Background: Several proteins catalyze branch migration (BM) of the Holliday junction.
Results: RAD54 is a robust BM protein capable of bypassing extensive regions of DNA heterology. RAD54, BLM, and RECQ1 drive BM in the 3′→5′ direction.
Conclusion: The displacement strand of joint molecules (JMs) defines the polarity of BM.
Significance: BM is mechanistically distinct from helicase activity of DNA translocating proteins.
Keywords: ATPases, DNA-binding Protein, DNA Helicase, DNA Recombination, DNA Repair, DNA-Protein Interaction, Homologous Recombination
Abstract
Several proteins have been shown to catalyze branch migration (BM) of the Holliday junction, a key intermediate in DNA repair and recombination. Here, using joint molecules made by human RAD51 or Escherichia coli RecA, we find that the polarity of the displaced ssDNA strand of the joint molecules defines the polarity of BM of RAD54, BLM, RECQ1, and RuvAB. Our results demonstrate that RAD54, BLM, and RECQ1 promote BM preferentially in the 3′→5′ direction, whereas RuvAB drives it in the 5′→3′ direction relative to the displaced ssDNA strand. Our data indicate that the helicase activity of BM proteins does not play a role in the heterology bypass. Thus, RAD54 that lacks helicase activity is more efficient in DNA heterology bypass than BLM or REQ1 helicases. Furthermore, we demonstrate that the BLM helicase and BM activities require different protein stoichiometries, indicating that different complexes, monomers and multimers, respectively, are responsible for these two activities. These results define BM as a mechanistically distinct activity of DNA translocating proteins, which may serve an important function in DNA repair and recombination.
Introduction
Homologous recombination is the evolutionarily conserved pathway that promotes the repair of DNA double-strand breaks, restart of stalled replication forks, and faithful chromosome segregation (1–3). Homologous recombination promotes the repair of DNA double-strand breaks or single-strand gaps by utilizing homologous DNA sequences as a template (4). The initial steps of homologous recombination, the search for homologous DNA sequences, and the invasion of broken DNA ends into the homologous duplex DNA are promoted by Rad51/RecA and its auxiliary proteins (5). The invasion leads to formation of the Holliday junction, the cross-like DNA structure that contains the exchange point between two recombining DNA molecules. The Holliday junction can undergo branch migration (BM),2 in which one DNA strand is progressively exchanged for another, affecting the amount of genetic information that can be transferred between two parental DNA molecules (6). At later steps, the two physically linked DNA molecules are separated by cleaving with structure-specific nucleases.
Several specialized helicase-like proteins have been identified that promote BM of Holliday junctions in an ATP hydrolysis-dependent manner (7). In prokaryotes, RuvAB and RecG were shown to promote BM of Holliday junctions (8, 9). In eukaryotes, several members of helicase superfamily 2 promote BM of Holliday junctions including Rad54 and Rad5, proteins of the Snf2 family of DNA translocases (10, 11); BLM, WRN, RECQ1, and RECQ5, members of the RecQ helicase family (12–15); and Fanconi anemia, complementation group M (FANCM), a member of the DEAH helicase family (16). All these helicase-like proteins (i) are capable of translocation on DNA in an ATPase-dependent manner and (ii) show high binding affinity to Holliday junctions or Holliday junction-like structures.
Biochemical studies provided important insights into the mechanisms of BM and specific functions of the BM proteins in DNA repair and recombination. It was shown that Rad54, BLM, or WRN can promote dissociation of D-loops (17–20), which may help to channel homologous recombination into the synthesis-dependent strand annealing pathway, which decreases the rate of somatic crossing over, thereby reducing loss of heterozygosity (21). Also, BLM in a complex with TOPO IIIα and BLAP75 catalyzes resolution of double-Holliday junctions into non-crossover intermediates through the decatenation mechanism (22, 23). In addition, RAD54, BLM, WRN, FANCM, Rad5, and RecG were shown to promote in vitro reversible regression of model replication forks into Holliday junctions, the process known as “chicken foot formation” that may play a role in DNA lesion bypass during replication in vivo (16, 24–29).
However, despite intensive biochemical studies of BM proteins, several fundamental questions remain unsolved. One such question is the polarity of protein-driven BM. dsDNA is a bipolar molecule with two strands of opposite polarity. For single-stranded DNA translocases, the polarity of the ssDNA provides the directionality of translocation. Many helicases bind preferably to the ssDNA region of tailed DNA substrates and then translocate into the duplex region by tracking the same ssDNA strand. Double-stranded DNA translocases determine their polarity also by tracking one of the two strands of the DNA duplex. In this case, an asymmetry in the initiation complex may play a critical role in defining polarity during translocation. Because the Holliday junction is a symmetrical structure, the polarity of BM proteins is difficult to define.
Most known BM proteins, including BLM, WRN, RecG, RuvAB, REQ5, and RECQ1, also possess a DNA helicase activity, i.e. the ability to separate complimentary strands of DNA. Still, a few other BM proteins, such as RAD54, do not have helicase activity. The relationship between BM and helicase activity and a specific role of the helicase activity in BM is currently unknown. Because homologous recombination involves similar but not identical DNA sequences, the passage of Holliday junctions through the sequence heterology must involve melting of DNA base pairs followed by formation of mismatches, insertions, or deletions in the newly formed heteroduplex DNA. Therefore, one may suggest that DNA helicase activity of BM proteins may facilitate the heterology bypass.
Here, to study BM activities of human RAD54, BLM, and RECQ1, we used the plasmid-length DNA substrates known as joint molecules (JMs) or α-structures (see Fig. 1A). The JMs with the displaced ssDNA strand of the opposite polarities were generated by DNA strand exchange promoted by human RAD51 and Escherichia coli RecA. We demonstrate that the polarity of the displaced ssDNA strand of JMs defines the directionality of BM of RAD54, BLM, RECQ1, and RuvAB. These results indicate the universal role of the displaced ssDNA strand in determining the polarity of BM proteins. Our results demonstrate that RAD54, BLM, and RECQ1 promote BM, both four-stranded and three-stranded, preferentially in the 3′→5′ direction, whereas RuvAB drives the reactions in the 5′→3′ direction relative to the displaced ssDNA strand of JMs. Our data indicate that the helicase activity of BM proteins does not play a role in the heterology bypass during BM of Holliday junctions. Thus, RAD54 that lacks canonical helicase activity is significantly more efficient in DNA heterology bypass than BLM or REQ1 helicases. Furthermore, we demonstrate that the BLM helicase and BM activities require different protein stoichiometries and reaction conditions, indicating that different protein complexes, monomers and multimers, respectively, are responsible for these two activities. These results define BM and DNA helicase as two mechanistically distinct activities of DNA translocating proteins and suggest that these two activities may serve different functions in DNA repair and recombination.
FIGURE 1.
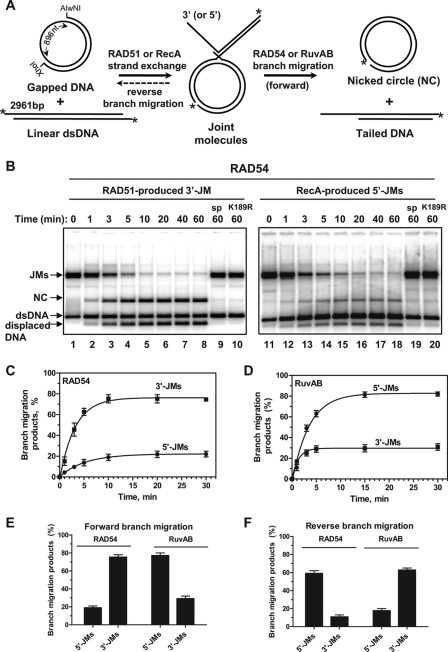
RAD54 and RuvAB promote BM of Holliday junctions in opposite directions. A, experimental design. JMs were produced in RAD51- or RecA-promoted DNA exchange between gapped DNA and pBSK(+) linear dsDNA. The asterisks indicate 32P label at either the 5′-end of XhoI-linearized or the 3′-end of AlwNI-linearized pBSK(+) DNA that was used for RAD51- or RecA-promoted DNA strand exchange, respectively. nt, nucleotide. B, the kinetics of four-stranded branch migration on 3′- and 5′-JMs (0.9 nm) by RAD54 (200 nm) (lanes 2–8 and 12–18) or RAD54K189R (200 nm) mutant protein (lanes 10 and 20) at 30 °C. Protein-independent branch migration (spontaneous (sp)) is shown after 60 min (lanes 9 and 19). DNA products were analyzed by electrophoresis on 1.5% agarose gels. C, the data from panel B are shown as a graph. D, the kinetics of four-stranded branch migration promoted by RuvAB (140 nm RuvA and 360 nm RuvB) at 37 °C (data shown in supplemental Fig. S1). E and F, the efficiencies of forward and reverse branch migration promoted by RAD54 and RuvAB on 3′-JMs and 5′-JMs shown as a graph. The error bars indicate the S.E.
