Background: Kv7.2 and Kv7.3 coassemble to form a heteromeric K+ channel that generates the M-current.
Results: Atomic force microscopy (AFM) imaging of Kv7.2/Kv7.3 heteromers in complex with subunit-specific antibodies reveals the channel architecture.
Conclusion: The Kv7.2/Kv7.3 heteromer assembles with a random subunit arrangement.
Significance: Unusually, heteromeric Kv7 channels have a variable architecture.
Keywords: Atomic Force Microscopy, Ion Channels, Membrane Proteins, Molecular Imaging, Potassium Channels, Protein Structure
Abstract
Voltage-gated K+ channels composed of Kv7.2 and Kv7.3 are the predominant contributors to the M-current, which plays a key role in controlling neuronal activity. Various lines of evidence have indicated that Kv7.2 and Kv7.3 form a heteromeric channel. However, the subunit stoichiometry and arrangement within this putative heteromer are so far unknown. Here, we have addressed this question using atomic force microscopy imaging of complexes between isolated Kv7.2/Kv7.3 channels and antibodies to epitope tags on the two subunits, Myc on Kv7.2 and HA on Kv7.3. Initially, tsA 201 cells were transiently transfected with equal amounts of cDNA for the two subunits. The heteromer was isolated through binding of either tag to immunoaffinity beads and then decorated with antibodies to the other tag. In both cases, the distribution of angles between pairs of bound antibodies had two peaks, at around 90° and around 180°, and in both cases the 90° peak was about double the size of the 180° peak. These results indicate that the Kv7.2/Kv7.3 heteromer generated by cells expressing approximately equal amounts of the two subunits assembles as a tetramer with a predominantly 2:2 subunit stoichiometry and with a random subunit arrangement. When the DNA ratio for the two subunits was varied, copurification experiments indicated that the subunit stoichiometry was variable and not fixed at 2:2. Hence, there are no constraints on either the subunit stoichiometry or the subunit arrangement.
Introduction
Voltage-gated Kv7 (otherwise known as KCNQ) K+ channels, of which there are five subtypes (Kv7.1–7.5), are of crucial importance in excitable tissues (1, 2). In the nervous system, Kv7.2 and Kv7.3 are the main components of the M-current, a voltage-dependent, non-inactivating K+ current that plays a central role in controlling the activity of both peripheral and central neurons (3–5). Consistent with this key role, spontaneous mutations in Kv7.2 and Kv7.3 cause human neonatal epilepsy (6, 7).
Coexpression of Kv7.2 and Kv7.3 leads to greater surface expression (8) and the generation of much larger currents (4, 9–12) than when either subunit is expressed alone. Additionally, channels produced following Kv7.2/Kv7.3 coexpression have ionic permeation and conductive properties distinct from those produced by individual expression of Kv7.2 or Kv7.3 (13). These observations suggest that Kv7.2 and Kv7.3 assemble to form a heteromeric channel, which has been assumed to be a tetramer, by analogy with other K+ channels (14). Experiments using fluorescence resonance energy transfer have pointed to an intimate interaction between Kv7.2 and Kv7.3 (15), and analysis of channels built from Kv7.3-Kv7.2 concatemers has shown that heteromers are functional (16), supporting the idea that the two subunits form a heteromeric channel. At present, however, the subunit stoichiometry and arrangement within heteromers generated after expression of individual Kv7.2 and Kv7.3 subunits is unclear.
We have developed a method, on the basis of AFM4 imaging, for determining the arrangement of subunits within multimeric proteins (17–19). The method involves engineering specific epitope tags onto each subunit and expressing the proteins in a suitable cell line (e.g. tsA 201). Crude membrane fractions from the transfected cells are solubilized in detergent, and the proteins are isolated by affinity chromatography. The isolated proteins are then imaged by AFM, and their mean molecular volume is compared with the molecular volume expected for the protein on the basis of its molecular mass. In this way, assembled multimers can be distinguished from unassembled subunits. The proteins are incubated with antibodies to the tags, and the resulting multimer-antibody complexes are imaged by AFM. Multimers with two bound antibodies are identified, and the angles between the antibodies are measured. A frequency distribution of these angles then reveals the structure of the multimer. In this study, we have used this method to determine the architecture of the Kv7.2/Kv7.3 heteromer. We show that Kv7.2 and Kv7.3 form a heterotetramer with a random subunit arrangement.
EXPERIMENTAL PROCEDURES
Cell Culture
tsA 201 cells (a subclone of human embryonic kidney 293 cells stably expressing the SV40 large T-antigen) were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in an atmosphere of 5% CO2/air.
Channel Constructs
DNA encoding human Kv7.2 bearing a Myc epitope tag at its N terminus was subcloned into the pSRC5 vector (20). Human Kv7.3 DNA, bearing either a double hemagglutinin (2xHA) or a FLAG/HA epitope tag at its N terminus, was subcloned into the same vector. Initial experiments demonstrated that the 2xHA tag could be decorated simultaneously by two anti-HA antibodies (data not shown). To circumvent this complication, we generated a new Kv7.3 construct that had a single HA tag for experiments involving decoration with anti-HA antibodies. (The FLAG tag was not used in these experiments.) It has been shown previously that addition of N-terminal tags does not affect the functional properties of either Kv7.2 or Kv7.3 (12). DNA encoding the rat P2X2 receptor subunit bearing a His6 epitope tag at its N terminus was subcloned into the pcDNA3.1 vector.
