Background: In AD, APP can be processed in lipid rafts, and γ-secretase-associated proteins (GSAPs) can affect Aβ production.
Results: We identify novel GSAPs in detergent-resistant membranes (DRMs).
Conclusion: VDAC1 and CNTNAP1 associate with γ-secretase in DRMs and affect APP processing with less effect on Notch processing.
Significance: Novel GSAPs that regulate Aβ production can be used as AD therapeutic targets.
Keywords: Alzheimers Disease, Brain, Lipid Raft, Mass Spectrometry (MS), Proteomics, Affinity Pulldown, Detergent-resistant Membranes, gamma-Secretase
Abstract
In Alzheimer disease, oligomeric amyloid β-peptide (Aβ) species lead to synapse loss and neuronal death. γ-Secretase, the transmembrane protease complex that mediates the final catalytic step that liberates Aβ from its precursor protein (APP), has a multitude of substrates, and therapeutics aimed at reducing Aβ production should ideally be specific for APP cleavage. It has been shown that APP can be processed in lipid rafts, and γ-secretase-associated proteins can affect Aβ production. Here, we use a biotinylated inhibitor for affinity purification of γ-secretase and associated proteins and mass spectrometry for identification of the purified proteins, and we identify novel γ-secretase-associated proteins in detergent-resistant membranes from brain. Furthermore, we show by small interfering RNA-mediated knockdown of gene expression that a subset of the γ-secretase-associated proteins, in particular voltage-dependent anion channel 1 (VDAC1) and contactin-associated protein 1 (CNTNAP1), reduced Aβ production (Aβ40 and Aβ42) by around 70%, whereas knockdown of presenilin 1, one of the essential γ-secretase complex components, reduced Aβ production by 50%. Importantly, these proteins had a less pronounced effect on Notch processing. We conclude that VDAC1 and CNTNAP1 associate with γ-secretase in detergent-resistant membranes and affect APP processing and suggest that molecules that interfere with this interaction could be of therapeutic use for Alzheimer disease.
Introduction
The pathological hallmarks of the Alzheimer disease (AD)4 brain are the senile plaques, composed of fibrils formed by the amyloid β-peptide (Aβ), and neurofibrillary tangles, containing hyperphosphorylated Tau protein (1). In particular, the longer variants of Aβ, Aβ42 and Aβ43, have a strong tendency to polymerize into amyloid fibrils and accumulate in the brain (2, 3). Several lines of evidence suggest that oligomeric Aβ species formed during the polymerization affect synapses and are pathogenic. Thus, strategies aimed at curing AD involve the development of compounds targeting the production of Aβ. The clinical AD trials, including those targeting Aβ, have so far failed. One reason for this may be that the patients were recruited in the later stages of the disease, when severe nerve cell loss has manifested in clinical symptoms. It has also been difficult to recruit homogenous groups and to reach efficient levels of the drug in the central nervous system. There is an increased awareness of these problems, and hopefully, novel biomarkers and imaging techniques might be helpful for improving the clinical trials (4–6). Not all Aβ variants are toxic, and it has been suggested that shorter variants such as Aβ40 could be protective (7). For instance, the pyramidal neurons that become affected in AD show elevated levels of Aβ42 but not Aβ40 (8). In accord with this notion are the familial presenilin mutations, which in many cases result in decreased total Aβ production but an increased Aβ42/40 ratio (9). These familial presenilin mutations also affect the processing of other γ-secretase substrates, and it is possible that such alterations could contribute to the disease (10).
Aβ is produced from a type 1 transmembrane glycoprotein, the Aβ precursor protein (APP), by two sequential cleavages. The initial cleavage, mediated by β-secretase, occurs in the lumen/extracellularly and generates soluble APP and a membrane-bound C-terminal fragment (C99). Next, C99 is cleaved in the transmembrane region by γ-secretase, generating the APP intracellular domain and Aβ (11). γ-Secretase is an aspartyl protease complex, containing at least four transmembrane proteins that appear to be necessary and sufficient for γ-secretase activity as follows: presenilin, nicastrin, anterior pharynx defective-1 (Aph-1), and presenilin enhancer-2 (Pen-2) (12, 13). γ-Secretase cleaves more than 60 different substrates, most of them type 1 transmembrane proteins, and therapeutic inhibitors of its activity may thus be hampered by side effects. Clinically, inhibition of γ-secretase activity has been found to be problematic as a treatment for AD because of side effects related to decreased γ-secretase-dependent Notch signaling (14). Like APP, Notch undergoes ectodomain shedding by a metalloprotease and is further cleaved by γ-secretase, releasing the Notch intracellular domain (NICD) (15). To avoid the Notch-related side effects, it is necessary to find novel modulators of γ-secretase that specifically affect APP processing. Interestingly, it is possible that γ-secretase-associated proteins (GSAPs) could be involved in substrate selection and have a regulatory role. For instance, recent studies reported TMP21, CD147, proteins involved in the tetraspanin web, and syntaxin1 as possible regulators of γ-secretase (16–19). More recently, pigeon homologue protein was found as a γ-secretase-activating protein although it also interacts with the C-terminal fragment of APP (20). APP-interacting protein, BRI2, was found to suppress Aβ production by inhibiting docking of γ-secretase (nicastrin) to C99 as well as restricting the α- and β-secretase cleavage sites on APP (21).