EXPERIMENTAL PROCEDURES
Proteins and DNA
Human RAD51, RAD54, RAD54 K189R, BLM, BLM K695R, RPA, and RECQ1 were purified as described (19, 30–32). E. coli RecA was a generous gift of E. Golub (Yale University). E. coli RuvA and RuvB were from BioAcademia Inc. Shrimp alkaline phosphatase and E. coli exonuclease VII were from USB Corp. Calf thymus terminal deoxynucleotidyl transferase, restriction endonucleases, and T4 polynucleotide kinase were from New England Biolabs. Oligonucleotides were from Integrated DNA Technologies, Inc. (supplemental Table S1). Synthetic partial X-junction (PX junction) DNA substrate with four mismatches was prepared by annealing of equimolar amounts of 32P-labeled forked DNA intermediates (oligonucleotide 71/169*)3 with 3′-tailed DNAs (oligonucleotide 201/460), as described in Refs. 10 and 33. Gapped DNA was prepared by annealing the pBSK(+) XhoI-AlwNI fragment (2065 bp) to pBSK(+) ssDNA and purified as described (33). The pBSK(+)-derived plasmids containing 2–24-bp heterologous DNA sequences in the vicinity of the NgoMIV site were constructed by PCR-directed mutagenesis as specified by the manufacturer (Agilent Technologies). P34 and P35, P36 and P37, P38 and P39, P61 and P62, and P65 and P66 primer pairs were used to generate 2, 4, 8, 16, and 24 mismatches, respectively (supplemental Table S1). The pBSK(+)-derived plasmids containing 48- and 96-bp heterologous DNA sequences were constructed by molecular subcloning into the unique DraIII and NgoMIV sites. A 108-bp wild type DraIII-NgoMIV fragment was replaced with a 108-bp synthetic DraIII-NgoMIV fragment containing 48- or 96-bp mismatches. The synthetic DraIII-NgoMIV fragments were produced by annealing of the following complementary strands: S77 and S78 for 48 mismatches and S75 and S76 for 96 mismatches (supplemental Table S1). The identity of each DNA construct was verified by sequencing. All DNA concentrations are expressed in molecule units.
Branch Migration on Plasmid-length JM by RAD54, BLM, RECQ1, and RuvAB
Deproteinized 3′- and 5′-JMs were prepared and deproteinized as described in Ref. 33. Briefly, 3′-JMs were prepared in Rad51-mediated DNA strand exchange reaction (90 μl). Nucleoprotein filaments were formed by incubating RAD51 protein (5 μm) with pBSK(+) gapped DNA (20 μm, nucleotide) in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 275 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, 1 mm DTT, and 100 μg/ml BSA for 10 min at 37 °C. RPA (0.4 μm) was added to the nucleoprotein filaments followed by a 10-min incubation. DNA strand-exchange reaction was initiated by the addition of 32P-5-end-labeled XhoI-linearized pBSK(+) dsDNA (20 μm, nucleotide). The reaction was carried out for 3 h at 37 °C.
To prepare 5′-JMs (90 μl), RecA protein (4 μm) was incubated with pBSK(+) gapped DNA (20 μm, nucleotide) in buffer containing 33 mm Tris-HCl, pH 7.5, 3 mm ATP, 15 mm MgCl2, 2 mm DTT, 10 mm phosphocreatine, and 10 units/ml creatine phosphokinase for 5 min at 37 °C. DNA strand exchange was initiated by the addition of 32P-3′-end-labeled AlwNI-linearized pBSK(+) dsDNA (17.2 μm, nucleotide) and Ssb (ssDNA-binding protein) (0.33 μm) to the nucleoprotein filaments. The reaction was carried out for 10 min at 37 °C. The JMs were deproteinized by incubation with SDS (0.8%) and proteinase K (1.6 mg/ml) for 15 min at 37 °C, and then EDTA was added to 2 mm. The JMs were purified by passage twice through an S-400 HR spin column (GE Healthcare) equilibrated with 25 mm Tris acetate, pH 7.5, and then magnesium acetate in concentrations optimal for RAD54-, BLM-, RECQ1-, and RuvAB-mediated branch migration was added to inhibit spontaneous branch migration. The concentrations of the purified JMs were estimated by comparing their radioactivity with a known amount of 32P-labeled dsDNA after gel electrophoresis in 1.5% agarose gels.
RAD54-mediated BM of the JMs (0.9 nm, molecules) was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 100 μg/ml BSA, 3 mm magnesium acetate, 2 mm DTT, 2 mm ATP, 20 mm phosphocreatine, and 30 units/ml creatine phosphokinase for the indicated periods of time at 30 °C. The reaction was initiated by the addition of RAD54 (200 nm). When indicated, RPA (25–200 nm) was added to the reaction. The BLM- and RECQ1-promoted BM reactions were performed under the same conditions, except that the temperature was 37 °C, the BLM concentration was 20 nm, and magnesium acetate concentrations were 1 and 1.5 mm for BLM and RECQ1-reactions, respectively.
RuvAB-promoted BM of the JMs (0.5 nm, molecules) was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 100 μg/ml BSA, 10 mm magnesium acetate, 2 mm DTT, 2 mm ATP, 10 mm phosphocreatine, and 10 units/ml creatine phosphokinase at 37 °C. RuvA (140 nm) and RuvB (360 nm) were added to initiate the reaction, which was carried out for the indicated periods of time.
The DNA products of BM were deproteinized by the addition of 1.5% SDS and proteinase K (1.6 mg/ml) for 15 min at 37 °C, mixed with a 0.10 volume of loading buffer (70% glycerol, 0.1% bromphenol blue), and analyzed by electrophoresis in 1.5% agarose gels in TAE buffer (40 mm Tris acetate, pH 8.3, and 1 mm EDTA) at 5 V/cm for 3 h. The gels were dried on DEAE-81 paper (Whatman) and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Removal of Displaced ssDNA of Joint Molecules with Exonuclease VII
32P-labeled joint molecules (0.3 nm, molecules) generated by RAD51 were treated with exonuclease VII (0.1 units/μl) in buffer, containing 50 mm Tris acetate buffer, pH 7.5, and 100 μg/ml BSA for 10 min at 37 °C. The exonuclease was inactivated by incubation for 10 min at 65 °C. The joint molecules were cooled to 30 °C and supplemented with 3 mm magnesium acetate, 2 mm DTT, 2 mm ATP, 8 mm phosphocreatine, and 8 units/ml creatine phosphokinase. Branch migration was initiated by the addition of RAD54 (50 nm) and carried out for the indicated periods of time.
BM on Oligonucleotide-based Substrates
BLM-promoted branch migration was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine and creatine phosphokinase (10 units/ml), 32P-labeled PX junction oligonucleotide 71/169*/201/460 (32 nm, molecules), and the indicated concentrations of magnesium acetate. Branch migration was initiated by the addition of BLM (20 nm) and carried out for 30 min at 37 °C. To determine the optimal amount of BLM helicase required for helicase and branch migration activities, its concentration was varied in the presence of 2 nm (molecules) PX junction. The reactions were carried out for 20 min at 37 °C.