Transient Transfection of tsA 201 Cells
Transient transfections of tsA 201 cells with DNA were carried out using the calcium phosphate precipitation method. A total of 250 μg of DNA (usually 125 μg for each Kv7 construct) was used to transfect cells in 5 × 162 cm2 culture flasks. After transfection, cells were incubated for 48 h at 37 °C to allow protein expression. Protein expression and intracellular localization were checked using immunofluorescence analysis of small-scale cultures. Cells were fixed, permeabilized, and incubated with appropriate primary antibodies (rabbit polyclonal anti-Myc (Abcam), mouse monoclonal anti-HA (Covance), and mouse monoclonal anti-V5 (Invitrogen) as a negative control), followed by either Cy3- or fluorescein isothiocyanate-conjugated goat secondary antibodies (Sigma). Cells were imaged by confocal laser scanning microscopy.
In Situ Proximity Ligation Assay
Cells growing on lysine- and collagen-coated glass coverslips in 3.5-cm diameter culture wells were cotransfected with 1 μg each of DNA encoding Myc-Kv7.2 and 2xHA-Kv7.3. All transfections also included pEGFP (0.5 μg of DNA) to identify transfected cells. In a control experiment, cells were cotransfected with DNA encoding Myc-Kv7.2 and His6-P2X2. Cells were incubated for 24 h at 37 °C to allow protein expression. Cells were fixed, permeabilized, and incubated with primary antibodies (rabbit polyclonal anti-Myc plus either mouse monoclonal anti-HA or mouse monoclonal anti-His6 (Invitrogen), all diluted 1:50) for a further 1 h. Cells were washed with phosphate-buffered saline and then incubated for 1 h with anti-mouse (+) and anti-rabbit (-) proximity ligation secondary antibodies, diluted 1:5, at 37 °C. These antibodies were obtained as part of a kit from Olink Bioscience that also included ligation and amplification buffers. Cells were washed with phosphate-buffered saline and incubated in T4 DNA ligase diluted 1:40 in ligation buffer for 30 min at 37 °C. Cells were washed with TBS-T (150 mm NaCl, 0.05% Tween 20, 10 mm Tris (pH 7.4)), and DNA amplification was performed by incubation with 1:80 DNA polymerase in amplification buffer for 100 min at 37 °C. This buffer also contained a fluorescent detection probe (excitation wavelength 554 nm, emission wavelength 576 nm). Cells were washed with 1× TBS (100 mm NaCl, 200 mm Tris (pH 7.5)) and then 0.01× TBS. Coverslips were mounted on slides and imaged by confocal laser scanning microscopy.
Solubilization and Purification of Epitope-tagged Proteins
Transfected cells were solubilized in 1% Triton X-100 for 1 h before centrifugation at 78,000 × g to remove insoluble material. To isolate Kv7, the solubilized extract was incubated with either anti-Myc- or anti-HA-agarose beads (Sigma) for 3 h. The beads were washed extensively, and bound proteins were eluted with Myc or HA peptide (100 μg/ml). Samples were analyzed by SDS-polyacrylamide gel electrophoresis, and proteins were detected both by silver staining and by immunoblotting using mouse monoclonal antibodies against Myc (Invitrogen) or HA.
AFM Imaging of Kv7 or Kv7-Antibody Complexes
Isolated channels were imaged either alone or following overnight incubation at 4 °C with a 1:2 molar ratio (∼0.2 nm channel protein concentration) of anti-epitope tag monoclonal antibody (Invitrogen). Proteins were diluted to a final concentration of 0.04 nm, and 45 μl of the sample was allowed to adsorb to freshly cleaved, poly-L-lysine-coated mica disks. After a 10-min incubation, the sample was washed with Biotechnology Performance Certified-grade water (Sigma) and dried under nitrogen. Imaging was performed with a Veeco Digital Instruments multimode atomic force microscope controlled by a Nanoscope IIIa controller. Samples were imaged in air using tapping mode. The silicon cantilevers used had a drive frequency of ∼300 kHz and a specified spring constant of 40 N/m (Olympus). The applied imaging force was kept as low as possible (As/A0 ∼0.85).
The molecular volumes of the protein particles were determined from particle dimensions on the basis of AFM images. After adsorption of the channels onto the mica support, the particles adopt the shape of a spherical cap. As described previously (21), the heights and radii were measured from multiple cross-sections of the same particle, and the molecular volume was calculated using the following equation:
where h is the particle height and r is the radius. Molecular volume on the basis of molecular mass was calculated using the equation
where M0 is the molecular mass, N0 is Avogadro's number, V1 and V2 are the partial specific volumes of particle (0.74 cm3/g) and water (1 cm3/g), respectively, and d is the extent of protein hydration (taken as 0.4 g water/g protein).
Selection of Binding Events
Several criteria were used to identify Kv7-antibody complexes. Heights and radii were measured for all particles, and the particle volumes were calculated. Bound particles needed to have between half and twice the predicted volume of 285 nm3 to be accepted as antibodies. A cross-section was drawn through the junction between the antibody and the adjacent Kv7 channel, and the height of the lowest point between antibody and channel was measured. This height needed to be greater than 0.3 nm for the antibody to be considered bound. Any particle was rejected if its length was greater than twice its width. To be considered a double binding event, all particles and both binding events needed to meet all of the above criteria.
Statistical Analysis
Histograms were drawn with bin widths chosen according to Scott's equation:
where σ is an estimate of the standard deviation, and n is the sample size (22). Where Gaussian curves were fitted to the data, the number of curves was chosen so as to maximize the r2 value while giving significantly different means using Welch's t test for unequal sample sizes and unequal variances (23).