The activity of proteins can be affected by the lipid membrane environment, for instance by ordered membrane microdomains called lipid rafts. These are enriched in cholesterol, sphingolipids, and certain proteins and are important for cell signaling, membrane protein sorting, and transport (22). Lipid rafts are partly resistant to detergents and can be biochemically isolated by treating cellular membranes with e.g. Triton X-100 or CHAPSO at 4 °C, followed by centrifugation. As a result, the detergent-resistant membranes (DRMs) float to the top of the gradient where they can be collected. Previously, we isolated CHAPSO-resistant DRMs and showed that active γ-secretase is localized to DRMs in human and rat brain (23). Size exclusion chromatography showed that the DRMs containing γ-secretase elutes in the void volume (around 2000 kDa) suggesting that the DRMs may contain several copies of γ-secretase complexes, other proteins, lipids, and CHAPSO. Thus, we hypothesized that potential GSAPs could interact with γ-secretase in DRMs.
By affinity purification, using a biotinylated γ-secretase inhibitor, in combination with LC-MS/MS, we have identified novel GSAPs in DRMs prepared from rat brain. The effect of the potential GSAPs on Aβ production was tested by small interfering RNA (siRNA)-mediated knockdown of gene expression. Treatment with siRNA directed to voltage-dependent anion channel 1 or contactin-associated protein 1 reduced Aβ production. Furthermore, the effect of the potential GSAPs on Notch processing was analyzed by measuring NICD levels. Interestingly, the NICD levels after knockdown of voltage-dependent anion channel 1 or contactin-associated protein 1 were not reduced to the same extent as Aβ levels.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: PS1-N-terminal fragment (NTF) (529591, Calbiochem), raised against amino acid residues 1–65 of human PS1; nicastrin (MAB5556, Chemicon, Billerica, MA), raised against synthetic peptide from mouse nicastrin; TMP21 (3999, Nordic BioSite, Täby, Sweden), raised against the 18 amino acid peptides from near the center of human TMP21; syntaxin1 (S0664, Sigma), raised against the synaptosomal plasma membrane fraction from adult rat hippocampus; cleaved Notch1 (Val-1744) (2421, Cell Signaling Technology, Danvers, MA), raised against a synthetic peptide corresponding to the sequence at the Val-1744 cleavage site of human Notch1; voltage-dependent anion channel 1 (VDAC1, sc-8828, Santa Cruz Biotechnology, Santa Cruz, CA), raised against a synthetic peptide from the N terminus of human VDAC1.
Rat Brain Material
Male Sprague-Dawley rats (200–250 g) were obtained from B&K Universal (Sollentuna, Sweden). Ethical permit was approved by the Animal Trial Committee of Southern Stockholm (Permit S80-08). The rats were sacrificed by carbon dioxide. The brains were dissected to remove blood vessels and white matter. Sprague-Dawley rat brains (8–12 weeks old) were also purchased from Rockland Immunochemicals (Gilbertsville, PA) and stored at −70 °C before use.
Preparation of Membranes
Membranes were prepared as described previously (24) with some modifications. Briefly, the brain material was homogenized by 25 strokes at 1500 rpm with a mechanical pestle homogenizer (RW20, IKA Laborteknik, Hattersheim, Germany) in lysis buffer (1 ml of buffer/0.2 g of tissue) containing 20 mm Hepes (pH 7.5), 50 mm KCl, 2 mm EGTA, and CompleteTM protease inhibitor mixture (Roche Applied Science). All the procedures were carried out on ice. The samples were centrifuged at 1000 × g for 10 min to remove nuclei and poorly homogenized material. The pellet was homogenized and centrifuged at 1000 × g for 10 min, and the post-nuclear supernatants were then pooled and centrifuged at 10,000 × g for 30 min to remove mitochondria. The supernatant was centrifuged once more, and the resulting supernatant was then centrifuged at 100,000 × g for 1 h to yield the final pellet (P3).