RAD54-promoted BM was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 2 mm DTT, BSA (100 μg/ml), 20 mm phosphocreatine and creatine phosphokinase (30 units/ml), 32P-labeled PX junction oligonucleotide 71/169*/201/460 (32 nm, molecules), and the indicated concentrations of magnesium acetate. The reactions were initiated by the addition of RAD54 (200 nm) and carried out for 30 min at 30 °C.
The BM reactions were stopped by the addition of 1.5% SDS and proteinase K (1 mg/ml), mixed with a 0.1 volume of loading buffer (70% glycerol, 0.1% bromphenol blue), and analyzed by electrophoresis in 9% polyacrylamide gels (17:1) in TBE buffer (89 mm Tris borate, pH 8.3, and 1 mm EDTA) at 135 V for 2 h. Gels were dried on DEAE-81 paper (Whatman), and the products were quantified using a Storm 840 PhosphorImager (GE Healthcare).
RESULTS
RAD54 and RuvAB Show Opposite Polarities of Branch Migration
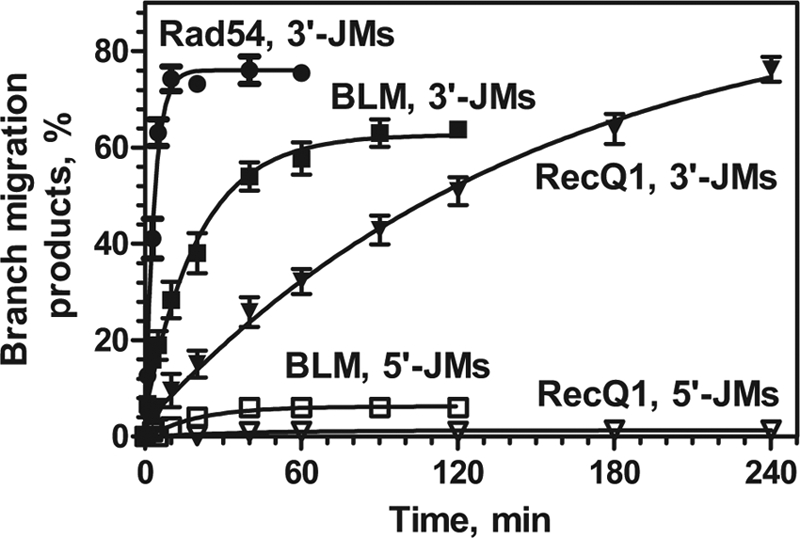
Here, we examined the polarity of BM promoted by human RAD54 using plasmid-length DNA JMs that contain displaced ssDNA strand with either 3′→5′ (3′-JMs) or 5′→3′ (5′-JMs) polarity (Fig. 1A). The 3′-JMs and 5′-JMs were generated using RAD51 or RecA, respectively (34). We found that RAD54 promoted BM on the 3′-JMs with an ∼4-fold higher efficiency than on the 5′-JMs (Fig. 1, B, C, and E). We then tested the polarity of BM by E. coli RuvAB, a canonical BM enzyme, and found that RuvAB promoted BM on the 5′-JMs with an ∼3-fold higher efficiency than on the 3′-JMs (Fig. 1, D and E; supplemental Fig. S1). Thus, RAD54 and RuvAB promote BM preferentially in the 3′→5′ and the 5′→3′ direction relative to the displaced ssDNA strand, respectively. The rate of BM promoted by RAD54 on the 3′-JMs was similar to that of RuvAB on the 5′-JMs (Fig. 1, C and D).
BM of JMs can proceed both in the forward direction (four-stranded reaction) and in the reverse direction (three-stranded reaction). The reverse BM leads to disruption of JMs into the original DNA substrates (32P-labeled linear dsDNA and unlabeled gapped DNA) (Fig. 1A). The extent of the reverse reaction was determined by the increase in the amount of 32P-labeled linear dsDNA (Fig. 1B). For RAD54, the reverse reaction was ∼6-fold more efficient on the 5′-JMs as compared with the 3′-JMs, consistent with the 3′→5′ polarity of BM relative to the displaced ssDNA strand (Fig. 1F). The opposite polarity bias was observed for RuvAB that promoted reverse BM on the 3′-JM with an ∼4-fold higher efficiency than on the 5′-JMs (Fig. 1F).
Because the presence of the ssDNA region significantly increases the affinity of RAD54 for branched DNA substrates (35), we suggested that the displaced ssDNA strand may play an important role for the overall efficiency of BM, e.g. by facilitating binding of BM proteins and assembly of active BM complexes. We tested this proposition for RAD54 by digesting the displaced ssDNA strand with exonuclease VII (Fig. 2). Indeed, we found that after exonuclease treatment, the efficiency of BM on both the 3′-JMs and 5′-JMs decreased significantly.
FIGURE 2.
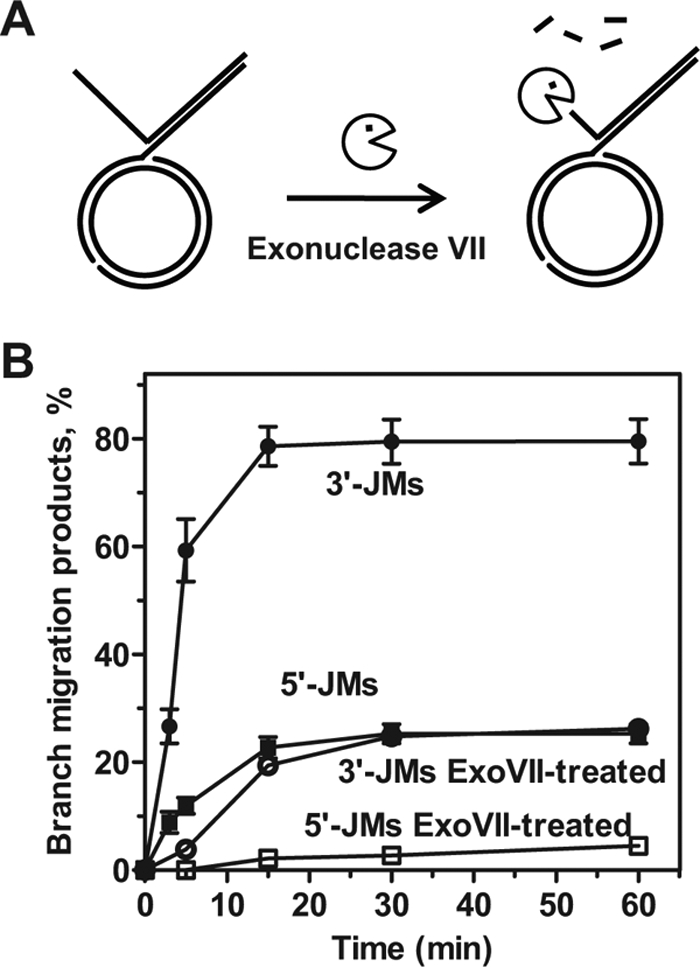
Displaced ssDNA on JMs stimulates BM promoted by RAD54. A, scheme of the ssDNA tail removal by an E. coli exonuclease VII. B, the kinetics of RAD54 (50 nm) branch migration JMs (0.3 nm) with 3′- and 5′-ssDNA tails that were either intact (closed circles and squares, respectively) or treated with exonuclease VII (ExoVII) (open circles and squares, respectively). The error bars indicate S.E.
Thus, RAD54 drives BM in the 3′→5′ direction relative to the displaced ssDNA strand. RuvAB promotes BM in the opposite, 5′→3′, direction.
