Background: The lack of a known phosphoserine phosphatase (PSP) gene has been a missing link in the serine biosynthetic pathway for a number of organisms.
Results: Proteins with PSP activity were purified from a bacterium that lacks a classical-type PSP.
Conclusion: Some cofactor-dependent phosphoglycerate mutase homologs function as novel-type PSPs.
Significance: This finding will help to identify the missing link in the serine biosynthetic pathway.
Keywords: Amino Acid, Bacteria, Metabolism, Phosphatase, Serine, Autotroph, Histidine Phosphatase Superfamily, Phosphoglycerate Mutase, Phosphoserine Phosphatase
Abstract
Phosphoserine phosphatase (PSP) catalyzes the dephosphorylation of phosphoserine to serine and inorganic phosphate. PSPs, which have been found in all three domains of life, belong to the haloacid dehalogenase-like hydrolase superfamily. However, certain organisms, particularly bacteria, lack a classical PSP gene, although they appear to possess a functional phosphoserine synthetic pathway. The apparent lack of a PSP ortholog in Hydrogenobacter thermophilus, an obligately chemolithoautotrophic and thermophilic bacterium, represented a missing link in serine anabolism because our previous study suggested that serine should be synthesized from phosphoserine. Here, we detected PSP activity in cell-free extracts of H. thermophilus and purified two proteins with PSP activity. Surprisingly, these proteins belonged to the histidine phosphatase superfamily and had been annotated as cofactor-dependent phosphoglycerate mutase (dPGM). However, because they possessed neither mutase activity nor the residues important for the activity, we defined these proteins as novel-type PSPs. Considering the strict substrate specificity toward l-phosphoserine, kinetic parameters, and PSP activity levels in cell-free extracts, these proteins were strongly suggested to function as PSPs in vivo. We also detected PSP activity from “dPGM-like” proteins of Thermus thermophilus and Arabidopsis thaliana, suggesting that PSP activity catalyzed by dPGM-like proteins may be distributed among a broad range of organisms. In fact, a number of bacterial genera, including Firmicutes and Cyanobacteria, were proposed to be strong candidates for possessing this novel type of PSP. These findings will help to identify the missing link in serine anabolism.
Introduction
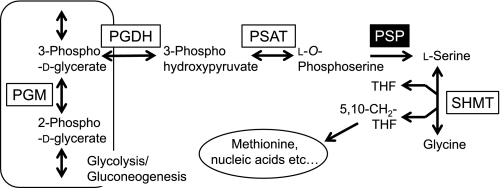
Serine biosynthesis is an important anabolic process for most organisms, particularly autotrophs, in which it is clearly essential. At least two serine biosynthetic reactions are known to function in vivo (Fig. 1) (1–3). The first involves the synthesis of serine from glycine and 5,10-methylenetetrahydrofolate, and is catalyzed by serine hydroxymethyltransferase (SHMT2; EC 2.1.2.1). This reaction is used by methanotrophic and methylotrophic bacteria for one-carbon assimilation (termed the serine pathway) (4). Higher plants also synthesize a portion of utilized serine from glycine (2). Because the SHMT reaction is reversible, many organisms also use SHMT for producing glycine from serine. A second serine biosynthetic reaction is catalyzed by phosphoserine phosphatase (PSP; EC 3.1.3.3), which forms serine from l-O-phosphoserine. This serine synthetic pathway, which functions in microorganisms (4) and higher plants (2), involves three steps with dedicated enzymes: 3-phosphoglycerate dehydrogenase (PGDH; EC 1.1.1.95), 3-phosphoserine aminotransferase (PSAT; EC 2.6.1.52), and PSP.
FIGURE 1.
General serine anabolic pathways. PGM, phosphoglycerate mutase; THF, tetrahydrofolate.
All known PSPs are Mg2+-dependent enzymes belonging to the haloacid dehalogenase-like hydrolase superfamily (referred to here as metal-dependent PSP (dPSP)) (5–7) and are distributed among the three domains of life: Bacteria, Archaea, and Eukarya. However, certain microorganisms lack orthologs of dPSP, although they possess those of PGDH and PSAT, according to the annotation of the Microbial Genome Database for Comparative Analysis (MBGD) (available on the World Wide Web), as shown in Table 1. In addition, it has been reported that although cyanobacteria lack a gene encoding dPSP, serine synthesis via phosphoserine is the predominant route in these organisms. PSP activity was also detected from the cell-free extracts (CFE) of cyanobacteria (8, 9). These findings suggest the possibility that some organisms may possess novel types of PSP involved in serine anabolism that are non-orthologous to dPSP.
TABLE 1.
Occurrence of orthologs involved in serine biosynthesis from 3-phosphoglycerate in Bacteria and Archaea
One strain was selected from each genus in the default mode of the MBGD, and the number of organisms analyzed within a phylum is given in parentheses. The numbers in brackets are the MBGD ID used for the analysis.