RESULTS
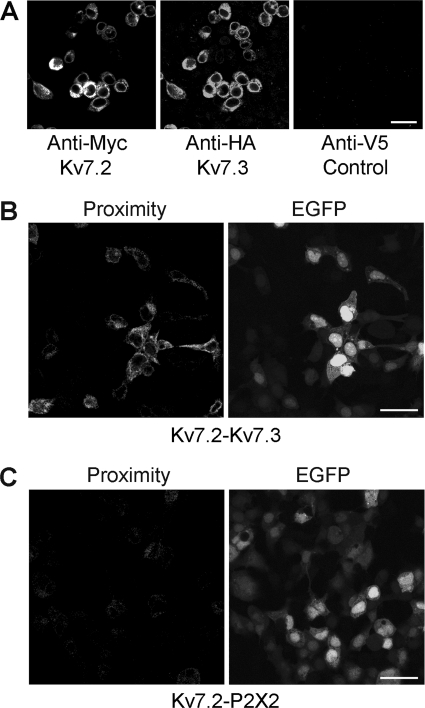
Initially, tsA 201 cells were transiently cotransfected with DNA encoding Myc-tagged Kv7.2, and 2xHA-tagged Kv7.3. Protein expression and localization was confirmed by immunofluorescence using appropriate anti-tag antibodies. As shown in Fig. 1A, cells gave positive immunofluorescence signals with anti-Myc and anti-HA antibodies, indicating the presence of the two Kv7 subunits. In contrast, an anti-V5 antibody gave only a background signal. The anti-Myc and anti-HA signals in doubly labeled cell populations overlapped extensively, indicating that the majority of transfected cells expressed both subunits. The reticular staining patterns suggest that proteins were localized predominantly in the endoplasmic reticulum.
FIGURE 1.
Expression and interaction between Myc-Kv7.2 and 2xHA-Kv7.3. A, immunofluorescence detection of epitope-tagged proteins in transiently transfected tsA 201 cells. Cotransfected cells were fixed, permeabilized, and incubated with appropriate anti-tag antibodies, as indicated, followed by fluorophore-conjugated secondary antibodies. Cells were imaged by confocal laser scanning microscopy. B, in situ proximity ligation assay for Kv7.2/Kv7.3 interaction. Cells were cotransfected with DNA encoding Myc-Kv7.2 and 2xHA-Kv7.3. Transfections also included pEGFP to identify transfected cells. Cells were fixed, permeabilized, and incubated with primary antibodies (rabbit polyclonal anti-Myc and mouse monoclonal anti-HA) followed by anti-mouse (+) and anti-rabbit (-) proximity ligation secondary antibodies. The proximity ligation assay was then carried out, and treated cells were imaged by confocal laser scanning microscopy. The left panel shows the proximity signal, and the right panel shows the EGFP signal. C, control experiment in which cells were transfected with Myc-Kv7.2 plus His6-P2X2 receptor. In this case, primary antibodies were rabbit polyclonal anti-Myc and mouse monoclonal anti-His6. Transfected cells are indicated by the EGFP signal (right panel). There is a considerable reduction in the proximity ligation signal (left panel). Scale bars = 20 μm.
To establish whether Kv7.2 and Kv7.3 coexpressed in the cells interact with each other, in situ proximity ligation assays were carried out (24). The assay uses two secondary antibodies, each bearing a short DNA strand. When the secondary antibodies are brought into close proximity (< 40 nm) by binding to their relevant primary antibodies, the DNA strands hybridize with an additional circle-forming oligodeoxynucleotide. Ligation then creates a complete circularized oligodeoxynucleotide, and rolling circle amplification increases the amount of circular DNA several hundred-fold. The DNA is then visualized using a fluorescent probe. Cells were cotransfected with Myc-Kv7.2 and 2xHA-Kv7.3, and pEGFP (enhanced green fluorescent protein) was used to reveal the transfected cells. Primary antibodies used to tag the subunits were rabbit anti-Myc (Kv7.2) and mouse anti-HA (Kv7.3). As shown in Fig. 1B, strong proximity signals were given by Myc-Kv7.2 + 2xHA-Kv7.3. In contrast, when cells expressed Myc-Kv7.2 + His6-P2X2 (plus pEGFP), only a very weak signal was generated using anti-Myc and anti-His6 antibodies (Fig. 1C). This result indicates that Kv7.2 and Kv7.3 come into close proximity within the cotransfected cells.
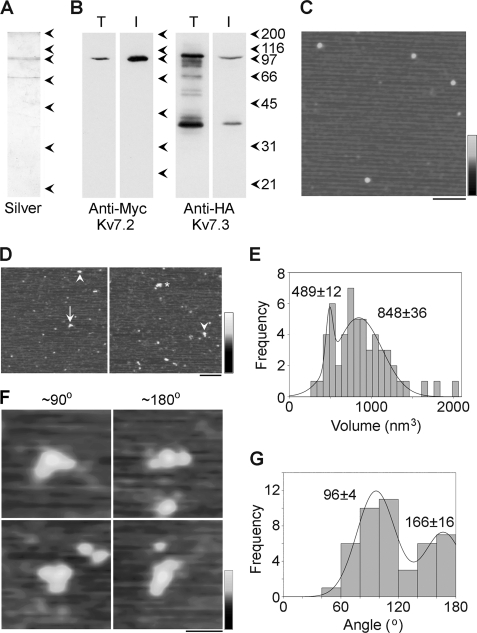
Cotransfected cells were solubilized in Triton X-100 detergent (1% w/v), and proteins were isolated through the binding of the 2xHA tag on Kv7.3 to anti-HA-agarose beads. A silver-stained gel of the isolated protein (Fig. 2A) shows a doublet at about 100 kDa, the expected size of the Kv7 subunit (12). The gel also shows an additional band at about 70 kDa that is likely a degradation product. Both the crude solubilized fraction and the isolated protein were subjected to immunoblotting using either anti-Myc (Fig. 2B, left panel) or anti-HA (Fig. 2B, right panel) antibodies. Both antibodies detected bands at about 100 kDa in both fractions, indicating that both Myc-Kv7.2 and 2xHA-Kv7.3 were successfully captured via the 2xHA tag on Kv7.3. This coisolation provides further evidence for an intimate interaction between the two subunits.