BLM and RECQ1 Promote Branch Migration with 3′→5′ Polarity
We extended our analysis of BM polarity to two other known BM proteins, BLM and RECQ1. In preliminary experiments, the reaction conditions for each of these proteins were optimized (Table 1). Then, using the 3′- and 5′-JMs generated by RAD51 and RecA, respectively, we examined the direction of four-stranded BM promoted by BLM and RECQ1. Our data demonstrated that BLM and RECQ1 have an even stronger preference for BM in the 3′→5′ direction as compared with RAD54 as these proteins promote the 5′→3′ four-stranded BM with a very low efficiency (Fig. 3; supplemental Fig. S2, A and B). Similarly, both RECQ1 and BLM promoted three-stranded (reverse) BM preferentially in the 3′→5′ direction (supplemental Fig. S2, A and B). Thus, all three tested eukaryotic proteins, RAD54, BLM, and RECQ1, promote BM with the 3′→5′ polarity, the same polarity as RAD51, but the opposite polarity than of RuvAB and RecA (34, 36). The results also show that RAD54 promotes BM significantly faster than BLM or RECQ1; the initial rate of the forward BM in the 3′→5′ direction promoted by RAD54 was 4- and 20-fold faster than in the reactions promoted by BLM and RECQ1, respectively (Fig. 3).
TABLE 1.
Optimal conditions for four-strand branch migration on plasmid-length 3′-JMs
See the reaction conditions under “Experimental Procedures.”
| Protein | RAD54 | BLM | RECQ1 |
|---|---|---|---|
| Optimal Mg2+ acetate concentration in the presence of 2 mm ATP and ATP regeneration system | 3 mm | 1 mm | 0.5–2 mm |
| Optimal protein monomer/α-structure binding stoichiometry | 150 | 20 | 120 |
| Optimal temperature | 30 °C | 37 °C | 37 °C |
| Time to complete branch migration of 1 nm (molecules) α-structures | 10 min | 90 min | 240 min |
FIGURE 3.
BLM and RECQ1 promote BM of Holliday junctions in 3′→5′ direction. The kinetics of branch migration promoted by BLM (20 nm) and RECQ1 (200 nm) on 3′- and 5′-JMs (0.9 nm) at 37 °C (data shown in supplemental Fig. S2) and by RAD54 (200 nm) on 3′-JMs (0.9 nm) at 30 °C (data shown in Fig. 1B) are shown. The error bars indicate S.E.
RAD54 Promotes Branch Migration through Large Regions of DNA Heterology
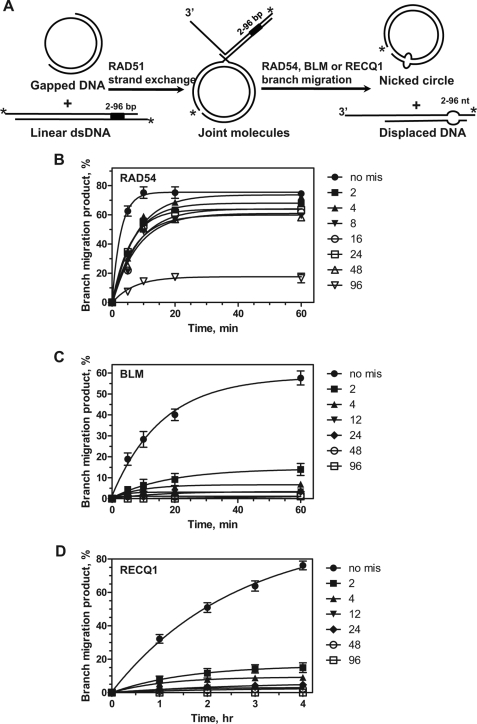
Although RAD54 is more efficient than BLM and RECQ1 at promoting BM on fully homologous JMs, one can suggest that the helicase activity of BLM or RECQ1 would make them more efficient than RAD54 when driving Holliday junctions through regions of sequence heterology. We constructed a series of plasmids in which 2-, 4-, 8-, 16-, 24-, 48-, and 96-bp DNA regions of pBSK(+) were replaced with heterologous sequences. The plasmids were linearized with XhoI endonuclease and used as substrates to produce 3′-JMs with pBSK(+) gapped DNA in RAD51-promoted DNA strand exchange (Fig. 4A). After deproteinization and purification, these JMs were used as DNA substrates to examine the efficiency of heterology bypass by RAD54, BLM, and RECQ. The DNA heterology was located at the distal part of the linear dsDNA so that BM would proceed through a 2.5-kb region of homology on JMs before encountering the heterologous sequences and then through an additional 0.3-kb region of homology before reaching the free dsDNA end. We found that Rad54 was able to efficiently bypass a 48-bp and even a 96-bp heterology region; the extents of the reactions were ∼80 and 20%, respectively, relative to the reaction with fully homologous JMs (Fig. 4B; Table 2; supplemental Fig. S3). Contrary to expectations, both BLM and RECQ1 helicases were far less efficient in heterology bypass than RAD54 (Fig. 4, C and D; Table 2; supplemental Fig. S3). Thus, BLM and RECQ1 BM were significantly inhibited by a heterology region as short as 2 bp (∼20% of the reaction efficiency on fully homologous JMs) and almost completely blocked by a 24-bp heterology region (3 and 6% of the reaction efficiency on fully homologous JMs, respectively). Thus, RAD54 that lacks canonical helicase activity showed significantly higher efficiency in heterology bypass during BM of Holliday junctions than BLM or RECQ1 helicases (Fig. 5).
FIGURE 4.
Kinetics of DNA heterology bypass by RAD54, BLM, and RecQ1. A, experimental design. RAD51-generated 3′-JMs with 2-, 4-, 8-, 16-, 24-, 48-, and 96-bp heterologies (denoted by the black box) were incubated with RAD54, BLM, or RECQ1. The asterisks indicate 32P-label. nt, nucleotide. B–D, the kinetics of branch migration by RAD54 (200 nm), BLM (20 nm), or RECQ1 (200 nm) on the deproteinized and purified JMs (0.9 nm) containing heterologies of the indicated length. The data are shown in supplemental Fig. S3. The error bars indicate S.E. no mis, no mismatches.
TABLE 2.
Efficiency of heterology bypass during four-strand branch migration on plasmid-length 3′-JMs
See the reaction conditions under “Experimental Procedures.”
| Heterology length, bp | Extent of reaction relative to homologous DNA control |
||
|---|---|---|---|
| RAD54 | BLM | RECQ1 | |
| % | |||
| 2 | 94 | 21 | 17 |
| 4 | 94 | 11 | 12 |
| 24 | 85 | 3 | 6 |
| 48 | 80 | 0 | 0 |
| 96 | 23 | 0 | 0 |
FIGURE 5.
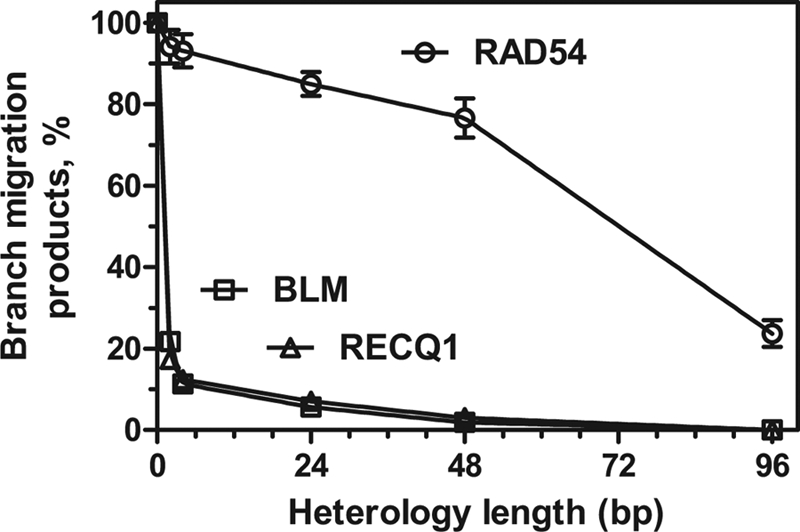
Efficiency of DNA heterology bypass by RAD54, BLM, and RECQ1. RAD51-generated 3′-JMs (0.9 nm) with DNA heterologies of the indicated lengths were incubated with RAD54 (200 nm) or BLM (20 nm) for 1 h or with RECQ1 (200 nm) for 4 h. The graph shows the extent of BM as a function of DNA heterology length (data shown in Table 2 and Fig. 4). 100% represents the yield of the branch migration products on fully homologous JMs (76, 60, and 77% for RAD54, BLM, and RECQ1, respectively). The error bars indicate S.E.