| Organism | PGDH [667] | PSAT [361] | PSP [1707] |
|---|---|---|---|
| Bacteria | |||
| Acidobacteria (3) | 3 | 3 | 1 |
| Actinobacteria (56) | 48 | 51 | 39 |
| Aquificae (6) | 6 | 6 | 0 |
| Bacteroifetes (27) | 19 | 25 | 16 |
| Chlamydiae (4) | 1 | 1 | 1 |
| Chlorobi (5) | 2 | 4 | 5 |
| Chloroflexi (7) | 7 | 7 | 0 |
| Cyanobacteria (12) | 12 | 12 | 0 |
| Deferribacteries (3) | 3 | 3 | 0 |
| Deinococcus-Thermus (5) | 5 | 5 | 0 |
| Dictyoglomi (1) | 1 | 1 | 0 |
| Elusimicrobia (2) | 1 | 2 | 0 |
| Fibrobacteres (1) | 0 | 1 | 0 |
| Firmicutes (51) | 35 | 42 | 5 |
| Fusobacteria (5) | 3 | 3 | 0 |
| Gemmatimonadetes (1) | 1 | 1 | 1 |
| Nitrospirae (2) | 2 | 2 | 0 |
| Planctomycetes (3) | 3 | 3 | 1 |
| Proteobacteria (204) | 158 | 194 | 166 |
| Spirochaetes (5) | 3 | 3 | 2 |
| Synergistetes (2) | 0 | 2 | 0 |
| Tenergistetes (5) | 0 | 0 | 0 |
| Thermotogae (5) | 5 | 5 | 0 |
| Verrucomicrobia (4) | 4 | 4 | 0 |
| Archaea | |||
| Crenarchaeota (15) | 11 | 14 | 10 |
| Euryarchaeota (39) | 38 | 39 | 32 |
| Korarchaeota (1) | 1 | 0 | 1 |
| Nanoarchaeota (1) | 0 | 0 | 0 |
| Thaumarchaeota (2) | 2 | 2 | 2 |
Hydrogenobacter thermophilus TK-6 is a thermophilic and hydrogen-oxidizing bacterium belonging to the order Aquificales (10, 11). Because H. thermophilus is an obligate chemolithoautotroph that assimilates CO2 through the reductive tricarboxylic acid cycle (12), it must possess serine biosynthetic ability. Evidence for the existence of a phosphoserine anabolic pathway in this bacterium has also been provided from the annotation of its complete genome sequence (13), which indicates that 3-phosphoglycerate can be synthesized by gluconeogenesis (Embden-Meyerhof-Parnas pathway) and that orthologs of PGDH and PSAT exist. Furthermore, PSAT activity has also been detected biochemically (14), and we confirmed the existence of phosphoserine by metabolome analysis.3 However, because H. thermophilus and other members of Aquificales lack an ortholog of dPSP (Table 1), we hypothesized that H. thermophilus may possess a novel type of PSP.
In the present study, we detected, purified, and characterized two novel PSPs from H. thermophilus. Because neither enzyme requires a metal ion for its activity and both belong to the histidine phosphatase superfamily, they were designated as metal-independent PSPs (iPSPs). The histidine phosphatase superfamily consists of proteins with considerably diverse functions, catalyzing reactions involving phosphatase or phosphomutase activities (15). Although the iPSPs identified here showed considerable primary structural similarity to cofactor-dependent phosphoglycerate mutase (dPGM; EC 5.4.2.1), which catalyzes the interconversion of 3-phosphoglycerate and 2-phosphoglycerate using 2,3-diphosphoglycerate as a cofactor, they lacked dPGM activity. To our knowledge, this represents the first report of dPGM-related proteins functioning as novel-type PSPs. The identification of novel PSPs may help to reveal the missing link in serine biosynthetic metabolism in other organisms. Further characterization of these PSPs may allow prediction of the role of histidine phosphatase superfamily proteins with unknown function.
EXPERIMENTAL PROCEDURES
Bacterial Strain and Growth Conditions
H. thermophilus TK-6 (IAM 12695, DSM6534) was cultivated in an inorganic medium at 70 °C under a gas phase of 75% H2, 10% O2, and 15% CO2. The composition of the medium was as described previously (16), except that 3 g liter−1 of (NH4)2SO4 was added in place of NH4Cl. Precultivation was performed using two 5-liter shaking flasks containing 1 liter of medium each. After ∼40 h, 2 liters of preculture was transferred into a 10-liter jar fermentor (Takasaki Kagaku Kikai Co.) containing 6 liters of fresh medium. The fermentor was continuously stirred at 1000 rpm and gassed with the above-described gas phase at 1.0–1.5 liters min−1. Cells were grown to the logarithmic phase (optimal density at 540 nm (A540) ≈ 2), harvested by centrifugation, washed once in 20 mm Tris-HCl buffer (pH 8.0), and then stored at −80 °C until needed for use.
Enzyme Assays
PSP activity was assayed by measuring the production of inorganic phosphate (17, 18). The reaction mixture contained 20 mm HEPES-NaOH (pH 8.0 at room temperature), 5 mm dl-O-phosphoserine, 1 mm mercaptoethanol, and an enzyme solution in a total volume of 50 μl. If necessary, 1 mm EDTA was added. The reaction mixture was routinely incubated at 70 °C for 5–20 min; however, when protein from Arabidopsis was used, the reaction mixture was incubated at 25 °C. The reaction was stopped by placing the tube in ice-cold water, followed by the addition of 450 μl of ferrous sulfate solution (0.008 g of FeSO4·7H2O in 1 ml of 7.5 mm H2SO4) and 37.5 μl of ammonium molybdate solution (0.066 g of (NH4)6Mo7O24·4H2O in 1 ml of 3.75 m H2SO4). After precipitated proteins were removed by centrifugation, the inorganic phosphate concentration was determined by measuring absorbance at 660 nm. A KH2PO4 solution was used for calibration. One unit of activity was defined as the amount of enzyme producing 1 μmol of inorganic phosphate/min.
dPGM activity was assayed spectrophotometrically (19). Reaction mixture contained 50 mm HEPES-NaOH (pH 7.5), 2.0 mm MgCl2, 1 unit ml−1 pyruvate kinase (from rabbit muscle; Sigma), 1 unit ml−1 l-lactate dehydrogenase (from rabbit muscle; Roche Applied Science), 1 unit ml−1 enolase (from Yeast; Wako), 0.5 mm ADP, 0.15 mm NADH, 5.0 mm 3-phosphoglyceric acid, 0.1 mm 2,3-diphosphoglyceric acid, and 2 μg ml−1 purified H. thermophilus PSPs. The reaction mixture was incubated at 37 °C.
Purification of native H. thermophilus PSPs
H. thermophilus PSPs were purified from 40 g of wet cells as follows. The cells were suspended in 120 ml of 20 mm Tris-HCl buffer (pH 8.0; buffer A) and disrupted by sonication, and cell debris was then removed by centrifugation at 100,000 × g for 1 h. The supernatant, designated CFE, was applied to a DE52 open column (25 × 15 cm; Whatman, Brentford, UK) equilibrated with buffer A. After the elution of bound proteins with buffer A containing 1 m NaCl, ammonium sulfate was added to the obtained fractions to give 30% saturation, and the samples were then applied to a Butyl-Toyopearl column (22 mm × 15 cm; Tosoh, Tokyo, Japan) equilibrated with buffer A supplemented with ammonium sulfate at 30% saturation. This and subsequent chromatography steps were performed using an ÄKTA purifier system (GE Healthcare) at room temperature. Proteins were eluted with a gradient of ammonium sulfate from 30 to 0% at a flow rate of 4 ml min−1. The active fractions were dialyzed against buffer A and then applied to a DEAE-Toyopearl column (22 mm × 15 cm; Tosoh) equilibrated with the same buffer. Proteins were eluted with a gradient of NaCl from 0 to 1 m at a flow rate of 4 ml min−1. The active fractions were diluted 5-fold in buffer A and were then applied to a MonoQ HR 5/5 column (bed volume, 1 ml; GE Healthcare) equilibrated with buffer A. Proteins were eluted with a gradient of NaCl from 0 to 300 mm at a flow rate of 0.5 ml min−1. The active fractions were pooled, concentrated using ultrafiltration spin columns (Vivaspin 2 ml, 10 kDa; Sartorius, Göttingen, Germany) and loaded onto a Superdex 75 (10/300) column (GE Healthcare). Proteins were eluted with buffer A supplemented with 150 mm NaCl.
