Background: Plants lack mammalian GalNAc-type protein O-glycosylation.
Results: Transient expression of a Glc(NAc) C4-epimerase and a polypeptide GalNAc-transferase in Nicotiana benthamiana resulted in O-glycosylation.
Conclusion: Mammalian O-glycosylation can be established in plants.
Significance: Plants may serve as host cells for recombinant production of custom-designed O-glycoproteins.
Keywords: Glycoprotein Biosynthesis, Mucins, Plant, Protein Expression, Vaccine Development, GalNAc, Glycoengineering, Nicotiana benthamiana, O-Glycosylation
Abstract
Mucin-type O-glycosylation is an important post-translational modification that confers a variety of biological properties and functions to proteins. This post-translational modification has a particularly complex and differentially regulated biosynthesis rendering prediction and control of where O-glycans are attached to proteins, and which structures are formed, difficult. Because plants are devoid of GalNAc-type O-glycosylation, we have assessed requirements for establishing human GalNAc O-glycosylation de novo in plants with the aim of developing cell systems with custom-designed O-glycosylation capacity. Transient expression of a Pseudomonas aeruginosa Glc(NAc) C4-epimerase and a human polypeptide GalNAc-transferase in leaves of Nicotiana benthamiana resulted in GalNAc O-glycosylation of co-expressed human O-glycoprotein substrates. A chimeric YFP construct containing a 3.5 tandem repeat sequence of MUC1 was glycosylated with up to three and five GalNAc residues when co-expressed with GalNAc-T2 and a combination of GalNAc-T2 and GalNAc-T4, respectively, as determined by mass spectrometry. O-Glycosylation was furthermore demonstrated on a tandem repeat of MUC16 and interferon α2b. In plants, prolines in certain classes of proteins are hydroxylated and further substituted with plant-specific O-glycosylation; unsubstituted hydroxyprolines were identified in our MUC1 construct. In summary, this study demonstrates that mammalian type O-glycosylation can be established in plants and that plants may serve as a host cell for production of recombinant O-glycoproteins with custom-designed O-glycosylation. The observed hydroxyproline modifications, however, call for additional future engineering efforts.
Introduction
Most recombinant produced biological therapeutics are glycoproteins, and selection of host cells for their production is critical for therapeutic effects and safety because the glycan moieties can interact with lectin scavenger receptors and immune cells. Traditionally, selection of host cells has aimed at producing glycoproteins with normal human glycosylation, i.e. mature glycans similar to those found on the natural glycoproteins, typically complex type N-glycans and core 1 O-glycans, all with sialic acid capping (1). However, aberrant glycosylation may be desirable, e.g. with respect to design of vaccines aimed at eliciting immunity to specific glycoforms found on virus particles and virus-infected cells or on cancer cells (2). We have previously shown that the cancer-associated mucin MUC1 contains immunodominant aberrant O-glycopeptide epitopes to which cancer-specific IgG antibodies can be elicited in man (3), and we have developed a chemoenzymatic strategy for synthesis of such vaccine glycopeptides (3). However, sustainable recombinant expression systems for production of vaccines targeting cancer cells with immature O-glycosylation or envelope virus glycoproteins are still in demand.
O-Glycosylation is controlled by a family of up to 20 UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (GalNAc-Ts)3; the repertoire of GalNAc-Ts present in a given cell defines the pattern and density with which proteins are decorated with O-glycans. Normal mammalian cells typically produce elongated and/or branched O-glycans capped with sialic acids or different blood group- related structures. The most frequently used host cell for recombinant expression of therapeutic proteins is CHO, which generally produces mono- and disialylated core 1 (Galβ1–3GalNAcα1-O-Ser/Thr) O-glycan structures. Recently, it was shown that CHO-K1 expresses four GalNAc-Ts (GalNAc-T2, -T7, -T11, and -T19) (4). Because all eumetazoan cells express multiple GalNAc-Ts and generally extend and cap O-glycans, it is desirable to develop a cell system for recombinant production in which the entire O-glycosylation pathway can be custom-built both with respect to the GalNAc-T repertoire for custom design of sites of O-glycosylation, as well as the repertoire of glycosyltransferases involved in extending and capping O-glycans.
Yeast and plants offer cell systems suitable for recombinant expression without the capacity for mammalian GalNAc O-glycosylation (5, 6). Neither species contain genes homologous to the GalNAc-T gene family that initiates O-glycosylation (7). Although GalNAc and the donor substrate UDP-GalNAc are not found in yeast, the presence of these compounds in plants is still disputed (5, 8–10). Most of the enzymes known to elongate O-glycans in mammals have no close homologs in plants (11–13). These features should make it possible to custom design and build a capacity for O-glycosylation from the bottom up in plants. In yeast, the endogenous protein O-mannosylation initiated in the ER is likely to compete for substrate sites with the Golgi-localized GalNAc O-glycosylation. Despite this, Narimatsu and co-workers (14) introduced O-GalNAc glycosylation into yeast and obtained recombinant production of reporter constructs with GalNAc and core1 O-glycosylation but also a degree of O-mannosylation. With the use of a rhodamine-3-acetic acid derivative added to the culture medium, O-mannosylation was reduced but not completely eliminated.
Plants do not have competing O-glycosylation pathways for Ser/Thr residues. Plants produce another type of protein O-glycosylation, whereby prolines are hydroxylated to yield hydroxyproline (Hyp), which may be substituted with various Hyp O-glycans (15). Hyp conversion occurs in the ER and extends into the Golgi apparatus (16, 17), and this modification is preferentially found at proline (Pro)-rich repetitive sequence motifs (15). Whereas the short Pro-rich hinge region of IgA1 in humans is substituted with mucin-type O-linked glycans, recombinant expression in plants results in several Hyp modifications and further substitution with short arabinosides (18). More recently, expression of a tandem repeat of the human mucin MUC1, which in humans is decorated with O-glycans, was also found to contain Hyp O-glycosylation (19). The key step in plant-specific Hyp O-glycosylation is regulated by 13 C4-hydroxylases (P4Hs), which individually, according to initial studies (17, 20–22), are not required for viability and growth; and in principle, one or more of these could be mutated to eliminate undesirable modifications on recombinant glycoproteins. It is therefore conceivable that plants can be engineered to eliminate specific unwanted Hyp modifications that are required by proteins of interest during recombinant expression. Plants should therefore offer a suitable and safe system for recombinant production of glycoprotein pharmaceuticals.
In a recent study Daskalova et al. (23) studied transient expression of a GalNAc-T in combination with a Glc(NAc) C4-epimerase and a UDP-Gal(NAc) transporter in Nicotiana benthamiana L plants. They used a chimeric reporter construct with a short sequence from the human mucin, MUC1, as acceptor for O-glycosylation, and they used questionable Vicia villosa agglutinin (VVA) lectin blotting results to conclude that GalNAc O-glycosylation could be established in plants with co-expression of GalNAc-T2 alone. They also concluded that co-expression of both an epimerase and a transporter were required for enhancing lectin reactivity with the reporter, whereas neither the epimerase nor the transporter alone improved lectin reactivity. The results were not corroborated by structural analysis, and it is known that lectin reactivity poses particular problems in plants (5, 24). Thus, the original proposal of the presence of sialic acids in plant glycoproteins (25, 26) was later shown likely to be due to contaminants (27). Furthermore, Daskalova and co-workers (28) more recently reported that the VVA lectin recognizes noncarbohydrate epitopes by Western blotting in plants, thus casting serious doubts on the conclusions drawn in their previous publication (23).
In this study, we have carefully tested different design strategies for the introduction of GalNAc mucin-type O-glycosylation into N. benthamiana plants. The results are essentially in complete disagreement with those reported by Daskalova et al. (23), which highlights the need for structural verification of products produced with glycoengineering. We used transient expression of enzymes and reporter substrates to identify requirements for establishing efficient glycosylation of mammalian O-glycoproteins (see Fig. 1). We found no evidence of GalNAc O-glycosylation when a reporter substrate was expressed in plants alone or with an appropriate GalNAc-T, GalNAc-T2. Basic O-glycosylation capacity was, however, achieved by introduction of a Glc(NAc) C4-epimerase and a GalNAc-T, which was validated through co-expression of three O-glycosylation target proteins. We did observe Hyp modifications in a MUC1-based substrate, which indicates that use of plants for production of O-glycoprotein therapeutics will require additional strategies to eliminate the corresponding endogenous proline hydroxylases.
FIGURE 1.