Preparation of DRMs
DRMs were prepared as described previously (23). In brief, to isolate DRMs from brain material, P3 was resuspended in 600 μl of buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 2.0% CHAPSO, and CompleteTM protease inhibitor mixture (Roche Applied Science). The samples were incubated with end-over-end rotation for 20 min at 4 °C. The sample was adjusted to 45% sucrose and placed at the bottom of a 14-ml Ultra-clearTM centrifuge tube (Beckman Coulter, Fullerton, CA). Then 6.9 ml of 35% sucrose followed by 2.3 ml of 5% sucrose was overlaid. The sample was centrifuged at 100,000 × g for 16 h at 4 °C in a SW40Ti rotor (Beckman Coulter). The fraction at the 5–35% interface was collected using a 5-ml syringe (CODAN, Hørsholm, Denmark).
Electron Microscopy
DRM fractions were mixed with 2% CHAPSO in PBS and centrifuged at 100,000 × g for 1 h at 4 °C in an SW40 rotor (Beckman Coulter). The bottom fraction was centrifuged once again at 100, 000 × g for 1 h in a TLA55 rotor (Beckman Coulter). The DRM pellets were fixed in 2% glutaraldehyde in 0.1 m sodium cacodylate buffer containing 0.1 m sucrose and 3 mm CaCl2 (pH 7.4) at 4 °C overnight, rinsed in 0.1 m phosphate buffer (pH 7.4) followed by postfixation in 2% osmium tetroxide in 0.1 m phosphate buffer (pH 7.4) at 4 °C for 2 h, dehydrated in ethanol followed by acetone, and embedded in LX-112 (Ladd, Burlington, VT). Sections were contrasted with uranyl acetate followed by lead citrate and examined in a Leo 906 transmission electron microscope at 80 kV (Leo, Oberkochen, Germany). Digital images were taken by using a Morada digital camera (Olympus Soft Imaging System, GmbH, Münster, Germany).
Thin Layer Chromatography (TLC)
Four DRM fractions were mixed with PBS and centrifuged at 100,000 × g for 1 h. The loose fraction at the bottom of the tube was resuspended in PBS and centrifuged at 100,000 × g for 30 min in a TLA55 rotor. The pelleted DRM fractions were dissolved in 200 μl of chloroform/methanol mixture (ratio 2:1). An additional volume of chloroform/methanol mixture was added up to 500 μl followed by addition of 100 μl of distilled water. The sample was vortexed and centrifuged at 1,000 × g for 10 min. The lipids were collected in the chloroform/methanol fraction. This fraction was dried using a vacuum centrifuge (Maxi Dry Lyo, Heto-Holten AIS, Allerød, Denmark) and dissolved in chloroform and methanol. Cholesterol and sphingomyelin in DRM fractions were separated on a TLC plate (Sigma) together with synthetic cholesterol and sphingomyelin standards (Sigma). The TLC plate was developed by bromothymol blue (Sigma) and air-dried. The concentrations of cholesterol and sphingomyelin in DRMs were quantified from the standards of cholesterol and sphingomyelin using a CCD camera (Bio-Rad).
SDS-PAGE and Western Blotting
Equal amounts of protein (20 μg) were mixed with SDS sample buffer and placed at room temperature for 20 min. The samples were loaded onto a NuPAGETM 4–12% BisTris gel (Invitrogen). The samples were electrophoresed and transferred to nitrocellulose membranes (Whatman), and the proteins of interest were detected by specific antibodies. Colloidal gold total protein stain solution (Bio-Rad) was used for the total protein staining.
Affinity Pulldown of γ-Secretase in DRMs
γ-Secretase was captured from DRM fractions by a γ -secretase inhibitor with a cleavable biotin group (GCB). Synthesis of GCB and affinity pulldown by GCB was described in detail elsewhere (19). In brief, CHAPSO was added to a final concentration of 0.5% (w/v) to DRM fractions. Samples were pre-cleared with streptavidin-conjugated magnetic beads (Dynabeads® M-280 Streptavidin, Invitrogen) overnight at 4 °C. The magnetic beads were collected by a magnet and the procedure was repeated once more. The supernatant was incubated for 30 min at 37 °C in the absence or presence of 50 μm of the γ-secretase inhibitor L-685,458 (Bachem, Torrance, CA). The samples were incubated further with 200 nm GCB for 1 h at 37 °C, followed by incubation with magnetic beads with end-over-end rotation for 2 h at 4 °C. The magnetic beads were collected by using a magnet. After washing three times with buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1.0% CHAPSO, and CompleteTM protease inhibitor mixture, the captured γ-secretase complexes were eluted by 100 mm DTT in SDS sample buffer (Invitrogen) for Western blotting. For the mass spectrometry analysis, the beads were washed twice with 10 mm ammonium bicarbonate (Ambic), and the samples were eluted with buffer containing 10 mm DTT, 0.01% RapiGest surfactant (Waters, Milford, MA), and 10 mm Ambic.