Construction of Expression Plasmids
pspA (HTH0103) and pspB (HTH0183) were PCR-amplified from H. thermophilus genomic DNA using the following primers: 5′-GTAGAATTCGGGCCATGGTAAAGC-3′ (PspA forward), 5′-TATCTGCAGAAATGAGGAAAAGCG-3′ (PspA reverse), 5′-AAAGAATTCATATGAAGCGTTTGTATTTGGTCAG-3′ (PspB forward), and 5′-ACAGTCGACGCTCGAGAAAATCAGGTAAGC-3′ (PspB reverse). The amplified fragments were first inserted into pUC19 plasmid using the EcoRI and PstI or SalI restriction sites introduced in the primers (single-underlined sequences). After the sequences of the inserts were confirmed, pspA and pspB were reinserted into the multicloning site 1 of pCDFDuet-1 or pET21c vector (Novagen) using NcoI and PstI or NdeI and XhoI (double-underlined sequences in the primers) restriction sites, respectively. The pET11 expression plasmid for Thermus thermophilus HB8 TTHA0368 (RDB6078) and cDNAs of Arabidopsis thaliana At3g05170 and At5g04120 (pda20245 and pdz61570, respectively) were provided by the RIKEN BRC through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan (20–22). Because At3g05170 contained an original NdeI site, a silent mutation was introduced into the site (His: CAT → CAC) as follows. The primers sets 5′-GTTTTCGAATTCCATATGAGTCCGGAC-3′ and 5′-CATGTGGTCGAAGAAATCACC-3′, 5′-TGATTTCTTCGACCACATGGC-3′ and 5′-TTTGTCGACTAATCTTCAGTGGCCTCAGTC-3′ were first used to amplify the upstream and downstream regions of the NdeI site, which was mutated at the end of each amplified fragment, from pda20245 by PCR. The primers 5′-GTTTTCGAATTCCATATGAGTCC-3′ and 5′-TTTGTCGACTAATCTTCAGTGGC-3′ were then used to amplify the complete gene containing the silent mutation using the two PCR fragments as template. The amplified fragment was inserted into pUC19 using EcoRI and SalI restriction sites introduced in the primers (single-underlined sequences). After the sequence was confirmed to be correct, At3g05170 was reinserted into pET21c plasmid using NdeI (double-underlined sequence in the primer) and SalI. Because sequence information in the 3′-terminal region of pdz61570 was unclear, we first sequenced the cDNA and confirmed that it contained the full-length sequence of At5g04120. At5g04120 was PCR-amplified using the primers 5′-GATCATATGGGTCATGAATGGATCG-3′ and 5′-GCCAAGCCCTAGAGAATTCGGTACCTCA-3′ and then inserted directly into pET21c using NdeI and EcoRI sites (underlined nucleotides). The inserted sequence was also confirmed to be free of errors.
Heterologous Protein Expression and Purification
E. coli BL21-Codon-Plus (DE3)-RIL was used for the individual and co-expression of pspA and pspB. E. coli BL21 (DE3) was used for the expression of TTHA0368, At3g05170, and At5g04120. The hosts transformed with the expression plasmids were inoculated into Luria-Bertani medium containing the appropriate antibiotic(s): 50 μg ml−1 streptomycin, 50 μg ml−1 ampicillin, and/or 34 μg ml−1 chloramphenicol. After cultivating the cells aerobically at 37 °C until the A600 reached ∼0.6, the induction of H. thermophilus PSP1, H. thermophilus PSP2, and TTHA0368-coded proteins was performed by the addition of 1 mm isopropyl thio-β-d-galactopyranoside to the medium, followed by further cultivation for 3 h at 37 °C. The expression of Arabidopsis proteins was induced with 1 mm isopropyl thio-β-d-galactopyranoside for 5 h at 30 °C. The harvested cells were resuspended in buffer A supplemented with 1 mm EDTA (4 ml per 1 g of wet cells), sonicated, and subjected to centrifugation, as described above, to obtain supernatants. H. thermophilus PSPs and T. thermophilus protein were purified from the supernatants as follows. The supernatants were first heat-treated at 80 °C for 10 min and centrifuged at 100,000 × g for 1 h, and the resulting supernatants were further purified using Butyl-Toyopearl and MonoQ columns, as described above.
N-terminal Amino Acid Sequencing
The N-terminal amino acid sequences of purified proteins were determined using a Procise 49X-cLC protein sequencer (Applied Biosystems) from a blotted PVDF membrane (0.2 μm; Bio-Rad).
Protein Assay
Protein concentrations were measured using a Bio-Rad protein assay dye (catalogue no. 500-0006). Bovine serum albumin was used as a standard.
Gel Filtration
For the estimation of molecular mass, gel filtration was performed using a Superdex 75 (10/300) column equilibrated with buffer A supplemented with 150 mm NaCl at a flow rate of 1 ml min−1. Gel filtration standard (Bio-Rad) was used as a molecular marker for calibration. Measurements for standards and samples were performed in triplicate and duplicate, respectively.
Phylogenetic Tree Construction
Evolutionary analyses were conducted using MEGA version 5.05 (23). Amino acid sequences of branch 1 and 2 were aligned separately using the MUSCLE program (24) and were then manually realigned according to the structure-based alignment presented by Rigden (15). After the removal of gap regions, phylogenetic trees were constructed by the maximum likelihood method based on the Whelan and Goldman + Freq. model (25), using the nearest neighbor interchange as the heuristic. The bootstrap was performed with 1000 replicates. A discrete γ distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 4.5538)).