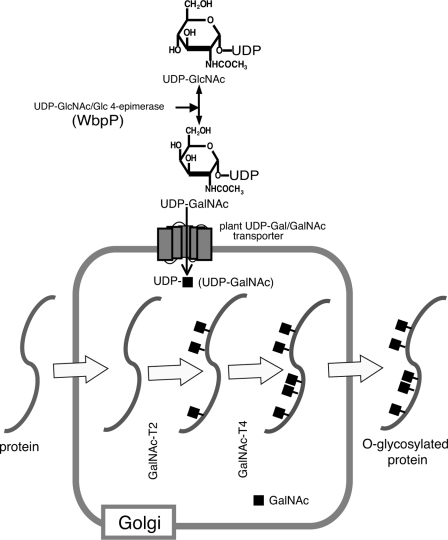
Depiction of the initiation process of mucin-type glycosylation in plants. The donor substrate UDP-GalNAc is synthesized from UDP-GlcNAc by P. aeruginosa C4-epimerase (WbpP) protein. UDP-GalNAc is transported into the Golgi lumen by endogenous plant sugar nucleotide transporter(s) where Golgi localized GalNAc-T2 and -T4 catalyze the transfer of GalNAc onto the target peptide(s) destined for secretion in the secretory pathway.
EXPERIMENTAL PROCEDURES
Inoculation and Growth Conditions of N. benthamiana
Growth conditions were as described in Egelund et al. (29). Agrobacterium tumefaciens strain C58C1 pGV3850 was used for agrobacterium-mediated expression in N. benthamiana, which was performed essentially as described by Sainsbury and Lomonossoff (30). In brief, agrobacteria were transformed with constructs by electroporation and selected with the appropriate antibiotics. Agrobacteria cultures were grown overnight in Luria-Bertani (LB) medium, harvested by centrifugation, and resuspended in a buffer containing 10 mm MES, 10 mm MgCl2, and 100 μm acetosyringone (A600 = 0.5) and left at 20 °C for 2 h. Leaves of 3–4-week-old N. benthamiana plants were infiltrated with the bacterial cell suspensions using 1-ml syringes, and leaf material was collected for analysis after 5–6 days. To increase expression of transgenes, all experiments included co-infiltration of agrobacteria carrying a p19 gene construct, a viral protein specifically inhibiting post-transcriptional gene silencing (31).
DNA Constructs for Plant Transformation and Transient Expression
Vector constructs used are depicted in Fig. 2A. Open source vectors used for Agrobacterium-mediated expression and transformation are as follows: pBI121 (GenBankTM accession number AY781296); pCAMBIA 2300 (GenBankTM accession number AF234315); pCAMBIA 1302 (GenBankTM accession number AF234298). Vectors used for Agrobacterium-mediated expression and transformation are as follows: pBI121 (GenBankTM accession number AF234315); pCAMBIA 1302 (GenBankTM accession number AF234298). For legacy of open source pCAMBIA and pBI121 binary vectors see online. pPS48 (32) is an intermediate Escherichia coli only vector, which contains the cauliflower mosaic virus 35S promoter followed by the 35S terminator interspaced by a multiple cloning site, into which genes of interest were cloned. Entire transcriptional units (35S-Pro-goi-35S-term) were then excised using XbaI or HindIII and ligated into the multiple cloning site of the pCAMBIA-derived plant expression plasmids. The Nicotiana tabacum ubiquitin promoter and terminator were synthesized by GenScript. O-Glycosylation target constructs were directed to the secretory pathway by N-terminal fusion of nucleotides encoding one of the following signal peptide sequences: 1) NtSP derived from N. tabacum proline-rich protein 3 (UniProt accession number Q40502), encoding the sequence MGKMASLFASLLVVLVSLSLA; and 2) PpSP derived from Physcomitrella patens aspartic protease (EMBL accession number AJ586914) encoding the sequence MGASRSVRLAFFLVVLVVLAALAEA.
FIGURE 2.
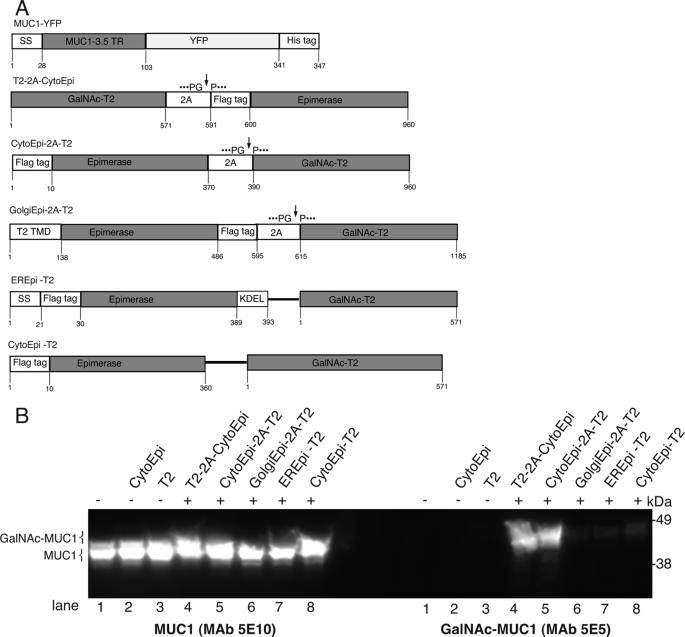
O-Glycosylation of the MUC1 reporter construct. A, O-glycosylation target protein MUC1-YFP was N-terminally fused to a signal peptide for direction into the secretory pathway, followed by a His6 tag. Gene constructs for introduction of O-glycosylation capacity into plants, comprising FLAG-tagged cytoplasmic, ER, or Golgi-targeted epimerase and native Golgi-targeted GalNAc-T2, either expressed as a single polycistronic protein, interspaced by the 2A self-splicing sequence (cleavage site indicated by an arrow), i.e. T2–2A-CytoEpi, CytoEpi-2A-T2, and GolgiEpi-2A-T2, or from separate promoters, i.e. EREpi-T2 and CytoEpi-T2. B, SDS-PAGE Western blot analysis of MUC1-YFP expressed alone (lane 1), together with cytoplasmic epimerase (lane 2), or with GalNAc-T2 (lane 3). MUC1-YFP co-expressed with either of the 2A-linked O-glycosylation machinery constructs (T2–2A-CytoEpi or CytoEpi-2A-T2) (lanes 4 and 5), 2A-linked Golgi-targeted epimerase, and GalNAc-T2 (lane 6), Golgi-targeted GalNAc-T2 with ER-targeted epimerase (EREpi) (lane 7), and cytoplasmic epimerase (lane 8) expressed from unlinked glycosylation machinery. Absence or presence of O-glycosylation machinery is indicated above the lanes with (−) or (+), respectively. Total protein extracts from transiently transformed N. benthamiana leaves were loaded and blots reacted with MUC1 specific MAbs 5E10 and 5E5, where 5E5 is specific for GalNAc-glycosylated MUC1 and does not react with unglycosylated MUC1. Approximately 30 μg of total protein was loaded in each lane.
The following constructs expressing mucins and other O-glycosylation target peptides were assembled or synthesized synthetically. The MUC1–3.5 tandem repeat (33) codon optimized for expression in Arabidopsis thaliana was synthesized with an N-terminal PpSP sequence (EUROFINS, MWG, Germany) and then utilized as a template for PCR amplification using the primers SPMUC1For and MUC1Rev (sequences of all primers are listed in supplemental Table S1). The resulting PCR product was then N-terminally fused to YFP by cloning into the pC2300u vector containing YFP (34), yielding pC2300u-35SPro-PpSP-MUC1–3.5TR-YFP(His)6-35STerm (MUC1-YFP). Two additional target peptides, under control of the ubiquitin promoter and terminator system, were created as follows: 1) pC2300D-UbiPro-NtSP-INFα2B-GM-UbiTerm (INFα2B) encoding interferon α2B (GenBankTM accession number AY255838.1), INFα2B, with an N-terminal signal peptide NtSP and C-terminal glyco-module ((SP)10), T7 (MASMTGGQQMG), and His6 tag; 2) pC2300D-UbiPro-NtSP-MUC16–1.2TR-UbiTerm (MUC16) encoding MUC16–1.2TR (33) (GenBankTM accession number AF414442.2) with an N-terminal signal peptide NtSP and C-terminal T7, and His6 tag was codon optimized for expression in Arabidopsis and synthesized by GenScript. Fragments encoding each mucin type peptide were subcloned into the HindIII site of pCAMBIA2300.