FIGURE 2.
Decoration of Myc-Kv7.2/2xHA-Kv7.3 channels with anti-Myc antibodies. A and B, samples of total cell extract (T) and protein isolated by affinity chromatography (I) were analyzed by SDS-polyacrylamide gel electrophoresis followed by either silver staining (A) or immunoblotting (B) using mouse monoclonal anti-Myc (Kv7.2, left panel) or anti-HA (Kv7.3, right panel) antibodies. Immunoreactive bands were visualized using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody followed by enhanced chemiluminescence. The arrowheads indicate molecular mass markers (kDa). C, low-magnification AFM image of a sample of isolated Kv7.2/Kv7.3. Scale bar = 200 nm; shade-height scale = 0–4 nm. D, low-magnification AFM images of a sample of Kv7.2/Kv7.3 that had been incubated with anti-Myc antibodies. The arrowheads indicate singly decorated particles, the arrow indicates a doubly decorated particle, and the asterisk indicates a triply decorated particle. Scale bar = 200 nm, shade-height scale = 0–4 nm. E, frequency distribution of particles that were multiply decorated by anti-Myc antibodies. The curve indicates the fitted Gaussian functions. The peaks of the distributions are indicated. F, gallery of zoomed images of Kv7.2/Kv7.3 particles that are decorated by two antibodies at angles of around 90° (left panels) and 180° (right panels). Scale bar = 50 nm, shade-height scale = 0–3 nm. G, frequency distribution of angles between pairs of bound antibodies. The curve indicates the fitted Gaussian functions. The peaks of the distribution are indicated.
Proteins isolated from cells expressing Myc-Kv7.2 and 2xHA-Kv7.3 were imaged by AFM. A representative low-magnification AFM image is shown in Fig. 2C. Several particles of approximately equal sizes can be seen. The isolated protein was then incubated with anti-Myc antibody and again imaged by AFM. In the representative images shown in Fig. 2D, large particles can be seen, some of which have been decorated by either one (arrowheads), two (arrow) or three (asterisk) smaller particles. The small particles had a molecular volume of around 240 nm3, close to the expected volume of 285 nm3 for an immunoglobulin G molecule, with a molecular mass of 150 kDa. Hence, these particles represent anti-Myc antibodies bound to the Myc epitopes on Kv7.2 channel.
Multiple binding events were identified, and the volumes of the central decorated particles were determined using Equation 1. A frequency distribution of these volumes had two peaks: a small peak at 489 ± 12 (n = 13) nm3 and a larger peak at 848 ± 36 (n = 42) nm3 (Fig. 2E). According to Equation 2, a Kv7 subunit with a molecular mass of 100 kDa should have a molecular volume of 190 nm3, so a Kv7 tetramer should have a volume of 760 nm3, close to the higher-volume peak. The two volume peaks, therefore, likely correspond to tetramers and dimers that may represent precursors or may have been produced by dissociation of the tetramers.
To quantitate decoration of the tetramers by anti-Myc antibodies, we counted particles that had volumes between 600 nm3 and 1500 nm3. (The lower limit was set to exclude dimers.) Of 1494 particles, 1101 (73.7%) were undecorated, 351 (23.5%) were singly decorated by anti-Myc antibodies, 41 (2.7%) were doubly decorated, and 1 (< 0.1%) was triply decorated. No quadruply decorated particles were seen. Corresponding percentages for a control anti-V5 antibody were 91.4% undecorated, 8.4% singly decorated, and 0.2% doubly decorated (n = 856 particles). Hence the vast majority of the decoration events seen were specific.
We identified Kv7 channels that had been decorated by two antibodies and measured the angles between the bound antibodies. This was done in each case by joining the highest point on the central particle (the Kv7 channel) to the highest points on the peripheral particles (the antibodies) by lines and then determining the angle between the two lines. Representative doubly decorated large particles are shown in Fig. 2F, and a frequency distribution of the angles obtained is shown in G. The angle distribution has two peaks: a large peak at 96 ± 4° (n = 30) and a smaller peak at 166 ± 16° (n = 14). The ratio of the numbers of particles in the two peaks is 2.1:1. This angle profile, with two peaks at around 90° and 180°, indicates that the Kv7 heteromer is indeed a tetramer. In contrast, a trimer would give a single angle peak at 120° (17), whereas a pentamer would give equally sized peaks at 72° and 144° (18).
The fact that we are isolating the channel via the 2xHA tag on Kv7.3 and decorating the Myc tag on Kv7.2 ensures that we are only decorating heteromers. However, the results shown in Fig. 2G do not allow us to distinguish between two possible subunit arrangements: the one shown in supplemental Fig. S1A, where the subunit stoichiometry is 2:2 but the subunit arrangement is random, and the one in supplemental Fig. S1B, where the subunit stoichiometry is 3:1, Kv7.2:Kv7.3. In both cases, the predicted ratio of 90°/180° angles is 2:1. Hence, the 2.1:1 ratio observed is consistent with either scenario.