RESULTS
Detection and Purification of Proteins with PSP Activity in H. thermophilus
In a CFE obtained from log phase H. thermophilus cells, we detected 0.090 unit mg−1 PSP activity. Although known dPSPs require Mg2+ for their activity and are inhibited up to 80% in the presence of 5 mm Ca2+ or Mn2+ (5), the PSP activity in H. thermophilus CFE was neither enhanced by the addition of Mg2+ nor inhibited by the presence of 10 mm EDTA. In addition, inhibition by Ca2+ or Mn2+ was not observed. Thus, the identification of proteins with PSP activity from H. thermophilus was performed by measuring the release of free phosphate from dl-phosphoserine in the absence of added metals.
The PSP activity was separated into two distinct fractions following the Butyl-Toyopearl column purification step (Table 2). One fraction was eluted under ∼15% saturated conditions (protein I), and the other was eluted under ∼0% saturated conditions (protein II). Notably, the two proteins were purified separately using the same procedure. From 40 g of wet cells, 3 μg of protein I and 7 μg of protein II were obtained (Table 2). Both proteins were also purified from late log to stationary phase cells at equal levels and ratios (data not shown).
TABLE 2.
Purification of PSPs from H. thermophilus
40 g of wet cells were subjected to purification.
| Enzyme | Fraction | Activity | Protein | Specific activity | Purification | Yield |
|---|---|---|---|---|---|---|
| units | mg | units/mg | -fold | % | ||
| iPSP1 | CFEa | 195 | 2176 | 0.090 | 1.0 | 100 |
| DE52a | 150 | 1120 | 0.134 | 1.5 | 77 | |
| Butyl-Toyopearl | 82 | 172 | 0.48 | 5.3 | 42 | |
| DEAE-Toyopearl | 54 | 13.9 | 3.9 | 44 | 28 | |
| MonoQ | 2.5 | 0.081 | 31 | 341 | 1.3 | |
| Superdex 75 | 0.17 | 0.003 | 58 | 644 | 0.1 | |
| iPSP2 | CFEa | 195 | 2176 | 0.090 | 1.0 | 100 |
| DE52a | 150 | 1120 | 0.134 | 1.5 | 77 | |
| Butyl-Toyopearl | 73 | 117 | 0.62 | 6.9 | 37 | |
| DEAE-Toyopearl | 43 | 17. 8 | 2.4 | 27 | 22 | |
| MonoQ | 4.2 | 0.192 | 22 | 242 | 2.1 | |
| Superdex 75 | 0.20 | 0.007 | 28 | 318 | 0.1 |
a H. thermophilus PSP1 and H. thermophilus PSP2 were purified simultaneously because they could not be separated until they were eluted from the Butyl-Toyopearl column.
Identification of Purified Proteins as Novel Type PSPs
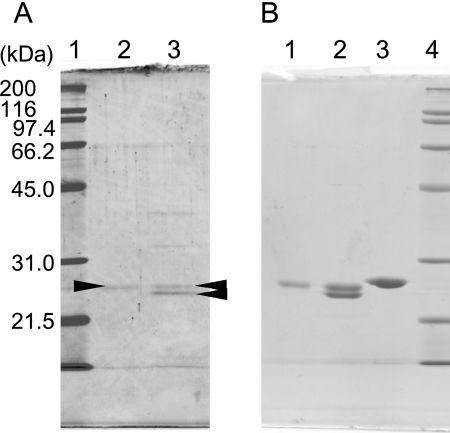
SDS-PAGE of purified protein I gave one main 24.3 kDa band, whereas protein II gave two main bands (24.0 and 24.5 kDa) (Fig. 2A). N-terminal amino acid sequences were determined from these bands, revealing that the sequences of protein I and the larger band of protein II were identical (MVKLILVRHA). Because only one gene encoding the identical N-terminal sequence was identified in the H. thermophilus genome (HTH0103) and the molecular masses of the protein bands were almost the same, we concluded that they were the identical protein. The sequence of the smaller band of protein II was determined to be MKRLYLVRHA, which agreed with the sequence of HTH0183-coded protein. Surprisingly, both HTH0103 and HTH0183 had been annotated as dPGM genes and displayed 37% identity with each other, as determined by a protein BLAST search of the National Center for Biotechnology Information (NCBI) database. The HTH0103- and HTH0183-encoded proteins were confirmed to belong the histidine phosphatase superfamily because the predicted sequence of each protein contained residues corresponding to the conserved catalytic core of this superfamily (Arg-8, His-9, Arg-58, and His-150; HTH0103 numbering) (15). In contrast, none of the residues that are reported to be important in substrate binding for dPGMs (Arg-90, Tyr-92, Arg-116, Arg-117, and Asn-186; E. coli dPGM coding numbers) (26, 27) were conserved in the HTH0103- or HTH0183-encoded proteins (supplemental Fig. S1). In fact, neither protein I nor protein II displayed dPGM activity. In addition, both proteins lacked clear primary structural similarities to dPSPs (the identity with E. coli PSP was less than 10%), which belong to the haloacid dehalogenase-like hydrolase superfamily. Therefore, we concluded that purified proteins I and II were novel-type PSPs and named them iPSP1 and iPSP2, respectively. In addition, the proteins encoded by HTH0103 and HTH0183 were named PspA and PspB, respectively.
FIGURE 2.
12% SDS-PAGE analysis of purified native iPSPs (A), heterologously expressed H. thermophilus PSPs, and an iPSP homolog from T. thermophilus (B). A, lane 1, molecular mass markers; lane 2, 0.2 μg of purified protein I (htPSP1); lane 3, 0.5 μg of purified protein II (htPSP2). The arrowheads indicate the bands corresponding to proteins I and II. B, heterologously expressed and purified proteins. Lane 1, 2 μg of purified protein I; lane 2, 2 μg of purified protein II; lane 3, 2 μg of TTHA0368-coded protein; lane 4, molecular mass markers.