O-Glycosylation machinery consisted of cytosolic Pseudomonas aeruginosa Glc(NAc) C4-epimerase WbpP (GenBankTM accession number AAF23998.1) (35), human GalNAc-T2 (UniProt accession number X85019), and GalNAc-T4 (GenBankTM accession number NP_003765.2). O-Glycosylation machineries were implemented as single constructs either with each gene driven by their own promoter or as a polycistronic transcript interspaced by the 2A auto-cleavable sequence (FMDV) (36, 37) under control of a single promoter driving expression of the resulting 2A-linked fusion protein. Construction of O-glycosylation machineries encoding C4-epimerase and GalNAc-Ts, with each gene brought under control of a separate promoter, was carried out as follow. 1) Cloning of the N-terminal FLAG-tagged (MDYKDDDD) epimerase into the pCAMBIA2300 vector (pC2300) involved PCR amplification using pET23-WbpP (35) as template and the primers PBY7For and PBY7Rev, which was subcloned into the SacI site of pPS48 under control of the 35S promoter and 35S terminator sequence. The entire transcriptional unit (35SPro-CytoEpi-35STerm) was then excised using HindIII and cloned into the HindIII site of pC2300, yielding pC2300–35SPro-CytoEpi-35Sterm (CytoEpi). ER-targeted epimerase was constructed by PCR amplification of CytoEpi using the primers ERWbpPFor and ERWbpPRev for addition of the secretion signal peptide and C-terminal KDEL ER retention signal, resulting in the construct EREpi, which was subcloned into pCAMBIA2300 using the SacI site. 2) Full-length GalNAc-T2 was excised from an existing pBKS-GalNAc-T2 containing plasmid (GenBankTM accession number X85019) (38) using EcoRI, blunted with Klenow, and inserted into the StuI site of pC1302, thus rendering GalNAc-T2 under the control of 35S promoter and terminator and resulting in the construct pC1302D-35SPro-T2–35STerm (T2). 3) O-Glycosylation machinery construct encoding cytosolic epimerase and Golgi-targeted GalNAc-T2 from separate transcripts was made to compare with the Golgi UDP-GalNAc pool with the cytosolic pool. This construct was assembled by inserting the XbaI-35SPro-CytoEpi-35STerm-XbaI of construct CytoEpi into the XbaI site of construct T2, resulting in pC1302D-35SPro-T2-35STerm;35SPro-CytoEpi-35STerm (CytoEpi-T2). 4) O-Glycosylation machinery construct encoding ER epimerase and Golgi-targeted GalNAc-T2 (EREpi-T2) was cloned same as the cloning procedure of CytoEpi-T2. 5) Full-length of GalNAc-T4 (codon optimized for Arabidopsis) was synthesized by EUROFINS, MWG, Germany, and subcloned into pCAMBIA2300 using the HindIII site, resulting in pC2300D-35SPro-T4–35STerm.
Three additional O-glycosylation machineries were made as 2A-linked polycistronic constructs. 1) GalNAc-T2 was PCR-amplified with primer-introduced N-terminal hemagglutinin tag (HA tag) using the primers HAT2For and HAT2Rev. N-terminal FLAG-tagged epimerase was PCR-amplified using the primer set PFwbppFor and PFwbppRev. The two PCR fragments were then ligated and interspaced with a sequence encoding the 2A sequence in pC130035Su in accordance to the USER cloning method delineated by Nour-Eldin et al. (34) and Geu-Flores et al. (39), yielding pC1302–35SPro-T2–2A-CytoEpi-35STerm (T2–2A-CytoEpi). The 2A linker region of T2–2A-CytoEpi encodes QGSGQTLNFDLLKLAGDVESNPG↓PMD, where underline designates the 2A sequence, and arrow indicates the auto-cleavage site (36, 37). 2) The reverse order, encoding (from N to C terminus) cytosolic epimerase followed by the 2A sequence and Golgi-targeted GalNAc-T2, was made by PCR-amplifying CytoEpi-2A-T2 fragment from construct pBI121–35SPro-MUC1–3.5TR-YFP-2A-GolgiEpi-2A-T2-NosTerm,4 with primers PwbppFor and PT2Rev. CytoEpi-2A-T2 was ligated into pBI121, using the XbaI and SacI sites, yielding pBI121–35SPro-CytoEpi-2A-T2-NosTerm (CytoEpi-2A-T2). 3) A third construct encoding polycistronic Golgi-targeted epimerase and Golgi-targeted GalNAc-T2 interspaced by the 2A sequence was PCR-amplified using pBI121–35SPro-MUC1–3.5TR-YFP-2A-GolgiEpi-2A-T2-NosTerm as template and the primers PFLAGFor and PT2Rev, yielding the fragment GolgiEpi-2A-T2, which was inserted back into pBI121 vector using the XbaI and SacI sites, yielding pBI121–35SPro-GolgiEpi-2A-T2-NosTerm (GolgiEpi-2A-T2).
Preparation of Leaf Total Protein Extracts
Approximately 1 g of freshly harvested leaves were frozen in liquid N2 and comminuted using a pestle and mortar with 2 ml of extraction buffer A (50 mm NaPO4, 250 mm NaCl, 5 mm imidazole, pH 8.0) containing Complete Proteinase Inhibitor (Roche Applied Science) and 1 mm phenylmethanesulfonyl fluoride. The sample was incubated for 10 min on ice, and insoluble material was pelleted by centrifugation (20,000 × g) for 10 min, and the supernatant was recovered and stored at −20 °C.
SDS-PAGE Western Blotting
SDS-PAGE Western blot analysis was performed as described previously (40). Monoclonal antibodies (mAbs) to the T7 tag, and M11 to MUC16 were obtained from Invitrogen and Dako, respectively. mAbs to MUC1 and GalNAc-MUC1, 5E10 and 5E5, respectively, have been described previously (41). Lectin blot analysis was performed essentially as Western blot analysis, except the antibody was HRP-conjugated Vicia villosa lectin (VVA, EY Laboratories).
Nickel Affinity Chromatography
Cleared supernatants (50 ml, 20,000 × g for 30 min) were incubated with 0.5 ml of nickel-nitrilotriacetic acid-agarose beads (Qiagen) for 2 h (4 °C) under gentle rolling. Beads were washed for 10 min with 20 ml of wash buffer (50 mm NaPO4, 250 mm NaCl, 20 mm imidazole, pH 8.0), and eluted five times with 1 ml of elution buffer (50 mm NaPO4, 250 mm NaCl, 250 mm imidazole, pH 8.0).
Endo-Asp Digestion of MUC-3.5TR and Sample Purification
Purified MUC1–3.5TR-YFP (∼25 μg) was incubated with 1 μg of endoproteinase Asp-N from Pseudomonas fragi (Roche Applied Science) in 300 μl of 100 mm Tris-HCl, pH 8.0, for 16 h at 37 °C with shaking. The digest was cleaned on a C18 Zip-Tip (Millipore). Briefly, the digestion mixture was dissolved in 20 μl of 0.1% TFA and drawn through the column, desalted with 0.5% formic acid, and eluted with acetonitrile. In some instances, the released 20-mer MUC1 tandem repeat peptide was further isolated by HPLC using a Dionex system. Trifluoroacetic acid (TFA) was added to samples (0.05% v/v) before application to a C12 column (150 × 4.6 mm Jupiter Proteo with 90-Å pore size, 4-μm particle size, Phenomenex). Chromatographic separation was obtained in a two-eluent system, where eluent A was 0.05% TFA in water, and eluent B was 0.05% TFA in acetonitrile, and the pump speed was a constant 0.5 ml min−1. From 0 to 5 min, the eluent was 5% B; from 5 to 35 min, eluent B increased in a linear gradient to 40%; and from 35–45 min eluent B increased to 100%.
Matrix-assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF)
Lyophilized peptides were dissolved in 20 μl of water. The MALDI matrix was 25 g/liter 2,5-dihydroxybenzoic acid (Sigma) dissolved in a 1:1 mixture of water and methanol. Samples were prepared for analysis by placing 0.5 μl of sample solution on a probe tip followed by 0.5 μl of matrix. All spectra were obtained in the linear mode and calibrated using external calibration. All mass spectra were acquired on a Voyager-Elite MALDI time of flight mass spectrometer (PerSeptive Biosystem Inc., Framingham, MA), equipped with delayed extraction.