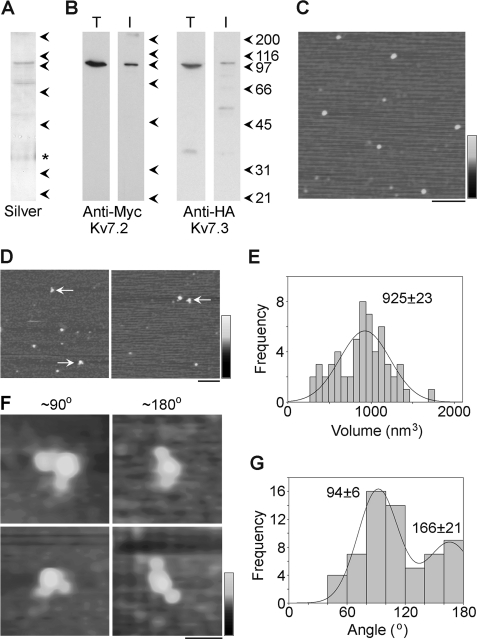
To distinguish between the two possible assembly states, we carried out the reverse experiment, isolating the channel via the Myc tag and decorating the HA tag. Cells were cotransfected with DNA encoding HA-Kv7.3 along with Myc-Kv7.2. Proteins were isolated through the binding of the Myc tag on Kv7.2 to anti-Myc-agarose beads. A silver stain of the isolated protein shows a doublet of bands at around 100 kDa (Fig. 3A), along with two contaminating bands. (Note that the asterisk indicates a band that appeared even in blank lanes that is likely a contaminant in the gel and not in the protein sample.) Immunoblot analyses using anti-Myc and anti-HA antibodies show decoration of bands at around 100 kDa (Fig. 3B). Low-magnification AFM images of isolated proteins showed a relatively homogenous distribution of particles (Fig. 3C), and when proteins were incubated with anti-HA antibodies, some particles became decorated by two antibodies (arrows in D).
FIGURE 3.
Decoration of Myc-Kv7.2/HA-Kv7.3 channels with anti-HA antibodies. A and B, samples of total cell extract (T) and protein isolated by affinity chromatography (I) were analyzed by SDS-polyacrylamide gel electrophoresis followed by either silver staining (A) or immunoblotting (B) using mouse monoclonal anti-Myc (Kv7.2, left panel) or anti-HA (Kv7.3, right panel) antibodies. Immunoreactive bands were visualized using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody followed by enhanced chemiluminescence. (Note that the asterisk in A indicates a band that appeared even in blank lanes that is likely a contaminant in the gel and not in the protein sample.) The arrowheads indicate molecular mass markers (kDa). C, low-magnification AFM image of a sample of isolated Kv7.2/Kv7.3. Scale bar = 200 nm, shade-height scale = 0–4 nm. D, low-magnification AFM images of a sample of Kv7.2/Kv7.3 that had been incubated with anti-HA antibodies. The arrows indicate doubly decorated particles. Scale bar = 200 nm, shade-height scale = 0–4 nm. E, frequency distribution of particles that were multiply decorated by anti-HA antibodies. The curve indicates the fitted Gaussian function. The peak of the distribution is indicated. F, gallery of zoomed images of Kv7.2/Kv7.3 particles that are decorated by two antibodies at angles of around 90° (left panels) and 180° (right panels). Scale bar = 50 nm, shade-height scale = 0–3 nm. G, frequency distribution of angles between pairs of bound antibodies. The curve indicates the fitted Gaussian functions. The peaks of the distribution are indicated.
To quantitate decoration of the tetramers by anti-HA antibodies, we counted particles that had volumes between 350 nm3 and 1500 nm3. Of 2862 large particles imaged, 2312 (80.8%) were undecorated, 492 (7.2%) were singly decorated by anti-HA antibodies, 56 (2.0%) were doubly decorated, and 2 (< 0.1%) were triply decorated. No quadruply decorated particles were seen. Corresponding percentages for a control anti-V5 antibody were 91.6% undecorated, 8.1% singly decorated, and 0.3% doubly decorated (n = 1228 particles). Hence, as for decoration by anti-Myc, the vast majority of the decoration events seen were specific.
A frequency distribution of the molecular volumes of doubly decorated particles (Fig. 3E) shows a single peak at 925 ± 23 (n = 58) nm3, close to the expected volume of a tetramer (760 nm3). Zoomed images (Fig. 3F) show angles between pairs of bound antibodies of around 90° (left panels) and around 180° (right panels), and the angle distribution (G) had two peaks at 94 ± 6° (n = 44) and 166 ± 21° (n = 18). The ratio of the numbers of particles within the two peaks is 2.4:1. The fact that a very similar angle distribution is observed irrespective of which antibody is used to isolate or decorate the heteromer strongly argues for the situation illustrated in supplemental Fig. S1A, where the channel generated by cotransfection with equal amounts of DNA for the two subunits has a predominant subunit stoichiometry of 2:2 and a random subunit arrangement.