Tryptic Digestion and Nanoscale Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
CaCl2 was added to the eluted samples to a final concentration of 2 mm, and the samples were boiled for 5 min. The samples were digested by trypsin at 37 °C overnight. To cleave RapiGest according to the manufacturer's instructions, 0.5 μl of 37% HCl was added to the samples. The samples were incubated for 1 h at 37 °C and centrifuged at 16,000 × g for 10 min, and the supernatant was transferred to siliconized tubes. The tryptic peptides were concentrated by using a ZipTip® C18 (Millipore, Billerica, MA) after equilibration according to the manufacturer's instructions. The peptides were washed twice with 10 μl of 0.1% formic acid followed by elution with 80% acetonitrile, 0.1% formic acid. The eluate was dried using a vacuum centrifuge (Maxi Dry Lyo, Heto-Holten AIS) and dissolved in 9 μl of 0.2% formic acid before the mass spectrometry analysis. The samples were analyzed on a 6330 Ion Trap LC/MS system (Agilent Technologies, Santa Clara, CA).
Protein Identification by SpectrumMill
MS/MS spectra were extracted from all experiments by using the data extractor of the SpectrumMill Proteomics Workbench Version A.03.03.078 (Agilent Technologies). Proteins were identified by searching the SwissProt protein data base. Human and rodent were set as species. The criteria for positive peptide hits were of a score >10 and scored peak intensity >70%.
Candidate Protein Selection
The mean intensity of the identified peptides was normalized to the total intensity of each sample to compensate for potential differences in sample recovery. The ratio was calculated by dividing the intensity value of −L-685,458 by the one of +L-685,458. To be selected as candidate proteins, a minimum of two unique peptides per protein was required.
RNA Interference
The pre-designed siRNA oligonucleotide-targeting candidates were purchased from Ambion (Austin, TX), Invitrogen, and Qiagen (Valencia, CA). Some siRNAs have been validated for reduced off-target effects of siRNAs. The following siRNA oligonucleotides were used: sortilin 1 (siRNA number 162, sense sequence 5′-GGUUCCUGGUGCAUCGAUAtt-3′ and antisense sequence 5′-UAUCGAUGCACCAGGAACCtt-3′, Ambion); synaptophysin (163, 5′-GUACCGAGAGAAUAACAAAtt-3′ and 5′-UUUGUUAUUCUCUCGGUACtt-3′, Ambion); contactin 1 (166, 5′-GGAUAUAAGGUACUCUACAtt-3′ and 5′-UGUAGAGUACCUUAUAUCC cg-3′, Ambion); voltage-dependent anion channel 1 (176, validated, 5′-GGAUACACUCAGACUCUAAtt-3′ and 5′-UUAGAGUCUGAGUGUAUCCta-3′, Ambion); syntaxin12 (179, 5′-GGAUUUGGAACUUAUUAAAtt-3′ and 5′-UUUAAUAAGUUCCAAAUCCtg-3′, and 180, 5′-GCAGGACUCAAGCAAGCUAtt-3′ and 5′-UAGCUUGCUUGAGUCCUGCtt-3′, Ambion); cytochrome c oxidase subunit IV isoform 1 (184, 5′-CGCAAAGCUUUGACAAAGAtt-3′ and 5′-UCUUUGUCAAAGCUUUGCGgg-3′, and 186, 5′-GUCGAGUUGUAUCGCAUUAtt-3′ and 5′-UAAUGCGAUACAACUCGACtt-3′, Ambion); contactin-associated protein 1 (237, validated, 5′-GCGUGUGAUGGAGACAGGAGUCAUU-3′ and 5′-AAUGACUCCUGUCUCCAUCACACGC-3′, Invitrogen); presenilin1 (5′-CGAUUUUGACAUACUUCAtt-3′ and 5′-UGAAGUAUGUUCAAAAUCGtt-3′, Ambion). siRNA oligonucleotides were transfected to human embryonic kidney (HEK)-293 APP695 cells (a kind gift from Dr. Eirikur Benedikz, Karolinska Institutet, Stockholm, Sweden), HEK-293 Notch ΔE cells (a kind gift from Dr. Helena Karlström, Karolinska Institutet, Stockholm, Sweden), or wild type (WT) HEK-293 cells at four different concentrations (6.0, 1.8, 0.6, and 0.18 pmol per well) by using LipofectamineTM RNAiMAX (Invitrogen), and cells were grown in a 24-well poly-d-lysine-coated plate (BD Biosciences) at 5% CO2, 95% air at 37 °C. A scrambled siRNA was used as negative control (AllStars Negative Control siRNA, Qiagen). After 2 days, the medium (Opti-MEM reduced serum media, Invitrogen) was replaced. The conditioned medium was harvested after 24 h, and Aβ40 and Aβ42 levels were measured by a commercial sandwich enzyme-linked immunosorbent assay (ELISA) (see below). To check cell viability, Alamar Blue (BIOSOURCE) was added to the cells and incubated for 30 min in 5% CO2, 95% air at 37 °C. The medium was transferred to a 96-well black polystyrene microplate (Corning Glass), and the fluorescence was read at 544 nm by FLUOstar galaxy (BMG Labtech, Offenburg, Germany).