Characterization of O-Glycosylation Sites by ETD-MS2
Products of O-glycosylation were characterized by electrospray ionization-linear ion trap-Fourier transform mass spectrometry in an LTQ-Orbitrap XL hybrid spectrometer (Thermo-Scientific) equipped for ETD for peptide sequence analysis by MS/MS (MS2) with retention of glycan site-specific fragments. Samples were dissolved in methanol/water (1:1) containing 1% formic acid and introduced by direct infusion via a TriVersa NanoMate ESI-Chip interface (Advion BioSystems) at a flow rate of ∼100 nl/min and 1.4-kV spray voltage. Mass spectra were acquired in positive ion FT mode using parameters similar to previous studies (42), except at a nominal resolving power of either 30,000 or 60,000. MS1 spectra, in which multiple charge states were observed, were deconvoluted for clarity of presentation using the Xtract function in the Xcalibur data analysis software package (Thermo Fisher). ETD-MS2 spectra were analyzed by comparison with theoretical c- and z-fragment m/z values calculated for all positional combinations of HexNAc residues distributed on all the potential Ser and Thr glycosylation sites in the sequence. Where potential hydroxylation of Pro was observed, positional combinations, including one or more Hyp residues, were also calculated. Calculations were performed using the web-based Protein Prospector MS-Product software routine.
RESULTS
Design of GalNAc O-Glycosylation Engineering in Plants
Depicted in Fig. 1 are the principle constituents needed for GalNAc O-glycosylation. Introduction of UDP-GalNAc synthesis required a Glc/GlcNAc C4-epimerase. High concentrations of nucleotide sugars in Golgi are maintained by transporters, and it has been suggested that plants lack a specific transporter (43, 44) for UDP-GalNAc (23). Most UDP-Gal transporters, however, function as UDP-Gal/GalNAc transporters (43, 45–47). We therefore first investigated whether introduction of an epimerase, P. aeruginosa cytoplasmic epimerase (WbpP) (35), and a human GalNAc-T, GalNAc-T2 (48), would be sufficient for O-glycosylation. A series of construct designs were prepared and tested to address the optimal order of coding regions, efficiency of introducing a 2A self-cleaving sequence, and effect of targeting the epimerase to ER, Golgi, or the cytoplasm (Fig. 2A). For expression and localization of O-glycosylation machinery components see supplemental Fig. S1.
Expression of a MUC1-YFP Reporter Substrate in Plants
We initially assessed O-glycosylation capacity by expressing a known acceptor substrate for GalNAc-T2, which was based on the tandem repeat sequence of MUC1 and linked to YFP (MUC1-YFP). When expressed alone, the reporter MUC1-YFP substrate migrated as an ∼45-kDa protein and reacted with mAb 5E10 (reactive with unglycosylated and all glycoforms of MUC1) but not with mAb 5E5 (reactive with GalNAc-glycosylated MUC1), indicating that the MUC1 reporter substrate was not substituted with GalNAc in wild type plants (Fig. 2B, lane 1). This was subsequently confirmed by mass spectrometric analysis of the MUC1 peptide enzymatically digested from the expressed MUC1-YFP protein (Fig. 3D), where a product corresponding to unmodified MUC1 (m/z 1887) was present, but no products corresponding to glycosylated MUC1 were observed. This analysis did uncover evidence of Hyp modification of MUC1. The presence of an ∼16-Da increment (m/z 1903) in the MS spectrum of MUC1-YFP corresponds to hydroxylation of a single proline. Electrospray ionization-electron transfer dissociation Fourier transform-MS2 analysis further demonstrated that Pro11 of the MUC1 tandem repeat (DTRPAPGSTAP11PAHGVTSAP) was hydroxylated.4 Based on peak sizes, this Hyp modification was a minor constituent, and evidence of other modifications, including Hyp O-glycosylation, was not observed (Fig. 3D).
FIGURE 3.
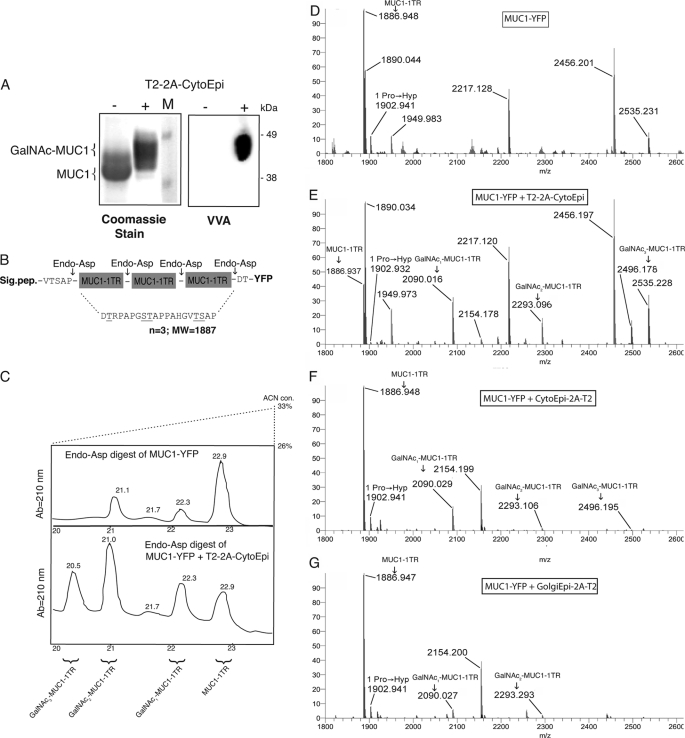
Structural analysis of MUC1-YFP expressed in N. benthamiana. A, purification of His-tagged MUC1-YFP. Left panel, SDS-PAGE Coomassie staining of MUC1-YFP expressed (−) alone and (+) together with 2A-linked glycosylation machinery (T2-2A-CytoEpi). Right panel, staining with Vicia villosa agglutinin (VVA) lectin for detection of GalNAc glycosylation. B, schematic illustration of endo-Asp-N digestion of O-glycosylated MUC1. C, analytical HPLC purification of endo-Asp-N-digested MUC1-YFP expressed alone (upper panel) or together with O-glycosylation machinery (lower panel) separating released 20-mer MUC1 repeat sequences with and without GalNAc-glycosylation. D–G, Orbitrap FT-MS analysis of purified endo-Asp-N MUC1-YFP products. D, MUC1-YFP expressed alone. E, MUC1-YFP co-expressed with the O-glycosylation machinery (T2-2A-CytoEpi). F, MUC1-YFP co-expressed with the inverse ordered glycosylation machinery (CytoEpi-2A-T2). G, MUC1-YFP co-expressed with 2A-linked Golgi targeted epimerase and GalNAc-T2 (GolgiEpi-2A-T2). Each spectrum is a deconvoluted average centroid m/z representation. Peaks consistent with the MUC1-YFP sequence are labeled with monoisotopic m/z. These correspond to DTRPAPGSTAPPAHGVTSAP (31–50, 51–70, or 71–90, calculated monoisotopic m/z 1886.9355; +1 × oxidation (i.e. Pro → Hyp), calculated monoisotopic m/z 1902.9304); DTLVNRIELKGIDFKE (220–235, calculated monoisotopic m/z 1890.0331); DGPVLLPDNHYLSYQSALSK (293–312, calculated monoisotopic m/z 2217.1186); DFFKSAMPEGYVQERTIFFK (185–204, +1 × oxidation (of Met or Pro), calculated monoisotopic m/z 2456.1955); DGNILGHKLEYNYNSHNVYITA (236–257, calculated monoisotopic m/z 2535.2263). Arrows denote peaks consistent with mono-, di-, and tri-glycosylated DTRPAPGSTAPPAHGVTSAP peptide (31–50, 51–70, or 71–90, calculated monoisotopic m/z 2090.0149, 2293.0943, and 2496.1736, respectively).
Construct Designs of O-Glycosylation Machinery
Co-expression of either C4-epimerase WbpP or GalNAc-T2 alone with the MUC1-YFP reporter substrate did not result in detectable O-glycosylation (Fig. 2B, lanes 2 and 3). In contrast, co-expression of MUC1-YFP with a combination of C4-epimerase and GalNAc-T2 resulted in detectable O-glycosylation as evidenced by reactivity with mAb 5E5 and a slight mobility shift (1–2 kDa) on SDS-PAGE-derived Western blotting (Fig. 2B, lanes 4–8). The 2A auto-cleaving motif is a 20-amino acid sequence derived from the foot-and-mouth virus, which through an intra-ribosomal skipping event during translation results in synthesis of two protein products (49). Insertion of the 2A sequence between two coding regions in a polyprotein construct allows for expression of two independent proteins under control of a single promoter, and it has been widely tested in many eukaryotic expression systems (49, 50). The 2A construct design is an attractive approach for achieving coordinated expression of multiple proteins. Tests of different construct designs for expression of the epimerase and GalNAc-T2 demonstrated that only the 2A-linked bicistronic constructs (T2–2A-CytoEpi and Cyto-Epi-2A-T2) produced efficient GalNAc O-glycosylation (Fig. 2B, lanes 4 and 5). The attempts to direct the epimerase to the Golgi (GolgiEpi-2A-T2) or to the ER (EREpi-T2) did not result in substantial glycosylation (Fig. 2B, lanes 6 and 7). Furthermore, expression of the cytoplasmic epimerase and GalNAc-T2 from separate promoters resulted in detectable but substantially lower efficiency in glycosylation (Fig. 2B, lane 8) indicating that the 2A sequence is essential for the vector design.