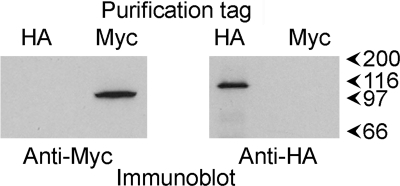
Two additional explanations of the apparent variability in subunit arrangement indicated by the results should be considered. One is the possibility that Kv7 tetramers might be self-associating during purification so that a Myc-Kv7.2 homotetramer, for instance, might be copurified on anti-HA-agarose beads with a Myc-Kv7.2/2xHA-Kv7.3 heterotetramer or, indeed, a 2xHA-Kv7.3 homotetramer. This possibility seems unlikely because there was no sign of dimers of Kv7 tetramers in the AFM images (unlike the situation for P2X receptors in a previous study, for example, where receptor dimers were relatively common (25)). Another possible scenario involves dissociation of Kv7 tetramers during the isolation procedure, followed by reassembly into a novel tetramer that does not exist in vivo. We carried out a further experiment to check these possibilities. Two sets of cells were singly transfected with DNA encoding either Myc-Kv7.2 or HA-Kv7.3. The two populations of transfected cells were mixed at the start of the purification procedure, and the combined detergent extract of the cells was later split into two batches for purification using either anti-Myc or anti-HA immunobeads. The two eluates were then immunoblotted using anti-Myc and anti-HA antibodies. If either of the above scenarios was occurring, then anti-Myc beads should have captured HA-Kv7.3 along with Myc-Kv7.2, and anti-HA beads should have captured Myc-Kv7.2 along with HA-Kv7.3. In fact, anti-Myc beads captured only Myc-Kv7.2, and anti-HA beads captured only HA-Kv7.3 (Fig. 4). We therefore conclude that the Kv7.2/Kv7.3 tetramer does indeed assemble with a variable subunit arrangement.
FIGURE 4.
Interaction of mixed populations of Kv7.2 and Kv7.3 homotetramers with anti-Myc and anti-HA immunobeads. Two sets of tsA 201 cells were singly transfected with DNA encoding either Myc-Kv7.2 or HA-Kv7.3. The two populations of transfected cells were mixed at the start of the purification procedure, and the combined detergent extract of the cells was later split into two batches for purification using either anti-Myc or anti-HA immunobeads. The two eluates were then immunoblotted using anti-Myc and anti-HA antibodies. Immunoreactive bands were visualized using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody followed by enhanced chemiluminescence. The arrowheads indicate molecular mass markers (kDa).
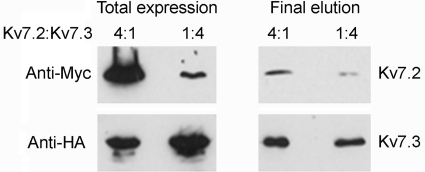
The fact that we observed triply decorated channels (as shown in Fig. 2D) must mean that some channels had a 3:1, Kv7.2:Kv7.3 stoichiometry. However, when equal amounts of DNA were used for the two subunits, these triple decoration events were very rare, indicating that the predominant stoichiometry is 2:2. The possibility does exist, however, that a greater proportion of heteromers of 3:1 stoichiometry can be generated when the expression levels of the two proteins are different. To test this possibility, we transfected cells with DNA encoding Kv7.2 and Kv7.3 in ratios of 4:1 and 1:4, which resulted in the expected differences in expression of Myc-Kv7.2 and HA-Kv7.3 in the transfected cells (Fig. 5, left panels). Protein was isolated from the cells by sequential capture on anti-HA and anti-Myc immunobeads. This procedure will necessarily result in the isolation of heteromers. The protein concentrations in the eluted samples from the two cotransfections were adjusted to produce bands of approximately equal intensity on an anti-HA (Kv7.3) immunoblot. If the Kv7.2:Kv7.3 stoichiometry was fixed at 2:2, one would expect the two anti-Myc (Kv7.2) bands also to be of equal intensity. In fact, the band in the 4:1 Kv7.2:Kv7.3 sample is significantly more intense than the band from the 1:4 sample (Fig. 5, right panels). This result indicates that the Kv7.2:Kv7.3 subunit stoichiometry is not fixed at 2:2 but rather varies in response to changes in relative expression levels.
FIGURE 5.
Effect of varying relative subunit expression levels on the subunit composition of the Kv7.2/Kv7.3 heteromer. tsA 201 cells were cotransfected with DNA encoding Kv7.2 and Kv7.3 in ratios of 4:1 and 1:4. Protein was isolated from the cells by sequential capture on anti-HA and anti-Myc immunobeads, resulting in the isolation of heteromers. The left panels show total cell extracts. The right panels show the final eluates from the immunobeads. The protein concentrations in the two eluates were adjusted to produce bands of approximately equal intensity on the anti-HA (Kv7.3) immunoblot. Immunoreactive bands were visualized using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody followed by enhanced chemiluminescence.
DISCUSSION
Our results indicate that the Kv7.2/Kv7.3 heteromer assembles as a tetramer with a random subunit arrangement. A variable subunit stoichiometry and arrangement was suggested several years ago on the basis of an electrophysiological analysis of channels expressed in tsA 201 cells (26). In a more recent study, Kv7.3-Kv7.2 concatemers were found to produce channel characteristics almost identical with those formed after coexpression of individual subunits (16). In this case, presumably, the channel would have had an alternating subunit arrangement. However, the properties of channels with a constrained adjacent subunit arrangement were not tested.
The variability in subunit arrangement seen with the Kv7.2/Kv7.3 channel is unusual. More often we have seen an invariant subunit arrangement in both ionotropic receptors, such as the 5-HT3 receptor (18) and the GABAA receptor (19), and ion channels such as the transient receptor potential (TRP) channels. Specifically, TRPP2 forms heteromers with both TRPC1 (27) and TRPV4 (28), which have an alternating subunit arrangement.