Heterologous Expression of iPSP1 and iPSP2
pspA and pspB were cloned into the expression vectors pCDFDuet-1 and pET21c, respectively, and overexpressed both individually and simultaneously in E. coli. The expression was checked using SDS-PAGE and N-terminal sequencing. When PspA was expressed solely, it was obtained in the soluble fraction, and PSP activity was detected. In contrast, the overexpression of PspB alone resulted in insoluble inclusion body formation, and no phosphatase activity toward phosphoserine or para-nitrophenyl phosphate was detected in the soluble fraction. When PspA and PspB were expressed simultaneously within the same cells, however, both proteins were obtained in the soluble fraction. Approximately 90% of the total PSP activity was present in fractions eluted under 0% saturated conditions from Butyl-Toyopearl columns, similar to the elution profile of iPSP2, whereas the remaining 10% of PSP activity was found in fractions eluted under similar conditions as iPSP1. This latter fraction contained only PspA. The purified proteins (Fig. 2B) exhibited PSP activity, confirming that iPSP1 consists of PspA and iPSP2 consists of both PspA and PspB subunits.
Subunit Composition of iPSP1 and iPSP2
The molecular masses of PspA and PspB calculated from the deduced amino acid sequences were 24.6 and 23.5 kDa, respectively. The calculated masses were consistent with those estimated by SDS-PAGE. The native molecular masses of iPSP1 and iPSP2 were estimated to be 46 and 37 kDa, respectively, using a Superdex 75 column (Table 3), suggesting that they formed dimers.
TABLE 3.
Subunit compositions, molecular masses, and kinetic parameters for l-phosphoserine of H. thermophilus PSPs
| Annotation | Subunit | Calculated molecular mass |
Kinetic parameter (l-phosphoserine) |
|||
|---|---|---|---|---|---|---|
| SDS-PAGE | Amino acid sequence | Gel filtration | Km | Vmax | ||
| kDa | mm | units mg−1 | ||||
| iPSP1 | HTH0103 (PspA) | 24.3 | 24.6 | 46 | 1.6 ± 0.3 | 56 ± 3 |
| iPSP2 | HTH0103 (PspA) | 24.5 | 24.6 | 37 | 1.5 ± 0.2 | 32 ± 1 |
| HTH0183 (PspB) | 24.0 | 23.5 | ||||
Enzymatic Characterization of iPSPs
The Km values of native iPSP1 and iPSP2 for l-phosphoserine were nearly identical (1.6 ± 0.3 and 1.5 ± 0.2 mm, respectively) (supplemental Fig. S2 and Table 3). In contrast, the Vmax of iPSP2 (32 ± 1 units mg−1) was nearly half of that of iPSP1 (56 ± 3 units mg−1), suggesting that the PspB subunit has no PSP activity or has PSP activity considerably lower than that of PspA. Heterologously expressed iPSPs showed Km values equivalent to those of the native proteins, but the estimated Vmax values were ∼2-fold higher than those of native proteins (116 ± 7 units mg−1 for iPSP1 and 46 ± 2 units mg−1 for iPSP2). This difference may be attributable to the inactivation of enzymes during purification.
Substrate specificity was also examined using native iPSPs, as shown in Table 4. The two iPSPs displayed strict substrate specificity for l-phosphoserine compared with human brain cell dPSP, which also has been reported to show considerable amounts of phosphatase activity toward d-phosphoserine and fructose 6-phosphate (5). iPSP2 showed slightly higher activity to p-nitrophenyl phosphate (4 units mg−1) than iPSP1 (<1 unit mg−1). Although this activity may be catalyzed by the PspB subunit, it is possible that binding to PspB increases the p-nitrophenyl phosphate phosphatase activity of PspA. Heterologously expressed iPSP1 and iPSP2 also showed the same specificities.
TABLE 4.
Substrate specificities of PSPs and their relative proteins
In our study, reaction mixture contains 5 mm substrate, 1 mm EDTA, and 1 mm mercaptoethanol. l-Ser(P), l-phosphoserine; d-Ser(P), d-phosphoserine; l-Tyr(P), l-phosphotyrosine; dl-Thr(P), dl-phosphothreonine; 3PGA, 3-phosphoglyceric acid; pNPP, para-nitrophenylphosphate; F6P, fructose 6-phosphate, R5P, ribose 5-phosphate; HAD, haloacid dehalogenase; NSAP, nonspecific acid phosphohydrolase.
| Organism | Annotation | Protein |
Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| l-Ser(P) | d-Ser(P) | l-Tyr(P) | dl-Thr(P) | 3PGA | pNPP | AMP | F6P | R5P | |||
| units/mg | |||||||||||
| HAD-like hydrolase superfamily | |||||||||||
| Human (brain) | dPSP | 36.72 | 23.87 | 14.69 | 1.06 | 12.96 | Ref. 5 | ||||
| E. coli | dPSP (SerB) class B | 113 | 0.25 | 14.7 | <0.05 | Ref. 28 | |||||
| E. coli | NSAP (AphA) | 0.49 | 8.95 | 0.37 | 12.53 | 1 | 0.53 | Ref. 40 | |||
| Histidine phosphatase superfamily | |||||||||||
| H. thermophilus | iPSP1 | 41 | 1 | 4 | <1 | <1 | <1 | <1 | This study | ||
| H. thermophilus | iPSP2 | 20 | 1 | 1 | <1 | 4 | <1 | <1 | This study | ||
| T. thermophilus | TTHA0368 | 0.5 | 0.4 | 2.7 | 2.7 | 0.5 | <0.2 | 0.3 | This study | ||
| B. stearothermophilus | PhoE (YhfR) | 27 | 16 | 5 | 1 | 0.3 | Ref. 26 | ||||
| A. thalianaa | At5g04120 | 0.087 | 0.079 | <0.006 | This study | ||||||
a At5g04120-overexpressed E. coli CFE was used for the analysis.
Phylogenetic Position
The order Aquificales consists of three families: Aquificaceae, Hydrogenothermaceae, and Desulfurobacteriaceae. According to the 16S rRNA-based phylogenetic tree, Desulfurobacteriaceae first diverged from the common ancestor, followed by the separation of Aquificaceae and Hydrogenothermaceae into different families (29). Protein BLAST searches revealed that all of the Aquificales organisms whose complete genomes are available possess putative pspA orthologs (the identities were ≥40% against PspA of H. thermophilus), whereas only members of Aquificaceae and Hydrogenothermaceae possess putative pspB orthologs. All identified organisms were found to possess a single copy of each gene, except for Hydrogenivirga sp. 128-5-R1-1, which had two putative pspA orthologs (pspA and pspA2). In addition to putative pspA and pspB, Hydrogenobaculum sp. Y04AAS1 and all Hydrogenothermaceae species contained genes that were also annotated as dPGM. These genes were thought to encode true dPGM proteins because they all contained conserved residues that are considered to be essential for substrate binding.