Real Time PCR (RT-PCR)
After measuring cell viability, HEK-293 APP695 cells were washed with cold PBS and lysed with lysis solution (TaqMan®Gene Expression Cells-to-CTTM kit, Ambion), and cDNA was obtained by reverse transcription according to the manufacturer's instructions. Gene expression levels were measured by real time PCR (7500 Fast Real Time PCR System, Ambion).
Sandwich ELISA
Aβ40 and Aβ42 levels were measured by a commercial sandwich ELISA (Immuno-Biological Laboratories, Gunma, and Wako Chemicals, Osaka, Japan) according to the manufacturer's instructions. The conditioned medium was harvested after a 24-h incubation from siRNA-transfected HEK-293 APP695 cells. All the measurements were performed in duplicate, and Aβ40 and Aβ42 levels were calculated from the synthetic Aβ (1–40 and 1–42) standard curve. For calculating the Aβ level after siRNA transfection, Aβ levels of each siRNA were adjusted to Lipofectamine control and divided by cell viability data, which was also adjusted to Lipofectamine control. Data from siRNAs treatments that gave less than 70% cell viability were omitted from the Aβ level calculation.
NICD Analysis
After measuring cell viability, HEK-293 Notch ΔE cells (transfected as described above) were washed with cold PBS, mixed with SDS sample buffer, and boiled at 95 °C for 5 min. The samples were loaded onto a NuPAGETM 4–12% BisTris gel in triplicate and transferred to Immun-BlotTM Polyvinylidene fluoride (PVDF) membranes (Bio-Rad). NICD bands were detected by using SuperSignal West Dura chemiluminescent substrate (Thermo Scientific, Rockford, IL) and a CCD camera (Fuji LAS3000, Tokyo, Japan). NICD levels of each siRNA were quantified by using the MultiGauge software version 3.0 and calculated in the same way as Aβ.
Overexpression of VDAC1
HEK-293 APP695 cells (see above) were transiently transfected with a pCMV6-VDAC1 TrueORF Gold vector (OriGene) by using TransIT LT-1 transfection agent (Mirus Bio). Cells were grown in 5% CO2, 95% air at 37 °C. 24 h after transfection, the medium was exchanged to Opti-MEM. After a further 24 h, the medium was analyzed for Aβ levels as described above, and the cell viability assay (Alamar Blue) was performed. The Aβ levels were normalized to cell viability data as described above.
RESULTS
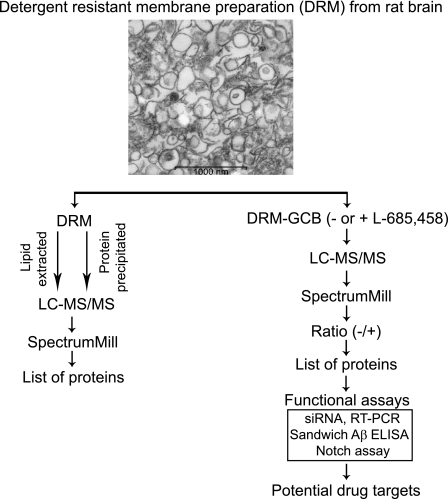
Previous studies using cultured cells or brain tissue have shown that APP, β-secretase, and especially γ-secretase localize to DRMs (23, 25–31), and it has been suggested that the clustering of these proteins in lipid rafts increase Aβ production. Here, we affinity-purified γ-secretase from DRMs, used LC-MS/MS to identify novel GSAPs, and studied their effect on Aβ production by using siRNA knockdown (Fig. 1, right).
FIGURE 1.