As mentioned above, the majority of the Kv7 proteins were localized in the endoplasmic reticulum, likely because of the overexpression occurring after transient transfection. We must, therefore, bear in mind the possibility that a quality control mechanism operates within the cells to restrict access to the cell surface to heteromers of a particular subunit arrangement. It has recently been shown that Kv7.3 homomers are retained in the endoplasmic reticulum (12). Whether such a mechanism could discriminate between different subunit arrangements within a heteromer remains to be seen.
Functional analysis of the behavior of point mutants and chimeras of Kv7.2 and Kv7.3 has identified the cytoplasmic C terminus as a major region involved in oligomerization of Kv7 subunits (11, 29–31). All Kv7 subunits contain two coiled coil domains, known as the A-domain head and tail, and the tail in particular is believed to be important for intersubunit recognition, channel assembly, and functional expression. Recently, the A-domain tail has been shown to crystallize as a left-handed coiled coil (32). This structure suggests a possible explanation for the inability of Kv7.3 to efficiently form homotetramers. Specifically, it would be problematic to pack four phenylalanine residues (position 622) within the core of the tetramer, and in addition, interhelical hydrogen bond and salt bridge interactions required to stabilize the tetramer would also be disrupted in Kv7.3. Replacement of the phenylalanine by leucine, as in Kv7.2, and improvement of the interhelical interactions would help to stabilize the tetramer.
Intriguingly, evidence has recently been presented that the pore region in Kv7.3, rather than the A domain, plays a key role in controlling channel assembly and surface expression (12). The Kv7.3 subunit is unusual in having an alanine residue in place of threonine in the pore vestibule (position 315). Although wild-type Kv7.3 is retained in the endoplasmic reticulum and requires coexpression of Kv7.2 to exit this compartment, an A315T Kv7.3 mutant is efficiently trafficked to the plasma membrane by itself and is functionally active and, therefore, fully assembled.
Similar to Kv7, the C-terminal domain of TRPP2 again contains two coiled coil domains, but here mutational analysis has shown that second of these domains functions as a heterodimerization domain essential for polycystin 1 binding but not for self-oligomerization of TRPP2 (33). In addition, there is evidence for the involvement of the cytoplasmic N terminus in TRPP2 channel assembly (34) and an intramembrane disulfide bond (35). Whether the distinct functions of the cytoplasmic domains in Kv7 and TRPP2 helps to explain the different levels of stringency in channel assembly in the two cases remains to be seen.
In conclusion, the variable subunit stoichiometry and arrangement demonstrated here raises the possibility that the overall behavior of the Kv7 channel in neurons can be manipulated through endogenous changes in subunit expression levels, potentially adding an extra layer of subtlety to the manner in which neurons control their excitability.
Supplementary Material
This work was supported by the Jean Shanks Foundation and the James Baird Fund of the University of Cambridge (to A. P. S.); the Department of Education, Universities, and Investigation of the Basque Government (to J. C. G. P.); and the Biotechnology and Biological Sciences Research Council (to R. D. M. L. and J. M. E.).

This article contains supplemental Fig. S1.
- AFM
- atomic force microscopy
- EGFP
- enhanced green fluorescent protein
- TRP
- transient receptor potential.
REFERENCES
- 1. Jentsch T. J. (2000) Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Rev. Neurosci. 1, 21–30 [DOI] [PubMed] [Google Scholar]
- 2. Brown D. A., Passmore G. M. (2009) Neural KCNQ (Kv7) channels. Br. J. Pharmacol. 156, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown D. A., Adams P. R. (1980) Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature 283, 673–676 [DOI] [PubMed] [Google Scholar]
- 4. Wang H. S., Pan Z., Shi W., Brown B. S., Wymore R. S., Cohen I. S., Dixon J. E., McKinnon D. (1998) KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 5. Shah M. M., Mistry M., Marsh S. J., Brown D. A., Delmas P. (2002) Molecular correlates of the M-current in cultured rat hippocampal neurons. J. Physiol. 544, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schroeder B. C., Kubisch C., Stein V., Jentsch T. (1998) Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature 396, 687–690 [DOI] [PubMed] [Google Scholar]
- 7. Maljevic S., Wuttke T. V., Lerche H. (2008) Nervous system Kv7 disorders: breakdown of a subthreshold brake. J. Physiol. 586, 1791–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwake M., Pusch M., Kharkovets T., Jentsch T. J. (2000) Surface expression and single channel properties of KCNQ2/KCNQ3, M-type K+ channels involved in epilepsy. J. Biol. Chem. 275, 13343–13348 [DOI] [PubMed] [Google Scholar]
- 9. Yang W. P., Levesque P. C., Little W. A., Conder M. L., Ramakrishnan P., Neubauer M. G., Blanar M. A. (1998) Functional expression of two KvLQT1-related potassium channels responsible for an inherited idiopathic epilepsy. J. Biol. Chem. 273, 19419–19423 [DOI] [PubMed] [Google Scholar]