Phylogenetic analysis was performed using the maximum likelihood method with the dPGM-related proteins of Aquificales and dPGMs whose activities have been confirmed biochemically (supplemental Fig. S3). Five proteins belonging to branch 2 of the histidine phosphatase superfamily were used as outgroups because dPGM and its related proteins belong to branch 1 (15). (Putative) PspAs and PspBs of Aquificales were placed in a different branch from true dPGMs. Because dPGMs are distributed among the three domains, the ancestor of iPSPs may have separated from the ancestor of dPGMs prior to the division of organisms into three domains. In addition, the separation of PspA and PspB on different branches suggested that these two proteins diverged from ancestral Psp after the separation of Desulfurobacteriaceae and before the separation of Aquificaceae and Hydrogenothermaceae. We predict that psp orthologs of Desulfurobacteriaceae possess PSP activity similar to PspA of H. thermophilus because of their higher primary structure similarity to PspA than PspB, and the physiological significance of PSP activity. For these reasons, we classified psp orthologs of Desulfurobacteriaceae as pspA.
Detection of PSP Activity among dPGM-related Proteins from Other Organisms
The distribution of the novel iPSPs identified here is of interest because many organisms possess genes encoding dPGM-related proteins with unknown physiological roles (26, 30, 31). Three proteins with significant homology to dPGMs but lacking conserved residues involved in substrate binding, similar to the case of PspA and PspB, were overexpressed in E. coli, and their PSP activity was evaluated.
The first examined protein was from T. thermophilus, a heterotrophic and thermophilic bacterium whose optimal growth temperature is around 70 °C. TTHA0368-coded protein which displayed the highest similarity (∼30% identity) to PspA of H. thermophilus, was expressed in the soluble fraction and successfully purified (Fig. 2B). The native mass of the protein was 41 kDa. Considering that the deduced amino acid sequence was calculated to be 23.6 kDa, the protein probably comprises a homodimer. Phosphatase activity toward various substrates was analyzed using the purified protein under the same conditions used to evaluate H. thermophilus PSPs (Table 4). The protein encoded by TTHA0368 showed low but distinct phosphatase activity toward l-phosphoserine (0.5 unit mg−1), and exhibited substrate specificity clearly distinct from that of H. thermophilus PSPs; it showed higher activity for dl-phosphothreonine and 3-phosphoglyceric acid than for phosphoserine.
The other two evaluated dPGM-related proteins were from A. thaliana, one of the most well studied higher plants. Although both At3g05170- and At5g0412-coded proteins were expressed in the soluble fraction, only the CFE from E. coli expressing At5g0412 product showed clear PSP activities (0.087 and 0.079 unit mg−1 for l- and d-phosphoserine, respectively) (Table 4) in a reaction mixture containing 1 mm EDTA and no Mg2+, which inhibits the dPSP activity of E. coli. The activities of At3g05170-coded protein were below the limit of detection (<0.006 unit mg−1).
DISCUSSION
In this study, we purified and characterized novel-type PSPs, which were previously annotated as dPGM, from H. thermophilus. Considering the PSP activity level in the CFE from H. thermophilus and the kinetic parameters, the two identified iPSPs in H. thermophilus may actually work as PSP in vivo, although further studies will be needed to confirm it. dPGM-like proteins have been reported to exist in a broad range of organisms and a few such proteins actually possess quantifiable phosphatase activity (26, 30, 31). For instance, Bacillus stearothermophilus PhoE (originally termed YhfR) shows phosphatase activity against a wide range of substrates (Table 4), with the highest activities toward 3-phosphoglyceric acid (27 units mg−1) and α-napthyl phosphate (21 units mg−1), although it lacks detectable dPGM activity (26). To date, however, the physiological role of dPGM-like proteins has not been conclusively determined. Therefore, the findings presented here shed light on the possible physiological role of dPGM-like proteins.
Because H. thermophilus possesses SHMT, another enzyme capable of synthesizing serine (32), a possibility exists that SHMT may be responsible for serine synthesis in vivo. However, we believe that H. thermophilus SHMT functions in vivo not for serine biosynthesis but rather for the production of glycine, because the single carbon released as 5,10-methylenetetrahydrofolate from serine appears to be essential for the growth of this obligately chemolithoautotrophic bacterium. This one-carbon unit is needed for the biosynthesis of methionine, purines, thymidylate, and other important biomolecules (33–35). In addition, it has been demonstrated that SHMT from H. thermophilus shows higher kcat and lower Km values to serine than to glycine (32). Interestingly, the gene encoding PspA (HTH0103) is located immediately downstream of the gene for PSAT (HTH0104) on the same strand and overlap by seven nucleotides. Therefore, these two genes are probably co-transcribed as an operon. Taking these observations into account, iPSPs may be the main enzymes synthesizing serine in members of the genus Hydrogenobacter. To clarify this speculation, flux analyses for amino acids and one-carbon metabolites are needed.
The reason why heterodimeric iPSP2 is expressed in H. thermophilus in addition to homodimeric iPSP1 is of interest. Because iPSP1 and iPSP2 display similar in vivo activities (the activities of iPSP1 and iPSP2 in Butyl-Toyopearl were 82 and 73 units, respectively; see Table 2) and Km values for l-phosphoserine, both proteins appear to actively function in vivo. However, our results strongly suggest that the PspB subunit has no PSP activity or has PSP activity considerably lower than that of PspA (Table 3). It has been reported that vertebrates have three dimeric isozymes of dPGM composed of two different subunits: type M, B, and MB (36). However, no remarkable differences exist in the kinetic constants or pH dependence of the three isozymes, suggesting that both subunits have similar catalytic properties (36). Furthermore, the sequences of dPGM-B and dPGM-M show ∼50% identity in protein BLAST searches of the NCBI database, whereas PspA and PspB of H. thermophilus show less than 40% identity. To our knowledge, this is the first identification of a dPGM-related protein forming a heteromer with subunits displaying distinct catalytic properties.