Structural Analysis of MUC1-YFP Expressed in Glycoengineered N. benthamiana
MUC1-YFP co-expressed with or without T2–2A-CytoEpi was purified by metal-affinity chromatography and analyzed via SDS-PAGE and VVA lectin blotting (Fig. 3A). The increase in molecular mass exhibited by MUC1-YFP when co-expressed with T2–2A-CytoEpi was readily apparent on a highly separated SDS-polyacrylamide gel (Fig. 3A, left panel). HRP-conjugated VVA lectin, specific for GalNAc moieties, reacted with MUC1-YFP co-expressed with T2–2A-Epi but not with MUC1-YFP expressed alone (Fig. 3A, right panel). Purified MUC1-YFP was enzymatically digested with endo-Asp-N to release individual MUC1 tandem repeats (Fig. 3B). Analysis of the products of this digest by HPLC enabled the separation of differentially modified MUC1 tandem repeats (Fig. 3C). The released MUC1 expressed in plants without O-glycosylation machinery eluted at 22.9 min, and MS analysis of the isolated product confirmed this.4 MS analysis of the total MUC1-YFP digest revealed the presence of unmodified MUC1, but no products corresponding to GalNAc modifications (Fig. 3D). The HPLC of digested MUC1-YFP from plants co-expressing the O-glycosylation machinery, T2–2A-CytoEpi, revealed additional products eluting between 20.5 and 22.3 min (Fig. 3C, lower panel). MS analysis of these products identified MUC1 tandem repeats with 1–3 GalNAc moieties (Fig. 3E). This HPLC and MS analysis revealed that a substantial amount of the tandem repeat sequence was modified by 1–3 mol of GalNAc, which is in agreement with the fact that GalNAc-T2 can attach GalNAc up to three of the five potential sites in the MUC1 tandem repeat sequence (Ser8, Thr9, and Thr17) (38). Two minor peaks at 21.1 and 22.3 min were found when MUC1-YFP was expressed alone (Fig. 3C, upper panel). MALDI-TOF analysis of these fractions showed the presence of MUC1–1TR with 1 Hyp (only in the 22.3-min fraction), YFP peptide fragments, and apparent contaminants. Analysis of MUC1-YFP co-expressed with the CytoEpi-2A-T2 or GolgiEpi-2A-T2 O-glycosylation machinery constructs confirmed incorporation of GalNAc, albeit at lower levels than observed with the T2–2A-CytoEpi construct (Fig. 3, F and G). Approximately 65% of total MUC1-YFP was glycosylated by the most efficient O-glycosylation machinery, T2–2A-CytoEpi, and MUC1-YFP was estimated to accumulate to ∼12 μg/g (fresh weight) equivalent to ∼1% of the total soluble protein.
Co-expression of GalNAc-T2 and -T4 Completes GalNAc O-Glycosylation of MUC1
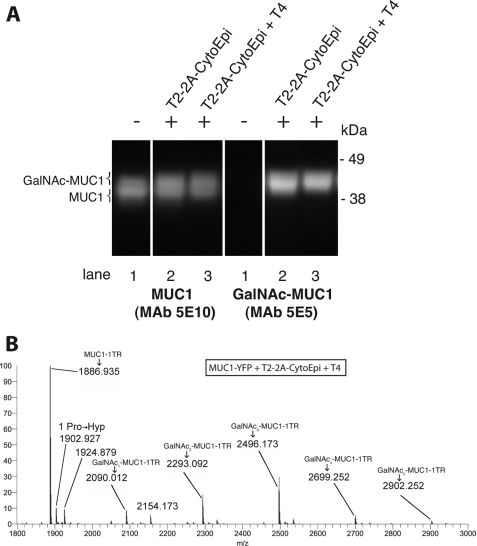
Complete O-glycosylation of all five potential sites of the MUC1 tandem repeat requires the coordinate action of GalNAc-T4 and GalNAc-T2 (38). We therefore co-expressed GalNAc-T4 (T4) with T2–2A-CytoEpi and MUC1-YFP, which resulted in a further minor shift in SDS-PAGE mobility of MUC1-YFP as evidenced by 5E5 mAb-mediated Western blot analysis (Fig. 4A). MS analysis of purified MUC1-YFP digested with endo-Asp-N confirmed incorporation of up to 5 mol of GalNAc per tandem repeat of MUC1 (m/z 2902) when co-expressed with GalNAc-T2 and -T4 (Fig. 4B). Again a minor amount of hydroxylation of Pro11 was observed (m/z 1903).
FIGURE 4.
Co-expression of GalNAc-T2 and -T4 completes GalNAc O-glycosylation of MUC1. A, Western blot analysis of target construct MUC1-YFP (lane 1) alone (−) or co-inoculated (+) with either of the 2A-linked glycosylation machinery constructs T2–2A-CytoEpi (lane 2) or T2–2A-CytoEpi and ectopically with GalNAc-T4 (lane 3). Co-expression of GalNAc-T4 with the 2A-linked glycosylation machinery (T2–2A-CytoEpi) conferred glycosylation of all five potential GalNAc O-glycosylation sites in MUC1–1TR. B, Orbitrap FT-MS analysis of endo-Asp-N-digested MUC1-YFP co-expressed with 2A-linked glycosylation machinery (T2–2A-CytoEpi) and GalNAc-T4, demonstrating glycosylation of the three GalNAc-T2-specific sites and the two additional GalNAc that are added by GalNAc-T4 (DTT4RPAPGST2TT2APPAHGVTT2ST4AP), i.e. complete GalNAc occupancy of sites.
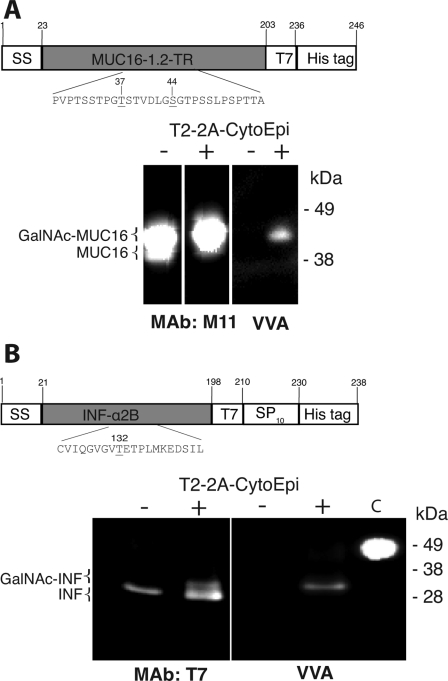
O-Glycosylation of Human MUC16 and INFα2B
Two other human O-glycoproteins were tested as substrates. A construct expressing 1.2 tandem repeats of MUC16 (MUC16-T7), i.e. encoding 223 amino acids with ∼30 putative O-glycosylation sites and 3 N-glycosylation sites, was co-expressed with the T2–2A-CytoEpi glycosylation machinery. A clear shift in SDS-PAGE mobility (∼45 to ∼50 kDa) of the T7-tagged protein combined with reactivity with VVA was observed with co-expression of T2–2A-CytoEpi (Fig. 5A). Co-expression of a construct encoding the full secreted INFα2B cytokine tagged with T7 and an Arabinogalactan Protein type (SP)10 glycomodule (INFα2B-T7-AGP) with the T2–2A-CytoEpi construct resulted in a mobility shift of a minor fraction of the protein (∼30 to ∼31 kDa) and reactivity with VVA (Fig. 5B). INFα2B has a single O-glycosylation site (GVGVT132ETPLM137), which is known to be O-glycosylated by GalNAc-T2 (51). The observed mobility shifts and VVA labeling are indicative of O-glycosylation, but further MS analysis is needed to confirm glycosylation sites.
FIGURE 5.