Effect of Protein Concentrations on Heterology Bypass
In studies on the BM activity of RuvAB, it was demonstrated that a probability of the heterology bypass is determined by both the length of the heterologous region and the lifetime of the stalled RuvAB complex (37). If the lifetime of the BM complex is shorter than the time required for the heterology bypass, then the reassembly of the complex would be required. Because the complex reassembly is generally stimulated by the increase in protein concentration, the heterology bypass is expected to show the same dependence. Alternatively, if the complex is intrinsically stable, the heterology bypass would not depend on the protein concentration. We examined the effect of the RAD54 concentration on the efficiency of BM on either fully homologous JMs (0.3 nm, molecules) or JMs that contained a 96-bp heterologous DNA region (0.3 nm, molecules) (Fig. 6A; supplemental Fig. S4A). We found that for both DNA substrates, the maximal extent of BM was achieved at the same RAD54 concentration (45 nm). We next examined the effect of protein concentration on the BLM-mediated heterology bypass. We used 3′-JMs with a 2-bp heterology region, on which the efficiency of BM was similar to that of RAD54 on 3′-JMs with a 96-bp heterology region. We found that more than a 2-fold higher BLM concentration was required to achieve the maximal extent of BM on these JMs than on fully homologous JMs, 14 and 6 nm (molecules), respectively (Fig. 6B and supplemental Fig. S4B).
FIGURE 6.
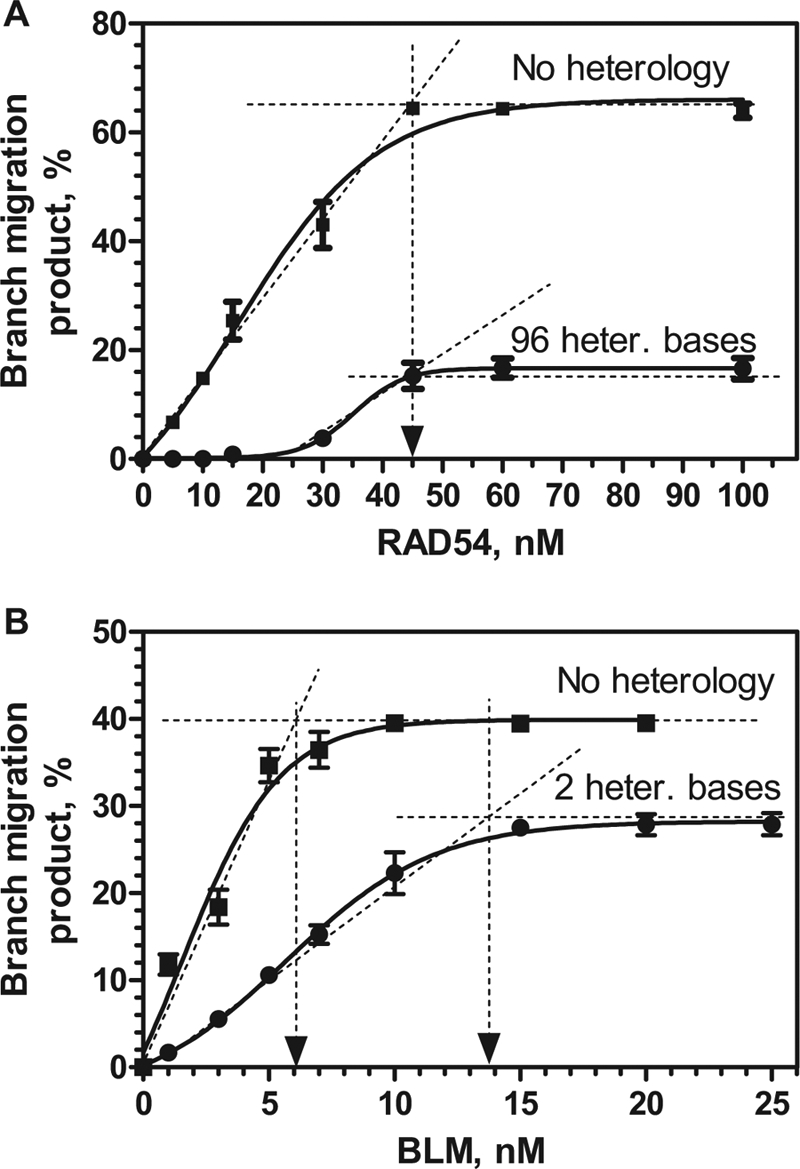
Effect of protein concentrations on heterology bypass by RAD54 and BLM. A and B, RAD54 and BLM in indicated concentrations were added to the 3′-JMs (0.3 nm), either fully homologous or containing heterology of the indicated length. The error bars indicate S.E. heter. bases, heterologous bases.
These results suggest formation of intrinsically stable RAD54 complexes with JMs, which may contribute to an efficient heterology bypass during BM of Holliday junctions. In contrast, BLM complexes with JMs appear to be less stable and may undergo a disassembly during bypass of even short regions of heterology.
Effect of RPA on Translocation through Sequence Heterology
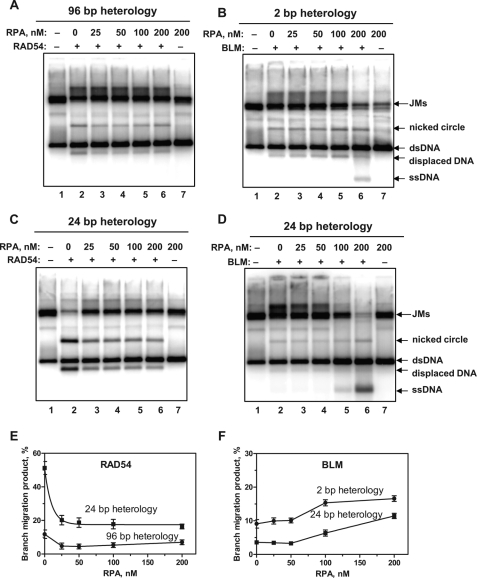
Previous works demonstrated that ssDNA-binding proteins, either prokaryotic Ssb protein or eukaryotic RPA, stimulate DNA unwinding activities of the RecQ family helicases, including BLM, either by trapping the individual ssDNA strands upon their separation or through direct interaction with a helicase (38–42). Here, we examined whether RPA exhibits a stimulatory effect on BM through the heterology regions promoted by RAD54 and BLM. We used 3′-JMs with the heterology regions of 96 and 2 bp, for RAD54 (200 nm) and BLM (20 nm), respectively, on which the efficiency of heterology bypass by these proteins was approximately the same (Table 2). We also used 3′-JMs with a heterology region of 24 bp for both proteins to exclude a possible effect of the heterology length on interaction with RPA. At the JM concentration used in these experiments (0.8 nm, molecules), ∼40 nm RPA was sufficient to saturate the ssDNA tail of JMs and the region of ssDNA bubble formed between noncomplementary DNA strands during the heterology bypass (Fig. 4A). In the range of RPA concentrations from 25 to 200 nm, we observe an ∼2-fold inhibition of either a 24-bp or a 96-bp heterology bypass by RAD54 (Fig. 7, A, C, and E). In the BLM-promoted reactions, RPA caused slight stimulation of heterology bypass at RPA concentration higher than 100 nm (Fig. 7, B, D, and F).
FIGURE 7.