Although we could not detect PSP enzymatic activity of the PspB subunit, we speculate that this subunit might have an important function for bacterial survival. This hypothesis is supported by the fact that pspB, in addition to pspA, was found in the genomes of all Aquificaceae and Hydrogenothermaceae species for which complete sequences are available. Based on our phylogenetic analyses, PspA and PspB are likely to have arisen from gene duplication events (supplemental Fig. S3). Generally, one member of a paralog pair has a small chance of acquiring a novel function after gene duplication (37, 38), although in most cases it becomes a pseudogene (39). Therefore, it is tempting to speculate that the PspB subunit might have an important function that is separate from catalyzing the PSP reaction. However, further studies are needed to determine the function of PspB.
TTHA0368- and At5g01420-coded proteins showed low but distinct PSP activities. Although their activities were too low to confirm that they function as iPSPs, it is clear that the potential to catalyze PSP reactions by histidine phosphatase superfamily proteins is distributed among bacteria, protozoa (31), and higher plants, at a minimum. These results suggest that the potential to catalyze PSP activity by dPGM-related proteins is not necessarily intrinsic to Aquificales but may be distributed among a wider range of organisms.
According to the annotation by the MBGD, eight phyla of the domain Bacteria (Chloroflexi, Cyanobacteria, Deferribacteries, Deinococcus-Thermus, Dictyoglomi, Firmicutes, Nitrospirae, Thermotogae, and Verrucomicrobia) also lack dPSP orthologs, although PGDH and PSAT orthologs are completely conserved, as in the case of Aquificae (Table 1). In addition, of 51 genera of Firmicutes, 35 and 42 genera possess orthologs of PGDH and PSAT, respectively, but dPSP orthologs are only found in five genera. These dPSP-lacking organisms are candidates for possessing novel-type PSPs like iPSPs. Among them, at least some Firmicutes, Chloroflexi, and Cyanobacteria possess dPGM-related proteins that show higher similarity to H. thermophilus PspA than the proteins encoded by TTHA0368 and At5g04120 (supplemental Fig. S1). These proteins are expected to have PSP activities equaling that of H. thermophilus PspA. In particular, dPGM-related proteins are strong candidates for PSPs in Cyanobacteria because CFE from these organisms have detectable PSP activity (8). Our findings may also help to reveal the missing link between phosphoserine and serine in organisms other than members of Aquificales.
Supplementary Material
Acknowledgment
We thank Masafumi Kameya for frequent and fruitful discussions.
This work was supported in part by Grant-in-aid for Scientific Research (A) 21248010 from the Japan Society for the Promotion of Science (JSPS) and Grant-in-aid for JSPS Fellows 23-3030.

This article contains supplemental Figs. S1–S3.
M. Kameya, Y. Chiba, Y. Sato, H. Kanbe, S. Anma, H. Arai, M. Ishii, and Y. Igarashi, unpublished data.
- SHMT
- serine hydroxymethyltransferase
- CFE
- cell-free extract(s)
- PSP
- phosphoserine phosphatase
- PGDH
- 3-phosphoglycerate dehydrogenase
- PSAT
- 3-phosphoserine aminotransferase
- dPSP
- metal-dependent PSP
- iPSP
- metal-independent PSP
- dPGM
- cofactor-dependent phosphoglycerate mutase
- MBGD
- Microbial Genome Database for Comparative Analysis.
REFERENCES
- 1. Snell K. (1984) Enzymes of serine metabolism in normal, developing, and neoplastic rat tissues. Adv. Enzyme. Regul. 22, 325–400 [DOI] [PubMed] [Google Scholar]
- 2. Ho C. L., Noji M., Saito K. (1999) Plastidic pathway of serine biosynthesis. Molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J. Biol. Chem. 274, 11007–11012 [DOI] [PubMed] [Google Scholar]
- 3. Hart C. E., Race V., Achouri Y., Wiame E., Sharrard M., Olpin S. E., Watkinson J., Bonham J. R., Jaeken J., Matthijs G., Van Schaftingen E. (2007) Phosphoserine aminotransferase deficiency. A novel disorder of the serine biosynthesis pathway. Am. J. Hum. Genet. 80, 931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helgadóttir S., Rosas-Sandoval G., Söll D., Graham D. E. (2007) Biosynthesis of phosphoserine in the Methanococcales. J. Bacteriol. 189, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veeranna, Shetty K. T. (1990) Phosphoserine phosphatase of human brain. Partial purification, characterization, regional distribution, and effect of certain modulators including psychoactive drugs. Neurochem. Res. 15, 1203–1210 [DOI] [PubMed] [Google Scholar]
- 6. Ho C. L., Saito K. (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino. Acids. 20, 243–259 [DOI] [PubMed] [Google Scholar]
- 7. Wang W., Cho H. S., Kim R., Jancarik J., Yokota H., Nguyen H. H., Grigoriev I. V., Wemmer D. E., Kim S. H. (2002) Structural characterization of the reaction pathway in phosphoserine phosphatase. Crystallographic “snapshots” of intermediate states. J. Mol. Biol. 319, 421–431 [DOI] [PubMed] [Google Scholar]
- 8. Colman B., Norman E. G. (1997) Serine synthesis in cyanobacteria by a nonphotorespiratory pathway. Physiol. Plant. 100, 133–136 [Google Scholar]
- 9. Knoop H., Zilliges Y., Lockau W., Steuer R. (2010) The metabolic network of Synechocystis sp. PCC 6803. Systemic properties of autotrophic growth. Plant Physiol. 154, 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawasumi T., Igarashi Y., Kodama T., Minoda Y. (1980) Isolation of strictly thermophilic and obligately autotrophic hydrogen bacteria. Agric. Biol. Chem. 44, 1985–1986 [Google Scholar]
- 11. Kawasumi T., Igarashi Y., Kodama T., Minoda Y. (1984) Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int. J. Syst. Bacteriol. 34, 5–10 [Google Scholar]
- 12. Shiba H., Kawasumi T., Igarashi Y., Kodama T., Minoda Y. (1985) The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141, 198–203 [Google Scholar]