O-Glycosylation of human Mucin 16 (MUC16) reporter and interferon α2B. A, Western blot and lectin staining analysis of Mucin 16–1,2TR (MUC16); B, interferon α2B (INF-α2B), alone (−) or co-inoculated (+) with the 2A-linked glycosylation machinery construct T2–2A-CytoEpi. mAb M11 is a specific antibody to MUC16, and mAb T7 is a T7 tag antibody. Glycosylations of MUC16 and INF-α2B are shown by staining with VVA. 30 μg of total protein was loaded in each lane. The two constructs expressing MUC16 and INF-α2B are shown with the potential glycosylation sites (Ser or Thr) denoted by underlining.
DISCUSSION
Plants represent one eukaryotic cell system in which animal O-glycosylation can be built from scratch and custom-designed for a particular purpose. Because the capacity for GalNAc-type O-glycosylation is highly regulated and dynamic in mammalian cells, it is desirable to develop cell systems with defined capacity for O-glycosylation for production of recombinant therapeutics and vaccines. In this study, we established GalNAc O-glycosylation capacity in N. benthamiana plant cells by expression of a UDP-Glc(NAc) C4-epimerase and GalNAc-Ts, and we showed that three different co-expressed secreted substrates were O-glycosylated. In this study, we expressed GalNAc-T2 and epimerase transiently in plants using both monocistronic constructs and bicistronic 2A linked constructs to screen different permutations of optimal vector design for the glycosylation machinery. In the present transient implementations, the bicistronic 2A construct designs proved most efficient (Fig. 2B).
The GolgiEpi-2A-T2 and EREpi-T2 constructs were designed to test if ectopic expression of the C4-epimerase in the ER-Golgi lumen would convert ER-Golgi lumen UDP-GlcNAc to UDP-GalNAc in situ and allow GalNAc O-glycosylation. However, targeting epimerase to cytoplasm was most effective as evidenced by glycosylation of the co-expressed acceptor substrate MUC1 (Fig. 2B). Factors including local substrate (UDP-GlcNAc) and resulting product (UDP-GalNAc) concentrations, the presence of chaperones, and pH differences in various compartments may influence the glycosylation efficiency. Other factors may be inefficient 2A sequence cleavage and aberrant localization of the individual proteins. For example, de Felipe and Ryan (52) reported that a two times 2A construct encoding a Golgi-targeted cyan fluorescence protein, Golgi-targeted YFP, and a cytosolic puromycin resistance protein (i.e. GT-CFP-2A-GT-YFP-2A-PAC), resulted in Golgi-localized cyan fluorescence protein and YFP aberrantly localized in the mitochondria. Our attempts to exploit the ER/Golgi GlcNAc pools by targeting the epimerase to the ER or the Golgi proved less efficient than targeting it to the cytosol. This demonstrates that plant cells efficiently transport UDP-GalNAc into the secretory pathway and either do not produce GalNAc and UDP-GalNAc or if so only in minute amounts. The results provide a basis for further exploration of plant cells as glycoengineered hosts for recombinant expression of O-glycoproteins.
Considerable efforts have been devoted to glycoengineering host cells to accommodate human type glycosylation on recombinant expressed therapeutics, which have led to remarkable improvements especially for protein N-glycosylation. Human type N-glycosylation has been engineered in yeast with unprecedented control of homogeneity of N-glycan structures added to proteins (53). In contrast, engineering of the abundant GalNAc-type O-glycosylation has lagged behind. Yeast cells were originally engineered to enable formation of GalNAcα and Galβ1–3GalNAcα O-glycosylation (14). The engineered glycosylation machinery included a UDP-GlcNAc C4-epimerase, a UDP-Gal(NAc) transporter, and the glycosyltransferases GalNAc-T2 and C1GalT1. Although yeast is a desirable expression system, it suffers from competing O-mannosylation initiated in the ER. O-Mannosylation is essential for yeast (54), but O-mannosyl glycans are immunogenic in man (55, 56). Although it is not possible to knock out all O-mannosyltransferases, some reduction of O-mannosylation was achieved by adding a rhodamine-3-acetic acid derivative to the culture medium (14).
While this study was being completed, Daskalova et al. (23) reported on similar glycoengineering attempts in N. benthamiana. Based on VVA lectin labeling experiments, the authors concluded that co-expression of a GalNAc-T with a reporter substrate was sufficient to produce low levels of glycosylation, whereas co-expression of both a UDP-Gal(NAc) transporter and an C4-epimerase were required for enhanced O-glycosylation. These results are in complete disagreement with the results presented here, where efficient glycosylation was achieved by co-expression of a GalNAc-T with an epimerase. We can only reconcile the different findings with erroneous interpretation of VVA lectin reactivity as was in fact reported by Daskalova and co-workers only recently (28). In this paper, Daskalova and co-workers presented evidence of noncarbohydrate-specific binding of VVA lectin to tobacco protein extracts, which could not be inhibited by hapten sugar, such as d-GalNAc. Furthermore, the claim that a specific UDP-Gal(NAc) transporter is required for GalNAc O-glycosylation in plants (23) is controversial. Although UDP-GalNAc transport capacity has not been directly demonstrated in plants, many transporters are known to have UDP-Glc/Gal and UDP-GlcNAc/GalNAc transport capacities (57). Here we clearly demonstrate efficient O-glycosylation without exogenous introduction of a dedicated UDP-GalNAc transporter. This was also found with the additional functional expression of the GalNAc-T4 isoform, which is unique among the GalNAc-T isoforms by being required for complete GalNAc O-glycosylation of all five potential sites in the MUC1 tandem repeat sequence as well as having a very high Km values for UDP-GalNAc (38).
Plants have a unique hydroxyproline-linked O-glycosylation, which has been found on recombinant expressed human IgA1 (18) and most recently MUC1 (19). This modification is initiated by a family of prolyl 4-hydroxylases (P4Hs), and the resulting Hyps may subsequently be glycosylated with arabinogalactan or shorter arabinosides. P4Hs are membrane-bound type II proteins found in the ER and extending into the Golgi apparatus (16). Thirteen putative P4Hs have been identified in Arabidopsis (58). In this study a minor amount of hydroxylation of Pro11 of the MUC1 tandem repeat was observed. Other post-translational modifications, including hydroxyproline-linked O-glycosylation, were not encountered. The recent findings of Pinkhasov et al. (19) showed that either Pro11 or Pro12 of the MUC1 tandem repeat was further substituted with three l-arabinofuranoses. This hydroxyproline linked O-glycosylation may be rationalized by the use of different promoter systems driving the transient expression of the MUC1 reporter, where Pinkhasov et al. (19) employed a potent virus-based expression system, perhaps causing increased contact with the endogenous plant post-translational modification machinery.
Our preliminary studies suggest that the number of Pro residues in the MUC1 tandem repeat undergoing hydroxylation and the degree of hydroxylation appear to increase in Arabidopsis plants and tobacco suspension BY-2 cells stably transformed with the same MUC1 reporter construct used in this study.4 In plants, protein O-glycosylation is primarily found in Hyp-rich glycoproteins of the cell wall, where Ser and in particular Hyp residues may be O-glycosylated. Dipeptidyl proline sequences in angiosperm Hyp-rich glycoproteins prone to hydroxylation were recently summarized by Kieliszewski et al. (59). Consistently hydroxylated sequences included the motifs AP, SP, PP, and the latter extensin-type motif is found in the APPA motif of the MUC1 tandem repeat. Several reports suggest involvement of the three P4Hs, AtP4H2, AtP4H5, and AtP4H13, in hydroxylation of consecutive Pro motifs in particular (17, 20–22), perhaps pointing to these enzymes as first targets for engineering.
Glycoengineering of host cells generally seeks to achieve human glycosylation produced by normal cells, which involves complex-type N-glycans and/or core1 O-glycans capped with sialic acids to ensure circulation of injectable therapeutics for therapeutic effect. However, another important purpose of engineering O-glycans is to modulate immunogenic glycoforms produced by cancer cells and virus-infected cells. Changes in O-glycosylation are a hallmark of cancer cells, and immunogenic short immature aberrant O-glycans are termed pan-carcinoma antigens (60). Our interest in engineering O-glycosylation from scratch stems from our previous identification of immunodominant aberrant O-glycopeptide epitopes in the cancer-associated MUC1 mucin, which are not covered by immunological tolerance (41, 61, 62). Vaccination with MUC1 glycopeptides with the truncated GalNAc-Ser/Thr O-glycoform produced by GalNAc-T2 and -T4 in combination produce IgG antibodies with cancer-specific reactivity (61), and spontaneous IgG antibodies to the same epitope are found in many cancer patients at time of diagnosis (3, 63). Recombinant production of such vaccines will require a host cell that produces similar truncated O-glycans, and glycoengineered plants as reported here may provide such a system. Interestingly, the acquisition of Hyp in the MUC1 repeat may not adversely affect immunogenicity, but rather stimulate immunity as recently reported for unglycosylated MUC1 vaccine produced in plants (19).
