cells and their niche: there's no place like home. Stem Cells 23, 150–165 [DOI] [PubMed] [Google Scholar]
- 3. Sancho E., Batlle E., Clevers H. (2004) Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20, 695–723 [DOI] [PubMed] [Google Scholar]
- 4. Nusse R. (2005) Wnt signaling in disease and in development. Cell Res 15, 28–32 [DOI] [PubMed] [Google Scholar]
- 5. Mosimann C., Hausmann G., Basler K. (2009) β-Catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10, 276–286 [DOI] [PubMed] [Google Scholar]
- 6. Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 7. Bottomly D., Kyler S. L., McWeeney S. K., Yochum G. S. (2010) Identification of {β}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 38, 5735–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao B., Li L., Lei Q., Guan K. L. (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harvey K. F., Pfleger C. M., Hariharan I. K. (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 [DOI] [PubMed] [Google Scholar]
- 10. Pantalacci S., Tapon N., Léopold P. (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 [DOI] [PubMed] [Google Scholar]
- 11. Wu S., Huang J., Dong J., Pan D. (2003) Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 [DOI] [PubMed] [Google Scholar]
- 12. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Ji J. Y., Yu M., Overholtzer M., Smolen G. A., Wang R., Brugge J. S., Dyson N. J., Haber D. A. (2009) YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 11, 1444–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 [DOI] [PubMed] [Google Scholar]
- 15. Lu L., Li Y., Kim S. M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M. J., Lee J. S., Johnson R. L. (2010) Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U.S.A. 107, 1437–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song H., Mak K. K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R. M., Wang C. Y., Gao B., Jiang J., Yang Y. (2010) Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 107, 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou D., Zhang Y., Wu H., Barry E., Yin Y., Lawrence E., Dawson D., Willis J. E., Markowitz S. D., Camargo F. D., Avruch J. (2011) Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. U.S.A. 108, E1312-E1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai J., Zhang N., Zheng Y., de Wilde R. F., Maitra A., Pan D. (2010) The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steinhardt A. A., Gayyed M. F., Klein A. P., Dong J., Maitra A., Pan D., Montgomery E. A., Anders R. A. (2008) Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 39, 1582–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yochum G. S., Sherrick C. M., Macpartlin M., Goodman R. H. (2010) A beta-catenin/TCF-coordinated chromatin loop at MYC integrates 5′ and 3′ Wnt responsive enhancers. Proc. Natl. Acad. Sci. U.S.A. 107, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yochum (2011) Multiple Wnt/ß-catenin responsive enhancers align with the MYC promoter through long-range chromatin loops. PLoS ONE 6, e18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sansom O. J., Meniel V. S., Muncan V., Phesse T. J., Wilkins J. A., Reed K. R., Vass J. K., Athineos D., Clevers H., Clarke A. R. (2007) Myc deletion rescues Apc deficiency in the small intestine. Nature 446, 676–679 [DOI] [PubMed] [Google Scholar]
- 24. Archbold H. C., Yang Y. X., Chen L., Cadigan K. M. (2012) How do they do Wnt they do?: Regulation of transcription by the Wnt/β-catenin pathway. Acta Physiol. 204, 74–109 [DOI] [PubMed] [Google Scholar]
- 25. Hatzis P., van der Flier L. G., van Driel M. A., Guryev V., Nielsen F., Denissov S., Nijman I. J., Koster J., Santo E. E., Welboren W., Versteeg R., Cuppen E., van de Wetering M., Clevers H., Stunnenberg H. G. (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 28, 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yochum G. S., McWeeney S., Rajaraman V., Cleland R., Peters S., Goodman R. H. (2007) Serial analysis of chromatin occupancy identifies β-catenin target genes in colorectal carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 104, 3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., Tjon-Pon-Fong M., Moerer P., van den Born M., Soete G., Pals S., Eilers M., Medema R., Clevers H. (2002) The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 [DOI] [PubMed] [Google Scholar]
- 28. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X., George J., Deb S., Degoutin J. L., Takano E. A., Fox S. B., Bowtell D. D., Harvey K. F. (2011) The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene 30, 2810–2822 [DOI] [PubMed] [Google Scholar]
- 30. Hall C. A., Wang R., Miao J., Oliva E., Shen X., Wheeler T., Hilsenbeck S. G., Orsulic S., Goode S. (2010) Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 70, 8517–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. da Costa L. T., He T. C., Yu J., Sparks A. B., Morin P. J., Polyak K., Laken S., Vogelstein B., Kinzler K. W. (1999) CDX2 is mutated in a colorectal cancer with normal APC/β-catenin signaling. Oncogene 18, 5010–5014 [DOI] [PubMed] [Google Scholar]
- 32. Kim N. G., Koh E., Chen X., Gumbiner B. M. (2011) E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U.S.A. 108, 11930–11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson C. B., Neufeld K. L., White R. L. (2002) Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc. Natl. Acad. Sci. U.S.A. 99, 8683–8688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clevers H. (2006) Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 35. Liu A. M., Poon R. T., Luk J. M. (2010) MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun. 394, 623–627 [DOI] [PubMed] [Google Scholar]
- 36. Silvis M. R., Kreger B. T., Lien W. H., Klezovitch O., Rudakova G. M., Camargo F. D., Lantz D. M., Seykora J. T., Vasioukhin V. (2011) α-Catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal 4, ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R., Camargo F. D. (2011) Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deleted in proof
- 39. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 40. Varelas X., Miller B. W., Sopko R., Song S., Gregorieff A., Fellouse F. A., Sakuma R., Pawson T., Hunziker W., McNeill H., Wrana J. L., Attisano L. (2010) The Hippo pathway regulates Wnt/β-catenin signaling. Dev. Cell 18, 579–591 [DOI] [PubMed] [Google Scholar]
- 41. Hergovich A., Hemmings B. A. (2010) TAZ-mediated crosstalk between Wnt and Hippo signaling. Dev. Cell 18, 508–509 [DOI] [PubMed] [Google Scholar]
- 42. Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L., Martin J. F. (2011) Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barker N., Clevers H. (2006) Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug. Discov. 5, 997–1014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.