Schematic overview of the method. DRMs were prepared from rat brain and analyzed by LC-MS/MS after either lipid extraction or protein precipitation. The data from three experiments were analyzed and compared by using the SpectrumMill software. Proteins found in two out of three experiments were considered to be valid hits and listed. For identification of novel γ-secretase-associated proteins, DRMs were incubated in the absence or presence of the γ-secretase inhibitor, L-685,458, followed by incubation with GCB. After pulldown by streptavidin-conjugated magnetic beads, the samples were analyzed by LC-MS/MS. MS/MS spectra from three independent experiments were analyzed and compared by using the SpectrumMill software. Proteins that were found three times or two out of three times only in the absence of L-685,458 are listed. For those unique proteins, functional assays such as siRNA, sandwich Aβ ELISA, and Notch assay were performed. RT-PCR was used to confirm knockdown in gene levels. Electron microscopy showed vesicle-like structures in DRM preparations from rat brain. Scale bar = 1000 nm.
Preparation and Characterization of DRMs
DRMs were prepared from rat brain as described previously (23). Electron microscopy showed vesicle-like structures in the prepared DRMs (Fig. 1). Bicinchoninic acid (BCA) protein assay and TLC indicated that one DRM fraction, prepared from 600 μg of microsomal membranes, contained around 40 μg of protein, 3 μg of cholesterol, and 0.4 μg of sphingomyelin (data not shown). To find out what proteins were present in DRMs, we used LC-MS/MS. Because LC-MS/MS analysis was difficult due to the high lipid and CHAPSO content in the DRMs, we isolated DRM proteins either by lipid extraction using the Folch method or by acetone precipitation (Fig. 1, left). The samples were injected three times into the LC-MS/MS system, and the resulting MS/MS spectra were analyzed using the SpectrumMill software and the SwissProt rodent data base. In total, 212 proteins were identified after lipid extraction (supplemental Table S1a and 1b) and 242 proteins after protein precipitation (supplemental Table S1a and 1c). Of these, 173 proteins were found by both methods (supplemental Table S1a). Lipid raft marker proteins such as flotillin-1 and flotillin-2 were found among them. Regarding previously reported GSAPs, we could only identify basigin (CD 147) and syntaxin1 (supplemental Table S1a). None of the four core γ-secretase complex components, PS1, nicastrin, Aph-1aL, and Pen-2, was identified by LC-MS/MS, but Western blot analysis confirmed that these four proteins were localized to DRMs (23). In addition, APP (and APP metabolites), Notch (and Notch metabolites), and BACE1 were not identified in the DRM proteome by LC-MS/MS, although we previously reported that the minor amount of APP, APP-CTFs, and BACE1 were associated with DRMs shown by Western blotting (23). We tried Western blot analysis on DRM fractions but could not detect Notch nor Notch-CTFs (data not shown).
Affinity Purification of γ-Secretase and GSAPs from DRMs
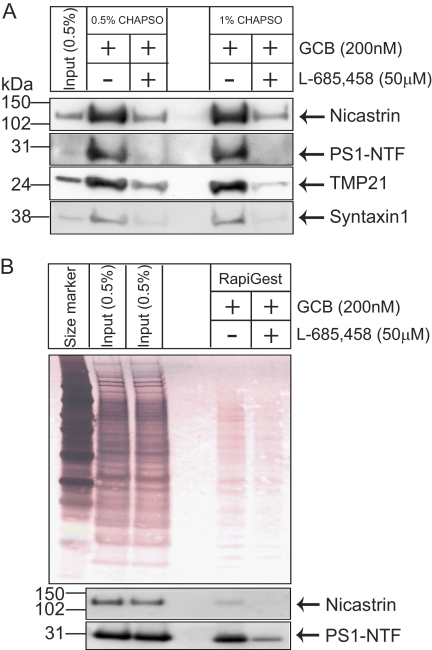
In our previous study, GCB was used for affinity purification of γ-secretase and GSAPs from microsomal membrane fractions prepared from brain (19). Here, we used a similar approach to isolate γ-secretase and GSAPs from DRMs. In brief, DRM fractions were pre-cleared with streptavidin-conjugated magnetic beads. The samples were incubated in the absence or presence of 50 μm of the competing inhibitor L-685,458 (−L-685,458 and +L-685,458, respectively), followed by incubation with 200 nm GCB. Streptavidin-conjugated magnetic beads were added, collected by a magnet after incubation, and repeatedly washed. Washing with 1% CHAPSO reduced background more efficiently than 0.5% CHAPSO (Fig. 2A). After washing, bound components were eluted with buffer containing 10 mm DTT, 0.01% RapiGest, and 10 mm Ambic. The efficiency of the purification and elution procedures was evaluated by using colloidal gold staining and Western blot (Fig. 2B). Western blot analysis showed that nicastrin and PS1-NTF were captured by GCB in the absence of L-685,458, and only a minor amount of PS1-NTF was captured in the presence of the competing inhibitor L-685,458 (Fig. 2B). Previously reported GSAPs, TMP21 and syntaxin1, were also specifically captured by GCB in DRMs. Thus, we conclude that the present approach is suitable for our purpose.