In this study we have elucidated requirements for engineering plants with the capacity to perform initiation of human type O-glycosylation. O-Glycosylation capacities may be engineered in e.g. tobacco with humanized N-glycosylation (64–69), which may serve as a more general platform for transient (70) as well as stable expression of glycoprotein pharmaceuticals. The results highlight that additional engineering is needed for plants to become versatile production platforms of therapeutics.
Supplementary Material
Acknowledgment
pET23-WbpP was a generous gift from J. S. Lam.
This work was supported by Danish Council for Strategic Research Grant DSF 09-063090, Danish Council for Independent Research Grant DFF 09-066624/274-09-0314, and a program of excellence from the University of Copenhagen.

This article contains supplemental Table S1, Fig. S1, “Experimental Procedures,” and additional references.
Z. Yang and B. L.. Petersen, unpublished results (information available upon request).
- GalNAc-T
- UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase
- P4H
- prolyl 4-hydroxylase
- Hyp
- hydroxyproline
- ETD
- electron transfer dissociation
- VVA
- V. villosa agglutinin
- ER
- endoplasmic reticulum
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Elliott S., Lorenzini T., Asher S., Aoki K., Brankow D., Buck L., Busse L., Chang D., Fuller J., Grant J., Hernday N., Hokum M., Hu S., Knudten A., Levin N., Komorowski R., Martin F., Navarro R., Osslund T., Rogers G., Rogers N., Trail G., Egrie J. (2003) Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 21, 414–421 [DOI] [PubMed] [Google Scholar]
- 2. Tarp M. A., Clausen H. (2008) Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 1780, 546–563 [DOI] [PubMed] [Google Scholar]
- 3. Wandall H. H., Blixt O., Tarp M. A., Pedersen J. W., Bennett E. P., Mandel U., Ragupathi G., Livingston P. O., Hollingsworth M. A., Taylor-Papadimitriou J., Burchell J., Clausen H. (2010) Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 70, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., Andersen M. R., Neff N., Passarelli B., Koh W., Fan H. C., Wang J., Gui Y., Lee K. H., Betenbaugh M. J., Quake S. R., Famili I., Palsson B. O., Wang J. (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerlach J. Q., Kilcoyne M., Eaton S., Bhavanandan V., Joshi L. (2011) Noncarbohydrate-mediated interaction of lectins with plant proteins. Adv. Exp. Med. Biol. 705, 257–269 [DOI] [PubMed] [Google Scholar]
- 6. Gomord V., Faye L. (2004) Post-translational modification of therapeutic proteins in plants. Curr. Opin. Plant Biol. 7, 171–181 [DOI] [PubMed] [Google Scholar]
- 7. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2011) Control of mucin-type O-glycosylation. A classification of the polypeptide GalNAc-transferase gene family. Glycobiology doi: 10.1093/glcob/cwr1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kishimoto T., Watanabe M., Mitsui T., Hori H. (1999) Glutelin basic subunits have a mammalian mucin-type O-linked disaccharide side chain. Arch. Biochem. Biophys. 370, 271–277 [DOI] [PubMed] [Google Scholar]
- 9. Kilcoyne M., Shah M., Gerlach J. Q., Bhavanandan V., Nagaraj V., Smith A. D., Fujiyama K., Sommer U., Costello C. E., Olszewski N., Joshi L. (2009) O-Glycosylation of protein subpopulations in alcohol-extracted rice proteins. J. Plant Physiol. 166, 219–232 [DOI] [PubMed] [Google Scholar]
- 10. Alonso A. P., Piasecki R. J., Wang Y., LaClair R. W., Shachar-Hill Y. (2010) Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry. Plant Physiol. 153, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimoto K., Tokimatsu T., Kawano S., Yoshizawa A. C., Okuda S., Goto S., Kanehisa M. (2009) Comprehensive analysis of glycosyltransferases in eukaryotic genomes for structural and functional characterization of glycans. Carbohydr. Res. 344, 881–887 [DOI] [PubMed] [Google Scholar]
- 12. Henrissat B., Surolia A., Stanley P. (2009) in Essentials of Glycobiology (Varki A., ed) 2nd Ed., pp. 89–99, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Séveno M., Bardor M., Paccalet T., Gomord V., Lerouge P., Faye L. (2004) Glycoprotein sialylation in plants? Nat. Biotechnol. 22, 1351–1353 [DOI] [PubMed] [Google Scholar]
- 14. Amano K., Chiba Y., Kasahara Y., Kato Y., Kaneko M. K., Kuno A., Ito H., Kobayashi K., Hirabayashi J., Jigami Y., Narimatsu H. (2008) Engineering of mucin-type human glycoproteins in yeast cells. Proc. Natl. Acad. Sci. U.S.A. 105, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamport D. T., Kieliszewski M. J., Chen Y., Cannon M. C. (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41, 81–94 [DOI] [PubMed] [Google Scholar]
- 17. Velasquez S. M., Ricardi M. M., Dorosz J. G., Fernandez P. V., Nadra A. D., Pol-Fachin L., Egelund J., Gille S., Harholt J., Ciancia M., Verli H., Pauly M., Bacic A., Olsen C. E., Ulvskov P., Petersen B. L., Somerville C., Iusem N. D., Estevez J. M. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403 [DOI] [PubMed] [Google Scholar]
- 18. Karnoup A. S., Turkelson V., Anderson W. H. (2005) O-Linked glycosylation in maize-expressed human IgA1. Glycobiology 15, 965–981 [DOI] [PubMed] [Google Scholar]
- 19. Pinkhasov J., Alvarez M. L., Rigano M. M., Piensook K., Larios D., Pabst M., Grass J., Mukherjee P., Gendler S. J., Walmsley A. M., Mason H. S. (2011) Plant Biotechnol. J. 9, 991–1001 [DOI] [PubMed] [Google Scholar]
- 20. Hieta R., Myllyharju J. (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor α-like peptides. J. Biol. Chem. 277, 23965–23971 [DOI] [PubMed] [Google Scholar]
- 21. Keskiaho K., Hieta R., Sormunen R., Myllyharju J. (2007) Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tiainen P., Myllyharju J., Koivunen P. (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280, 1142–1148 [DOI] [PubMed] [Google Scholar]
- 23. Daskalova S. M., Radder J. E., Cichacz Z. A., Olsen S. H., Tsaprailis G., Mason H., Lopez L. C. (2010) Engineering of N. benthamiana L. plants for production of N-acetylgalactosamine-glycosylated proteins. Toward development of a plant-based platform for production of protein therapeutics with mucin-type O-glycosylation. BMC Biotechnol. 10, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mitsui T., Kimura S., Igaue I. (1990) Isolation and characterization of Golgi membranes from suspension-cultured cells of rice (Oryza sativa L.) Plant Cell Physiol. 31, 15–25 [Google Scholar]
- 25. Shah M. M., Fujiyama K., Flynn C. R., Joshi L. (2003) Sialylated endogenous glycoconjugates in plant cells. Nat. Biotechnol. 21, 1470–1471 [DOI] [PubMed] [Google Scholar]
- 26. Joshi L., Lopez L. C. (2005) Bioprospecting in plants for engineered proteins. Curr. Opin. Plant Biol. 8, 223–226 [DOI] [PubMed] [Google Scholar]
- 27. Zeleny R., Kolarich D., Strasser R., Altmann F. (2006) Sialic acid concentrations in plants are in the range of inadvertent contamination. Planta 224, 222–227 [DOI] [PubMed] [Google Scholar]
- 28. Reaves M. L., Lopez L. C., Daskalova S. M. (2011) Noncanonical interactions between plant proteins and lectins cause false positives in lectin blots. Res. Plant Biol. 1, 49–54 [Google Scholar]
- 29. Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., Petersen B. L. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64, 439–451 [DOI] [PubMed] [Google Scholar]
- 30. Sainsbury F., Lomonossoff G. P. (2008) Extremely high level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 [DOI] [PubMed] [Google Scholar]
- 32. Odell J. T., Nagy F., Chua N. H. (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812 [DOI] [PubMed] [Google Scholar]
- 33. Pedersen J. W., Bennett E. P., Schjoldager K. T., Meldal M., Holmér A. P., Blixt O., Cló E., Levery S. B., Clausen H., Wandall H. H. (2011) Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J. Biol. Chem. 286, 32684–32696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A. (2006) Advancing uracil-excision based cloning towardan ideal technique for cloning PCR fragments. Nucleic Acids Res. 34, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Creuzenet C., Belanger M., Wakarchuk W. W., Lam J. S. (2000) Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275, 19060–19067 [DOI] [PubMed] [Google Scholar]