- 13. Arai H., Kanbe H., Ishii M., Igarashi Y. (2010) Complete genome sequence of the thermophilic, obligately chemolithoautotrophic hydrogen-oxidizing bacterium Hydrogenobacter thermophilus TK-6. J. Bacteriol. 192, 2651–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kameya M., Arai H., Ishii M., Igarashi Y. (2010) Purification of three aminotransferases from Hydrogenobacter thermophilus TK-6. Novel types of alanine or glycine aminotransferase. Enzymes and catalysis. FEBS J. 277, 1876–1885 [DOI] [PubMed] [Google Scholar]
- 15. Rigden D. J. (2008) The histidine phosphatase superfamily. Structure and function. Biochem. J. 409, 333–348 [DOI] [PubMed] [Google Scholar]
- 16. Suzuki M., Cui Z. J., Ishii M., Igarashi Y. (2001) Nitrate respiratory metabolism in an obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus TK-6. Arch. Microbiol. 175, 75–78 [DOI] [PubMed] [Google Scholar]
- 17. Shapiro B. M., Stadtman E. R. (1971) Glutamine synthetase (Escherichia coli). Methods Enzymol. 17A, 910–922 [Google Scholar]
- 18. Kameya M., Arai H., Ishii M., Igarashi Y. (2006) Purification and properties of glutamine synthetase from Hydrogenobacter thermophilus TK-6. J. Biosci. Bioeng. 102, 311–315 [DOI] [PubMed] [Google Scholar]
- 19. D'Alessio G., Josse J. (1975) Phosphoglycerate kinase and phosphoglyceromutase from Escherichia coli. Methods Enzymol. 41, 139–144 [DOI] [PubMed] [Google Scholar]
- 20. Yokoyama S., Hirota H., Kigawa T., Yabuki T., Shirouzu M., Terada T., Ito Y., Matsuo Y., Kuroda Y., Nishimura Y., Kyogoku Y., Miki K., Masui R., Kuramitsu S. (2000) Structural genomics projects in Japan. Nat. Struct. Biol. 7, 943–945 [DOI] [PubMed] [Google Scholar]
- 21. Seki M., Carninci P., Nishiyama Y., Hayashizaki Y., Shinozaki K. (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720 [DOI] [PubMed] [Google Scholar]
- 22. Seki M., Narusaka M., Kamiya A., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., Muramatsu M., Hayashizaki Y., Kawai J., Carninci P., Itoh M., Ishii Y., Arakawa T., Shibata K., Shinagawa A., Shinozaki K. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145 [DOI] [PubMed] [Google Scholar]
- 23. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5. Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whelan S., Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- 26. Rigden D. J., Bagyan I., Lamani E., Setlow P., Jedrzejas M. J. (2001) A cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus is actually a broad specificity phosphatase. Protein. Sci. 10, 1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rigden D. J., Walter R. A., Phillips S. E., Fothergill-Gilmore L. A. (1999) Sulfate ions observed in the 2.12 Å structure of a new crystal form of S. cerevisiae phosphoglycerate mutase provide insights into understanding the catalytic mechanism. J. Mol. Biol. 286, 1507–1517 [DOI] [PubMed] [Google Scholar]
- 28. Kuznetsova E., Proudfoot M., Gonzalez C. F., Brown G., Omelchenko M. V., Borozan I., Carmel L., Wolf Y. I., Mori H., Savchenko A. V., Arrowsmith C. H., Koonin E. V., Edwards A. M., Yakunin A. F. (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 281, 36149–36161 [DOI] [PubMed] [Google Scholar]
- 29. Hügler M., Huber H., Molyneaux S. J., Vetriani C., Sievert S. M. (2007) Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae. Evidence for two ways of citrate cleavage. Environ. Microbiol. 9, 81–92 [DOI] [PubMed] [Google Scholar]
- 30. Bourgis F., Botha F. C., Mani S., Hiten F. N., Rigden D. J., Verbruggen N. (2005) Characterization and functional investigation of an Arabidopsis cDNA encoding a homologue to the d-PGMase superfamily. J. Exp. Bot. 56, 1129–1142 [DOI] [PubMed] [Google Scholar]
- 31. Hills T., Srivastava A., Ayi K., Wernimont A. K., Kain K., Waters A. P., Hui R., Pizarro J. C. (2011) Characterization of a new phosphatase from Plasmodium. Mol. Biochem. Parasitol. 179, 69–79 [DOI] [PubMed] [Google Scholar]
- 32. Chiba Y., Terada T., Kameya M., Shimizu K., Arai H., Ishii M., Igarashi Y. (2012) Mechanism for folate-independent aldolase reaction catalyzed by serine hydroxymethyltransferase. FEBS J. 279, 504–514 [DOI] [PubMed] [Google Scholar]
- 33. Ogawa H., Gomi T., Fujioka M. (2000) Serine hydroxymethyltransferase and threonine aldolase. Are they identical? Int. J. Biochem. Cell Biol. 32, 289–301 [DOI] [PubMed] [Google Scholar]
- 34. Szebenyi D. M., Musayev F. N., di Salvo M. L., Safo M. K., Schirch V. (2004) Serine hydroxymethyltransferase. Role of Glu-75 and evidence that serine is cleaved by a retroaldol mechanism. Biochemistry 43, 6865–6876 [DOI] [PubMed] [Google Scholar]
- 35. Florio R., di Salvo M. L., Vivoli M., Contestabile R. (2011) Serine hydroxymethyltransferase. A model enzyme for mechanistic, structural, and evolutionary studies. Biochim. Biophys. Acta 1814, 1489–1896 [DOI] [PubMed] [Google Scholar]
- 36. Bartrons R., Carreras J. (1982) Purification and characterization of phosphoglycerate mutase isozymes from pig heart. Biochim. Biophys. Acta 708, 167–177 [DOI] [PubMed] [Google Scholar]
- 37. Prince V. E., Pickett F. B. (2002) Splitting pairs. The diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837 [DOI] [PubMed] [Google Scholar]
- 38. Ohno S. (1970) Evolution by Gene Duplication, Springer, New York [Google Scholar]
- 39. Oshima K., Kakizawa S., Arashida R., Ishii Y., Hoshi A., Hayashi Y., Kagiwada S., Namba S. (2007) Presence of two glycolytic gene clusters in a severe pathogenic line of Candidatus Phytoplasma asteris. Mol. Plant Pathol. 8, 481–489 [DOI] [PubMed] [Google Scholar]
- 40. Thaller M. C., Schippa S., Bonci A., Cresti S., Rossolini G. M. (1997) Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol. Lett. 146, 191–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.