FIGURE 2.
Affinity purification of DRM-associated γ-secretase by GCB pulldown. A, two different washing conditions showed washing with 1% CHAPSO reduced background compared with 0.5% CHAPSO. Western blotting showed that nicastrin, PS1-NTF, TMP21, and syntaxin1 were specifically pulled down in the absence of L-685,458. B, bound proteins were eluted with buffer containing 10 mm DTT, 0.01% RapiGest, and 10 mm Ambic. As shown by colloidal gold staining for total proteins as well as by Western blot analysis for γ-secretase components, the elution by DTT/RapiGest was efficient. Western blotting showed that nicastrin and PS1-NTF were specifically pulled down by GCB in the absence of L-685,458.
LC-MS/MS Analysis of γ-Secretase-associated Proteins in DRMs
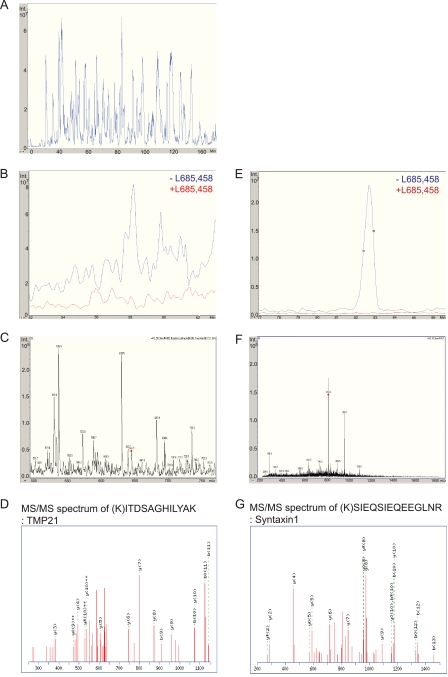
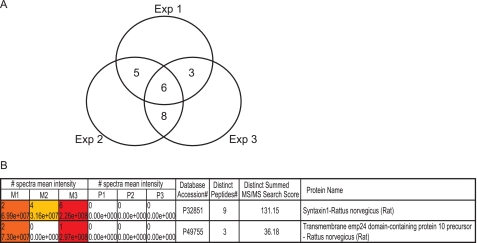
The samples were eluted in 10 mm DTT, 0.01% RapiGest, and 10 mm Ambic and digested with trypsin, and the tryptic peptides were concentrated by using a Zip Tip® C18. The samples were injected into the LC-MS/MS system, eluted by an acetonitrile gradient, and MS and MS/MS spectra were automatically sampled. A representative base peak chromatogram of −L-685,458 samples is shown in Fig. 3A. Data from three independent experiments (three samples of −L-685,458 and three from +L-685,458) were analyzed and compared by using the SpectrumMill software. The spectra mean intensity of each identified peptide was normalized to the total intensity of all peptides identified in the sample to compensate for variations in sample recovery. The intensity ratio was then calculated by dividing the spectra mean intensity of the specific binding proteins (found in the absence of a competing inhibitor) by the one of the nonspecific binding proteins (found in the presence of a competing inhibitor). To select candidate proteins for the further functional studies, we set the following protein identification criteria. First, it should have a minimum protein score of 10 to be a significant identification (equivalent to a score at around 40 in a Mascot search). Second, each protein should have a minimum of two unique peptides identified. Using these parameters, we identified 189 possible GSAPs. Third, we chose proteins identified only in the absence of L-685,458 in at least two out of three experiments. Six proteins were uniquely found in all three independent experiments. Sixteen proteins were uniquely found in two and not at all in one of three experiments (Fig. 4A and Tables 1 and 2). Most of these are trafficking proteins, transporters or channel proteins. The rest are mitochondria related proteins, signaling proteins, and chaperones. Among the identified proteins were the two previously reported GSAPs, TMP21 (SpectrumMill score = 36) and syntaxin1 (score = 131) (Figs. 3, B–D and E–G, and 4B) (16, 19). The extracted ion chromatograms of m/z = 644.9 (TMP21) (Fig. 3B) and m/z = 816.6 (syntaxin1) (Fig. 3E) showed that these proteins were specifically found in the −L-685,458 sample (in blue), while they were absent in the presence of L-685,458 (in red) indicating the specificity of the affinity purification. The comparison between three samples of −L-685,458 (M1, M2, and M3, M stands for minus L-685,458) and three from +L-685,458 (P1, P2, and P3, P stands for plus L-685,458) by using the SpectrumMill software also showed that TMP21 and syntaxin1 were only identified in the −l-685,458 samples (M1, M2, and M3 spectra mean intensities) (Fig. 4B). MS spectra from which the peptides automatically were selected for MS/MS analysis are shown in Fig. 3, C and F. The peaks labeled with red diamonds were identified as peptides from TMP21 (ITDSAGHILYAK) and syntaxin1 (SIEQSIEQEEGLNR), Fig. 3, D and G, respectively. As in the case of the DRM preparation, the core γ-secretase components were not identified by mass spectrometry. Next, additional candidates were selected based on the spectra mean intensity ratio. Candidates that were found uniquely in −L-685,458 more than one time and found in both samples (minus and plus L-685,458), but with higher spectra mean intensities in −L-685,458, were added. Four proteins fulfilled these criteria as follows: contactin-associated protein 1 (SpectrumMill score = 139); solute carrier family 6, member 17 (score = 55); contactin1 (score = 173); and synaptophysin (score = 70) (Table 3). We also included sortilin1, which was identified uniquely once (SpectrumMill score = 16), because it is similar to sortilin-related receptor (SORL1), which showed a genetic association with AD in a previous study (32) (Table 3). In summary, we have identified 27 potential GSAPs in DRMs.