Effect of human RPA on heterology bypass promoted by BLM and RAD54. A–D, various amounts of human RPA were added to the 3′-JMs (0.9 nm) containing heterologous DNA regions of the indicated lengths, prior to the addition of RAD54 (200 nm) (A and C) or BLM (20 nm) (B and D). The RAD54-promoted BM with JMs containing 24- and 96-bp heterology regions were carried out for 10 and 20 min, respectively, at 30 °C; the BLM-promoted BM was carried out for 1 h at 37 °C. E and F, the data from panels A–D are shown as graphs. The error bars indicate S.E.
As expected, we observed strong stimulation of the BLM helicase activity by RPA. Thus, in the presence of 100–200 nm RPA, an ssDNA band appeared in the agarose gel, which resulted from DNA strand separation of the JM duplex region of 2065 bp (Fig. 7B, lane 6; Fig. 7D, lanes 5 and 6). In contrast, RPA caused only weak, if any, stimulation of the BM activity of BLM or inhibited the BM activity of RAD54.
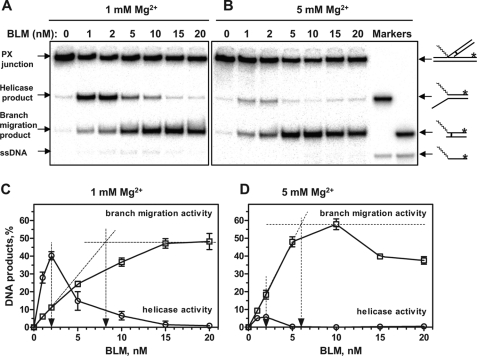
Branch Migration and Helicase Activities of BLM Require Different Reaction Conditions and Occur at Different Protein:DNA Ratios
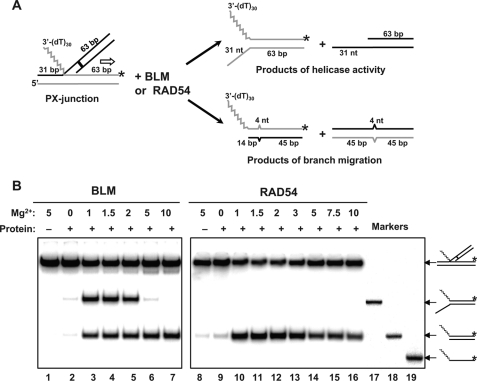
Here, we wanted to explore further the relationship between the helicase and the BM activities by examining the effect of different reaction conditions on these two activities of BLM. To detect both the helicase and the BM activity of BLM, we used the synthetic PX junction as a substrate (Fig. 8A). To block spontaneous BM, four mismatches were incorporated in one duplex DNA branch of the PX junction. Using this PX junction, the products of the helicase and BM activities could be easily separated by electrophoresis on a 9% polyacrylamide gel (Fig. 8B). First, we determined the optimal Mg2+ concentrations for both BLM helicase and BM activity. Using the PX junction, we found that the BLM helicase activity reached the maximum at 1.0–1.5 mm Mg2+ (Fig. 8B, lanes 3 and 4). The BLM BM activity showed different requirements for Mg2+, reaching the maximum at 5–10 mm Mg2+ (Fig. 8B, lanes 6 and 7). As expected, in the case of RAD54, only the product of BM, and not that of the DNA helicase activity, was observed (Fig. 8B, lanes 9–16); the RAD54 BM activity reached its maximum at 1.5–2.0 mm Mg2+.
FIGURE 8.
Effect of Mg2+ concentration on BM activities of BLM and RAD54 and helicase activity of BLM. A, the experimental scheme. The zigzagged line denotes the poly(dT)30 region of the PX junction (oligonucleotide 71/169*/201/460). The 4-bp heterology region (denoted by the solid box) was designed to block protein-independent BM of the PX junction. The direction of BM is shown by the black arrow. The asterisks indicate 32P-label. nt, nucleotide. B, BLM (20 nm) and RAD54 (200 nm) were added to PX junction (32 nm) in the presence of the indicated magnesium acetate concentrations. The reactions were carried out for 30 min at 37 and 30 °C for BLM and RAD54, respectively. The products of the helicase (forked DNA oligonucleotide 71/169*) and branch migration (3′-tailed DNA oligonucleotide 169*/460) reactions were separated in a 9% polyacrylamide gel (17:1).
Using the PX junction (2 nm, molecules), we then measured the stoichiometries of BLM required for helicase and BM activities at 1 and 5 mm Mg2+, the optimal concentrations for each activity, respectively (Fig. 9). We found that at both Mg2+ concentrations, the helicase activity reached the maximum at the stoichiometric ratio of one BLM monomer per one PX junction molecule. However, the extent of the helicase reaction at 1 mm Mg2+ was ∼10-fold higher than at 5 mm. The BM activity required higher BLM concentrations, four and three BLM monomers per one PX junction molecule at 1 and 5 mm Mg2+, respectively. Previously, we reported that the RAD54 BM activity required ∼10–12 protein monomers per one PX junction molecule (35).
FIGURE 9.
DNA helicase and BM activities require different BLM stoichiometries. A and B, BLM in the indicated concentrations was added to the PX junction (2 nm) (shown in Fig. 7A) in the presence of 1 (A) or 5 mm (B) magnesium acetate. The reactions were carried out for 20 min at 37 °C. C and D, the data from panels A and B are shown as graphs. The error bars indicate S.E.
Thus, the BLM BM and helicase activities showed different requirements in Mg2+ and different protein-to-DNA ratios. These results indicate that BM and DNA helicase represent two mechanistically distinct activities of BLM.
DISCUSSION
Previously, we found that RAD54, an Snf2 protein, possesses DNA BM activity (10, 24, 35, 43). Here, we further characterize the BM activity of RAD54, focusing on the polarity of BM and on DNA heterology bypass. Also, we compared the properties of RAD54-promoted BM with the reactions promoted by BLM and RECQ1.
The polarity of BM proteins has been a controversial issue. Previously, it was shown that RuvAB promotes BM of Holliday junctions on 5′-JMs made by RecA in both the forward and the reverse direction, i.e. without apparent polarity (44). However, the forward and reverse reactions on JM substrates are mechanistically different (Fig. 1A). Although the forward reaction proceeds through four-stranded BM, the reverse reaction involves three-stranded BM that normally occurs at a faster rate (45). Here, to define the polarity of BM, we employed a pair of JM substrates produced by RAD51 and by RecA, which have opposite polarities of the displaced ssDNA strand, 3′→5′ and 5′→3′, respectively (34). This pair of JMs enabled us to compare mechanistically identical reactions, either four-stranded or three-stranded, occurring in opposite directions relative to the displaced ssDNA strand. The results show that RAD54 drives both the three-stranded and the four-stranded BM in the 3′→5′ direction with an ∼4–6-fold higher preference than in the 5′→3′ direction. BLM and RECQ1 also show even stronger preference for the 3′→5′ direction of BM, whereas RuvAB shows the opposite bias, promoting BM in the 5′→3′ direction with 3–4-fold greater efficiently than in the 3′→5′ direction. Thus, for all three proteins tested, BLM, RECQ1, and RuvAB, which possess both DNA BM and helicase activities, the polarity of BM coincides with the polarity of DNA helicase activity, implying a fundamental dependence of both these activities on the underlying polarity of DNA translocation by these enzymes. Interestingly, RAD54 and RuvAB show the same polarity preferences of BM as their cognate DNA recombinases, RAD51 and RecA (34), with which they interact physically and functionally, as in the case of RAD54 and RAD51, or at least functionally, as in case of RuvAB and RecA.