- 10. Selyanko A. A., Hadley J. K., Brown D. A. (2001) Properties of single M-type KCNQ2/KCNQ3 potassium channels expressed in mammalian cells. J. Physiol. 534, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maljevic S., Lerche C., Seebohm G., Alekov A. K., Busch A. E., Lerche H. (2003) C-terminal interaction of KCNQ2 and KCNQ3 K+ channels. J. Physiol. 548, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gómez-Posada J. C., Etxeberría A., Roura-Ferrer M., Areso P., Masin M., Murrell-Lagnado R. D., Villarroel A. (2010) A pore residue of the KCNQ3 potassium M-channel subunit controls surface expression. J. Neurosci. 30, 9316–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prole D. L., Marrion N. V. (2004) Ionic permeation and conduction properties of neuronal KCNQ2/KCNQ3 potassium channels. Biophys. J. 86, 1454–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacKinnon R. (1991) Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature 350, 232–235 [DOI] [PubMed] [Google Scholar]
- 15. Bal M., Zhang J., Zaika O., Hernandez C. C., Shapiro M. S. (2008) Homomeric and heteromeric assembly of KCNQ (Kv7) K+ channels assayed by total internal reflection fluorescence/fluorescence resonance energy transfer and patch clamp analysis. J. Biol. Chem. 283, 30668–30676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hadley J. K., Passmore G. M., Tatulian L., Al-Qatari M., Ye F., Wickenden A. D., Brown DA. (2003) Stoichiometry of expressed KCNQ2/KCNQ3 potassium channels and subunit composition of native ganglionic M channels deduced from block by tetraethylammonium. J. Neurosci. 23, 5012–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrera N. P., Ormond S. J., Henderson R. M., Murrell-Lagnado R. D., Edwardson J. M. (2005) Atomic force microscopy imaging demonstrates that P2X2 receptors are trimers but that P2X6 receptor subunits do not oligomerize. J. Biol. Chem. 280, 10759–10765 [DOI] [PubMed] [Google Scholar]
- 18. Barrera N. P., Herbert P., Henderson R. M., Martin I. L., Edwardson J. M. (2005) Atomic force microscopy reveals the stoichiometry and subunit arrangement of 5-HT3 receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 12595–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barrera N. P., Betts J., You H., Henderson R. M., Martin I. L., Dunn S. M., Edwardson J. M. (2008) Atomic force microscopy reveals the stoichiometry and subunit arrangement of the α4β3δ GABAA receptor. Mol. Pharmacol. 73, 960–967 [DOI] [PubMed] [Google Scholar]
- 20. Etxeberria A., Aivar P., Rodriguez-Alfaro J. A., Alaimo A., Villacé P., Gómez-Posada J. C., Areso P., Villarroel A. (2008) Calmodulin regulates the trafficking of KCNQ2 potassium channels. FASEB J. 22, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 21. Schneider S. W., Lärmer J., Henderson R. M., Oberleithner H. (1998) Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflueger's Arch. Eur. J. Physiol. 435, 362–367 [DOI] [PubMed] [Google Scholar]
- 22. Scott D. W. (1979) On optimal and data-based histograms. Biometrika 66, 605–610 [Google Scholar]
- 23. Welch B. L. (1947) The generalisation of student's problems when several different population variances are involved. Biometrika 34, 28–35 [DOI] [PubMed] [Google Scholar]
- 24. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 25. Antonio L. S., Stewart A. P., Xu X. J., Varanda W. A., Murrell-Lagnado R. D., Edwardson J. M. (2011) P2X4 receptors interact with both P2X2 and P2X7 receptors in the form of homotrimers. Br. J. Pharmacol. 163, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shapiro M. S., Roche J. P., Kaftan E. J., Cruzblanca H., Mackie K., Hille B. (2000) Reconstitution of muscarinic modulation of the KCNQ2/KCNQ3 K+ channels that underlie the neuronal M current. J. Neurosci. 20, 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobori T., Smith G. D., Sandford R., Edwardson J. M. (2009) The transient receptor potential channels TRPP2 and TRPC1 form a heterotetramer with a 2:2 stoichiometry and an alternating subunit arrangement. J. Biol. Chem. 284, 35507–35513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stewart A. P., Smith G. D., Sandford R. N., Edwardson J. M. (2010) Atomic force microscopy reveals the alternating subunit arrangement of the TRPP2-TRPV4 heterotetramer. Biophys. J. 99, 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwake M., Jentsch T. J., Friedrich T. (2003) A carboxy-terminal domain determines the subunit specificity of KCNQ K+ channel assembly. EMBO Rep. 4, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwake M., Athanasiadu D., Beimgraben C., Blanz J., Beck C., Jentsch T. J., Saftig P., Friedrich T. (2006) Structural determinants of M-type KCNQ (Kv7) K+ channel assembly. J. Neurosci. 26, 3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakajo K., Kubo Y. (2008) Second coiled-coil domain of KCNQ channel controls current expression and subfamily-specific heteromultimerization by salt bridge networks. J. Physiol. 586, 2827–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howard R. J., Clark K. A., Holton J. M., Minor D. L., Jr. (2007) Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron 53, 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giamarchi A., Feng S., Rodat-Despoix L., Xu Y., Bubenshchikova E., Newby L. J., Hao J., Gaudioso C., Crest M., Lupas A. N., Honoré E., Williamson M. P., Obara T., Ong A. C., Delmas P. (2010) A polycystin-2 (TRPP2) dimerization domain essential for the function of heteromeric polycystin complexes. EMBO J. 29, 1176–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng S., Okenka G. M., Bai C. X., Streets A. J., Newby L. J., DeChant B. T., Tsiokas L., Obara T., Ong A. C. (2008) Identification and functional characterization of an N-terminal oligomerization domain for polycystin-2. J. Biol. Chem. 283, 28471–28479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng S., Rodat-Despoix L., Delmas P., Ong A. C. (2011) A single amino acid residue constitutes the third dimerization domain essential for the assembly and function of the tetrameric polycystin-2 (TRPP2) channel. J. Biol. Chem. 286, 18994–19000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.