- 36. El Amrani A., Barakate A., Askari B. M., Li X., Roberts A. G., Ryan M. D., Halpin C. (2004) Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol. 135, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., Vanin E. F., Vignali D. A. (2004) Correction of multigene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 [DOI] [PubMed] [Google Scholar]
- 38. Bennett E. P., Hassan H., Mandel U., Mirgorodskaya E., Roepstorff P., Burchell J., Taylor-Papadimitriou J., Hollingsworth M. A., Merkx G., van Kessel A. G., Eiberg H., Steffensen R., Clausen H. (1998) Cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273, 30472–30481 [DOI] [PubMed] [Google Scholar]
- 39. Geu-Flores F., Nour-Eldin H. H., Nielsen M. T., Halkier B. A. (2007) USER fusion. A rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petersen B. L., Egelund J., Damager I., Faber K., Jensen J. K., Yang Z., Bennett E. P., Scheller H. V., Ulvskov P. (2009) Assay and heterologous expression in Pichia pastoris of plant cell wall type-II membrane-anchored glycosyltransferases. Glycoconj. J. 26, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tarp M. A., Sørensen A. L., Mandel U., Paulsen H., Burchell J., Taylor-Papadimitriou J., Clausen H. (2007) Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17, 197–209 [DOI] [PubMed] [Google Scholar]
- 42. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., Clausen H. (2010) O-Glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3. Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 285, 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berninsone P., Hirschberg C. B. (1998) Nucleotide sugars, nucleotide sulfate, and ATP transporters of the endoplasmic reticulum and Golgi apparatus. Ann. N.Y. Acad. Sci. 842, 91–99 [DOI] [PubMed] [Google Scholar]
- 44. Hirschberg C. B., Robbins P. W., Abeijon C. (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49–69 [DOI] [PubMed] [Google Scholar]
- 45. Berninsone P. M., Hirschberg C. B. (2000) Nucleotide sugar transporters of the Golgi apparatus. Curr. Opin. Struct. Biol. 10, 542–547 [DOI] [PubMed] [Google Scholar]
- 46. Segawa H., Kawakita M., Ishida N. (2002) Human and Drosophila UDP-galactose transporters transport UDP-N-acetylgalactosamine in addition to UDP-galactose. Eur. J. Biochem. 269, 128–138 [DOI] [PubMed] [Google Scholar]
- 47. Handford M., Rodriguez-Furlán C., Orellana A. (2006) Nucleotide-sugar transporters. Structure, function, and roles in vivo. Braz. J. Med. Biol. Res. 39, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 48. White T., Bennett E. P., Takio K., Sørensen T., Bonding N., Clausen H. (1995) Purification and cDNA cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 270, 24156–24165 [DOI] [PubMed] [Google Scholar]
- 49. Halpin C. (2005) Gene stacking in transgenic plants. The challenge for 21st century plant biotechnology. Plant Biotechnol. J. 3, 141–155 [DOI] [PubMed] [Google Scholar]
- 50. Donnelly M. L., Luke G., Mehrotra A., Li X., Hughes L. E., Gani D., Ryan M. D. (2001) Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip.” J. Gen. Virol. 82, 1013–1025 [DOI] [PubMed] [Google Scholar]
- 51. DeFrees S., Wang Z. G., Xing R., Scott A. E., Wang J., Zopf D., Gouty D. L., Sjoberg E. R., Panneerselvam K., Brinkman-Van der Linden E. C., Bayer R. J., Tarp M. A., Clausen H. (2006) GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology 16, 833–843 [DOI] [PubMed] [Google Scholar]
- 52. de Felipe P., Ryan M. D. (2004) Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic 5, 616–626 [DOI] [PubMed] [Google Scholar]
- 53. Gerngross T. U. (2004) Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 22, 1409–1414 [DOI] [PubMed] [Google Scholar]
- 54. Willer T., Valero M. C., Tanner W., Cruces J., Strahl S. (2003) O-Mannosyl glycans. From yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13, 621–630 [DOI] [PubMed] [Google Scholar]
- 55. Yip C. L., Welch S. K., Klebl F., Gilbert T., Seidel P., Grant F. J., O'Hara P. J., MacKay V. L. (1994) Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. U.S.A 91, 2723–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuroda K., Kobayashi K., Kitagawa Y., Nakagawa T., Tsumura H., Komeda T., Shinmi D., Mori E., Motoki K., Fuju K., Sakai T., Nonaka K., Suzuki T., Ichikawa K., Chiba Y., Jigami Y. (2008) Efficient antibody production upon suppression of O-mannosylation in the yeast Ogataea minuta. Appl. Environ. Microbiol. 74, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu L., Xu Y. X., Hirschberg C. B. (2010) The role of nucleotide sugar transporters in development of eukaryotes. Semin. Cell Dev. Biol. 21, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liang Y., Faik A., Kieliszewski M., Tan L., Xu W. L., Showalter A. M. (2010) Identification and characterization of in vitro galactosyltransferase activities involved in arabinogalactan-protein glycosylation in tobacco and Arabidopsis. Plant Physiol. 154, 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kieliszewski M. J., Lamport D. T., Tan L., Cannon M. C. (2010) in Annual Plant Reviews: Plant Polysaccharides, Biosynthesis and Bioengineering (Ulvskov P., ed) pp. 321–342, Wiley-Blackwell, Oxford [Google Scholar]
- 60. Springer G. F. (1984) T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 [DOI] [PubMed] [Google Scholar]
- 61. Sabbatini P. J., Ragupathi G., Hood C., Aghajanian C. A., Juretzka M., Iasonos A., Hensley M. L., Spassova M. K., Ouerfelli O., Spriggs D. R., Tew W. P., Konner J., Clausen H., Abu Rustum N., Dansihefsky S. J., Livingston P. O. (2007) Pilot study of a heptavalent vaccine-keyhole limpet hemocyanin conjugate plus QS21 in patients with epithelial ovarian, fallopian tube, or peritoneal cancer. Clin. Cancer Res. 13, 4170–4177 [DOI] [PubMed] [Google Scholar]
- 62. Sørensen A. L., Reis C. A., Tarp M. A., Mandel U., Ramachandran K., Sankaranarayanan V., Schwientek T., Graham R., Taylor-Papadimitriou J., Hollingsworth M. A., Burchell J., Clausen H. (2006) Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16, 96–107 [DOI] [PubMed] [Google Scholar]
- 63. Blixt O., Bueti D., Burford B., Allen D., Julien S., Hollingsworth M., Gammerman A., Fentiman I., Taylor-Papadimitriou J., Burchell J. M. (2011) Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 13, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strasser R., Stadlmann J., Schähs M., Stiegler G., Quendler H., Mach L., Glössl J., Weterings K., Pabst M., Steinkellner H. (2008) Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 6, 392–402 [DOI] [PubMed] [Google Scholar]
- 65. Castilho A., Strasser R., Stadlmann J., Grass J., Jez J., Gattinger P., Kunert R., Quendler H., Pabst M., Leonard R., Altmann F., Steinkellner H. (2010) In planta protein sialylation through overexpression of the respective mammalian pathway. J. Biol. Chem. 285, 15923–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nagels B., Van Damme E. J., Pabst M., Callewaert N., Weterings K. (2011) Production of complex multiantennary N-glycans in Nicotiana benthamiana plants. Plant Physiol. 155, 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schoberer J., Strasser R. (2011) Subcompartmental organization of Golgi resident N-glycan processing enzymes in plants. Mol. Plant 4, 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaulfürst-Soboll H., Rips S., Koiwa H., Kajiura H., Fujiyama K., von Schaewen A. (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J. Biol. Chem. 286, 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yin B. J., Gao T., Zheng N. Y., Li Y., Tang S. Y., Liang L. M., Xie Q. (2011) Generation of glyco-engineered BY2 cell lines with decreased expression of plant-specific glycoepitopes. Protein Cell 2, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pogue G. P., Vojdani F., Palmer K. E., Hiatt E., Hume S., Phelps J., Long L., Bohorova N., Kim D., Pauly M., Velasco J., Whaley K., Zeitlin L., Garger S. J., White E., Bai Y., Haydon H., Bratcher B. (2010) Production of pharmaceutical-grade recombinant aprotinin and a monoclonal antibody product using plant-based transient expression systems. Plant Biotechnol. J. 8, 638–654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.