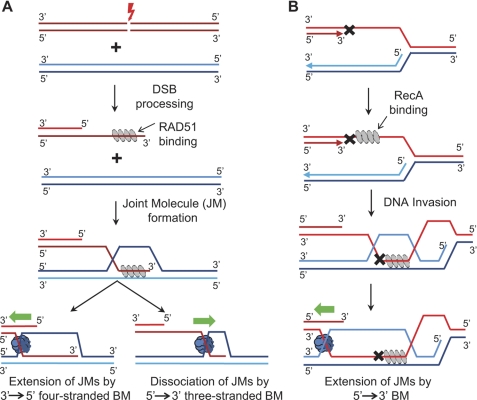
In eukaryotes, homologous recombination is initiated by DNA double-strand breaks (Fig. 10A). The DNA ends at the sites of breaks are then processed by exonucleases to generate ssDNA tails. The early step of homologous recombination involves invasion of ssDNA into homologous duplex DNA promoted by RAD51. This invasion results in formation of JMs, also known as D-loops. The JMs provide both the template and the primer for DNA synthesis, which leads to extension of the invaded ssDNA strand. Current results showed that the displaced ssDNA strand of JMs plays an important role in defining the polarity of BM of all tested BM proteins, RAD54, BLM, RECQ1, and RuvAB. This finding is consistent with our previous observation that RAD54 shows the highest affinity for the partial Holliday junction (PX junction) that structurally resembles JMs containing a displaced ssDNA strand (10, 35). The structural basis of the displaced ssDNA strand recognition by BM proteins remains to be elucidated. However, it is likely that the displaced ssDNA strand is responsible for establishing critical contacts with BM proteins. Structural studies of other superfamily 1 and 2 helicase proteins have shown that the conserved helicase domain makes extensive contacts with only one of the strands in the DNA duplex (46). These contacts define the DNA strand that the BM complex would track during DNA translocation.
FIGURE 10.
Effect of BM polarity on procession of recombination intermediates. A, in the RAD51 pathway, a DNA double-strand break (DSB) is processed by 5′ end resection to produce DNA with a 3′ ssDNA tail. RAD51 (gray ovals) forms a filament with an exposed 3′ ssDNA tail and promotes DNA invasion to form JMs. After DNA repair synthesis, JMs are then either extended or dissociated through BM by specialized enzymes (blue shape). B, in the RecA pathway, a lesion (X shape) in the leading strand of DNA replication results in a stalled replication fork, exposing a region of ssDNA. The RecA filament (gray ovals) forms on the exposed ssDNA and promotes invasion with the lagging strand template, forming JMs. Then RuvAB (blue shape) will extend JMs through BM in the 5′→3′.
The polarity of BM may have important biological implications. At the late step of homologous recombination, the JMs undergo BM, which causes either expansion or dissociation of JMs depending on the BM directionality. The expansion of JMs may facilitate capture of the second end of broken DNA, resulting in formation of double Holliday junctions for which resolution by specialized endonucleases leads to crossing over that is essential for accurate chromosome segregation during meiosis, but which can cause loss of heterozygosity in somatic cells (47). In contrast, dissociation of JMs leads to non-crossover recombinants, which are common in somatic cells. Thus, a specific bias in BM polarity of specialized BM proteins may contribute to a choice of the recombination pathways that lead to formation of either crossover or non-crossover recombinants (Fig. 10A). Our results demonstrate that all tested eukaryotic proteins including RAD54, BLM, and RECQ1 show the same polarity as RAD51. In the case when RAD51 promotes invasion of the 3′-ssDNA, this polarity of BM proteins would lead to preferential expansion of JMs. However, in the case when the 5′-ssDNA is used by RAD51 to form JMs, the 3′→5′ polarity of the BM proteins would help to dissociate these nonproductive recombination intermediates. Indeed, we showed recently that RECQ1 is especially effective in disrupting JMs formed as a result of 5′-ssDNA invasion promoted by RAD51 (14).
It also important to note that expansion of JMs involves the four-stranded BM, whereas their dissociation occurs through the three-stranded reaction (Fig. 10A). BM requires a transition of the DNA junction from the closed to the open conformation. This transition occurs more readily in the partial Holliday junction, an intermediate of the three-stranded reaction, than in the full Holliday junction in the four-stranded reaction (45). As a consequence, the three-stranded BM proceeds with a significantly higher efficiency than the four-stranded BM in both spontaneous and protein-driven reactions (14, 45). Therefore, the polarity bias of BM proteins may help to overcome this intrinsic difference in the reaction efficiencies and promote expansion of the JMs. At the same time, RAD54, BLM, and RECQ1 show different degrees of the 5′→3′ BM activity. These data agree with our previous observation that RAD54 can disrupt JMs generated by RAD51, which required three-stranded BM in the 5′→3′ direction relative to the displaced ssDNA strand (19). A similar role was earlier proposed for BLM (17, 21, 32).
The difference in the BM polarities between RecA and RAD51 is perplexing, but it is clear that the cognate BM proteins evolved to possess the same polarity as their recombinase. Previously, we suggested that RecA is primarily involved in the repair of the ssDNA gaps, e.g. formed in the leading DNA strand at replication forks stalled at DNA lesions (34). In this case, the 5′→3′ polarity of BM promoted by RecA and RuvAB is required for the extension of the DNA repair intermediates (Fig. 10B). Specialized BM proteins with the 5′→3′ polarity, which remains to be identified, may play a similar role in eukaryotes.
Our current results show that the use of the plasmid-length DNA substrates is essential for an accurate measurement of the polarity of BM proteins. Previously, no polarity bias of RAD54 or BLM was observed using short synthetic DNA substrates (10, 14). We suggest that an assembly of active BM complexes is less efficient on short DNA substrates and may represent the rate-limiting step that masks the polarity preference of seen in BM.
Despite the lack of canonical helicase activity, RAD54 shows high efficiency in driving Holliday junctions through stretches of DNA heterology. This RAD54 ability may help to complete homologous recombination initiated between DNA molecules containing heterologous sequences or damaged DNA bases. We found that RAD54 can bypass heterologous regions longer than 96 bp. Among BM proteins characterized so far, only RuvAB was documented to bypass longer DNA heterology regions (44). In contrast, BLM and RECQ1 helicases are much less efficient as a 2-bp DNA heterology caused an ∼5-fold decrease in the reaction yield, comparable with the effect of a 96-bp heterology on the RAD54-driven reaction. In comparison, BLM and RecQ1 possess helicase activities that disrupt at least 91-bp-long dsDNA (42, 48). These data indicate that the DNA helicase activity of BM proteins does not play a significant role during BM through the regions of heterology. Moreover, for BLM, we found that the BM and helicase activities require different Mg2+ concentrations and were differently affected by RPA that significantly stimulated only the helicase activity. The two activities also showed different dependence on protein:DNA stoichiometry; the DNA helicase activity required one BLM molecule per one PX junction, whereas the stoichiometry of 3–4 BLM molecules per one PX junction was optimal for BM activity. These results parallel previous observations that two oligomeric states of RECQ1 were responsible for different activities; monomers/dimers and pentamers/hexamers were required for DNA helicase activity and DNA strand annealing activity, respectively (49). Our current data suggest that a monomeric form of BLM is responsible for the DNA helicase and that an oligomeric form is required for the BM activity. Mg2+ may stimulate BM and suppress DNA unwinding by promoting BLM oligomerization.
Previously, it was shown that spontaneous four-stranded BM is strongly inhibited by regions of heterology as short as a single DNA base (45). The inhibition of BM was largely attributed to a thermodynamic barrier arising from the formation of unpaired bases in heteroduplex DNAs. Thus, Holliday junction that migrates through a single base heterology to create two mismatched duplexes very rapidly reverses direction to restore base-pairing (50). The BM protein complex that translocates unidirectionally along DNA would act as a reflective barrier that blocks this reverse BM, allowing bypass of the heterology. In accord with this model, the efficiency of the heterology bypass depends on the strength of this block. Thus, the processivity of the BM complex, not the helicase activity plays a critical role during the heterology bypass; the proteins that translocate via the distributive mechanism could not efficiently block the reversal of BM. We found that the heterology bypass by RAD54, in contrast to that of BLM, was not stimulated by an increase in protein concentration, indicating formation of stable RAD54-DNA complexes. The results of single-molecule studies also show high processivity of RAD54 translocation on DNA (51).
Overall, the current results show that RAD54 is a major BM protein in eukaryotes; RAD54 showed the highest rate of BM among tested proteins and was capable of driving BM through extensive regions of DNA heterology. The results also show that the displacement strand of JMs defines the polarity of BM of all examined proteins; RAD54 drives BM in the 3′→5′ direction, the same direction as that of BLM and RECQ1 and the opposite direction of RuvAB.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA100839 (to A. V. M.) and F31 AG033484 (to M. J. R.) This work was also supported by Leukemia and Lymphoma Society Scholar Award 1054-09 (to A. V. M.).

This article contains supplemental Table S1 and Figs. S1–S4.
The asterisks indicate the oligonucleotides that were 32P-labeled.
- BM
- branch migration
- JM
- joint molecule
- PX junction
- partial X-junction
- RPA
- replication protein A.
REFERENCES
- 1. Wyman C., Kanaar R. (2006) DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383 [DOI] [PubMed] [Google Scholar]
- 2. Moynahan M. E., Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krogh B. O., Symington L. S. (2004) Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 4. Pâques F., Haber J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kowalczykowski S. C. (2008) Structural biology: snapshots of DNA repair. Nature 453, 463–466 [DOI] [PubMed] [Google Scholar]
- 6. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 7. Mazin A. V., Mazina O. M., Bugreev D. V., Rossi M. J. (2010) Rad54, the motor of homologous recombination. DNA Repair 9, 286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y., West S. C. (2004) Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 5, 937–944 [DOI] [PubMed] [Google Scholar]
- 9. Rudolph C. J., Upton A. L., Briggs G. S., Lloyd R. G. (2010) Is RecG a general guardian of the bacterial genome? DNA Repair 9, 210–223 [DOI] [PubMed] [Google Scholar]
- 10. Bugreev D. V., Mazina O. M., Mazin A. V. (2006) Rad54 protein promotes branch migration of Holliday junctions. Nature 442, 590–593 [DOI] [PubMed] [Google Scholar]
- 11. Unk I., Hajdú I., Blastyák A., Haracska L. (2010) Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 9, 257–267 [DOI] [PubMed] [Google Scholar]
- 12. Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D. (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. U.S.A. 97, 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Constantinou A., Tarsounas M., Karow J. K., Brosh R. M., Bohr V. A., Hickson I. D., West S. C. (2000) Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bugreev D. V., Brosh R. M., Jr., Mazin A. V. (2008) RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 283, 20231–20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P. (2006) Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34, 5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A. (2008) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29, 141–148 [DOI] [PubMed] [Google Scholar]
- 17. van Brabant A. J., Ye T., Sanz M., German J. L., III, Ellis N. A., Holloman W. K. (2000) Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39, 14617–14625 [DOI] [PubMed] [Google Scholar]
- 18. Bachrati C. Z., Borts R. H., Hickson I. D. (2006) Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 34, 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bugreev D. V., Hanaoka F., Mazin A. V. (2007) Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 14, 746–753 [DOI] [PubMed] [Google Scholar]
- 20. Orren D. K., Theodore S., Machwe A. (2002) The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry 41, 13483–13488 [DOI] [PubMed] [Google Scholar]
- 21. Adams M. D., McVey M., Sekelsky J. J. (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265–267 [DOI] [PubMed] [Google Scholar]
- 22. Raynard S., Bussen W., Sung P. (2006) A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J. Biol. Chem. 281, 13861–13864 [DOI] [PubMed] [Google Scholar]
- 23. Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. (2006) BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. U.S.A. 103, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bugreev D. V., Rossi M. J., Mazin A. V. (2011) Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 39, 2153–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Machwe A., Karale R., Xu X., Liu Y., Orren D. K. (2011) The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) Holliday junctions to functional replication forks. Biochemistry 50, 6774–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ralf C., Hickson I. D., Wu L. (2006) The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281, 22839–22846 [DOI] [PubMed] [Google Scholar]
- 27. Machwe A., Xiao L., Groden J., Orren D. K. (2006) The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45, 13939–13946 [DOI] [PubMed] [Google Scholar]
- 28. Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., Haracska L. (2007) Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McGlynn P., Lloyd R. G., Marians K. J. (2001) Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. U.S.A. 98, 8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazina O. M., Mazin A. V. (2004) Human Rad54 protein stimulates DNA strand-exchange activity of hRad51 protein in the presence of Ca2+. J. Biol. Chem. 279, 52042–52051 [DOI] [PubMed] [Google Scholar]
- 31. Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 32. Bugreev D. V., Yu X., Egelman E. H., Mazin A. V. (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossi M. J., Mazina O. M., Bugreev D. V., Mazin A. V. (2010) Analyzing the branch migration activities of eukaryotic proteins. Methods 51, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossi M. J., Mazina O. M., Bugreev D. V., Mazin A. V. (2011) The RecA/RAD51 protein drives migration of Holliday junctions via polymerization on DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 6432–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazina O. M., Rossi M. J., Thomaä N. H., Mazin A. V. (2007) Interactions of human rad54 protein with branched DNA molecules. J. Biol. Chem. 282, 21068–21080 [DOI] [PubMed] [Google Scholar]
- 36. Murayama Y., Kurokawa Y., Mayanagi K., Iwasaki H. (2008) Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature 451, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 37. Dennis C., Fedorov A., Käs E., Salomé L., Grigoriev M. (2004) RuvAB-directed branch migration of individual Holliday junctions is impeded by sequence heterology. EMBO J. 23, 2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Umezu K., Nakayama H. (1993) RecQ DNA helicase of Escherichia coli: characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J. Mol. Biol. 230, 1145–1150 [DOI] [PubMed] [Google Scholar]
- 39. Brosh R. M., Jr., Orren D. K., Nehlin J. O., Ravn P. H., Kenny M. K., Machwe A., Bohr V. A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 274, 18341–18350 [DOI] [PubMed] [Google Scholar]
- 40. Brosh R. M., Jr., Li J. L., Kenny M. K., Karow J. K., Cooper M. P., Kureekattil R. P., Hickson I. D., Bohr V. A. (2000) Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275, 23500–23508 [DOI] [PubMed] [Google Scholar]
- 41. Harmon F. G., Kowalczykowski S. C. (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem. 276, 232–243 [DOI] [PubMed] [Google Scholar]
- 42. Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rossi M. J., Mazin A. V. (2008) Rad51 protein stimulates the branch migration activity of Rad54 protein. J. Biol. Chem. 283, 24698–24706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams D. E., West S. C. (1996) Bypass of DNA heterologies during RuvAB-mediated three- and four-strand branch migration. J. Mol. Biol. 263, 582–596 [DOI] [PubMed] [Google Scholar]
- 45. Panyutin I. G., Hsieh P. (1993) Formation of a single base mismatch impedes spontaneous DNA branch migration. J. Mol. Biol. 230, 413–424 [DOI] [PubMed] [Google Scholar]
- 46. Singleton M. R., Wigley D. B. (2002) Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol 184, 1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo G., Santoro I. M., McDaniel L. D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R. A., Bradley A. (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26, 424–429 [DOI] [PubMed] [Google Scholar]
- 48. Karow J. K., Chakraverty R. K., Hickson I. D. (1997) The Bloom's syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 272, 30611–30614 [DOI] [PubMed] [Google Scholar]
- 49. Muzzolini L., Beuron F., Patwardhan A., Popuri V., Cui S., Niccolini B., Rappas M., Freemont P. S., Vindigni A. (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 5, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biswas I., Yamamoto A., Hsieh P. (1998) Branch migration through DNA sequence heterology. J. Mol. Biol. 279, 795–806 [DOI] [PubMed] [Google Scholar]
- 51. Amitani I., Baskin R. J., Kowalczykowski S. C. (2006) Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23, 143–148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.