FIGURE 3.
LC-MS/MS analysis of DRM-associated γ-secretase. DRM-associated active γ-secretase was analyzed by LC-MS/MS after three independent GCB pulldowns. A, representative base peak chromatogram of −L-685,458 is shown in blue. The extracted ion chromatograms of TMP21 (retention time = 58.2 min) and syntaxin1 (retention time = 82.9 min) are shown in B and E. TMP21 and syntaxin1 were eluted in the −L-685,458 sample (in blue) but not in the +L-685,458 sample (in red). C and F, MS spectra, and D and G, MS/MS spectra of TMP21 (ITDSAGHILYAK) and syntaxin1 (SIEQSIEQEEGLNR) from −L-685,458 sample are shown.
FIGURE 4.
Data from three independent LC-MS/MS experiments were compared by using the SpectrumMill software. The signal intensity ratio of the −L-685,458 and +L-685,458 samples was calculated. A, proteins that were found three times or two out of three times only in the absence of L-685,458 are shown in a Venn diagram. B, by using the SpectrumMill software, it is possible to analyze all data in one batch and compare the results side by side. Previously reported GSAPs, syntaxin1, and TMP21 were identified only in the absence of L-685,458 (M1, M2, and M3 spectra mean intensities), while they were absent in the presence of L-685,458 (P1, P2, and P3 spectra mean intensities) shown by SpectrumMill software. M1/P1, M2/P2, and M3/P3 indicated the data from three independent experiments. SwissProt protein data base accession numbers, the distinct peptide numbers identified, and SpectrumMill scores identified for syntaxin1 and TMP21 are also shown.
TABLE 1.
List of DRM-GSAP candidate proteins found in three independent experiments
We applied the criteria of a minimum of two unique peptides per protein and a minimum significant score of 10. As a result, six proteins were uniquely found in three independent experiments. Protein accession numbers were from Uniprot; gene symbols, unique scores, numbers of unique peptides, and main functional category are also listed. Candidates tested for the functional analysis of Aβ lowering effects are marked in boldface italic.
| 3 out of 3 | |||||
|---|---|---|---|---|---|
| Accession no. | Gene symbol | Protein name | Score | Unique peptide | Main functional category |
| P32851 | Stx1a | Syntaxin-1A | 131 | 9 | Trafficking proteins |
| Q60932 | Vdac1 | Voltage-dependent anion-selective channel protein 1 | 79 | 6 | Transporter and channel proteins |
| Q66HF1 | Ndufs1 | NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | 53 | 4 | Mitochondrion-related proteins |
| P11951 | Cox6c2 | Cytochrome c oxidase polypeptide VIc-2 | 31 | 3 | Mitochondrion-related proteins |
| Q9ER00 | Stx12 | Syntaxin-12 | 29 | 2 | Trafficking proteins |
| P12369 | Prkar2b | cAMP-dependent protein kinase type II-β regulatory subunit | 26 | 2 | Signaling proteins |
TABLE 2.
List of DRM-GSAP candidate proteins found in two independent experiments
Sixteen proteins were uniquely found in two out of three experiments.
| 2 out of 3 | |||||
|---|---|---|---|---|---|
| Accession no. | Gene symbol | Protein name | Score | Unique peptide | Main functional category |
| O55143 | Atp2a2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 59 | 4 | Transporter and channel proteins |
| Q9JI12 | Slc17a6 | Vesicular glutamate transporter 2 | 52 | 4 | Transporter and channel proteins |
| P06576 | ATP5B | ATP synthase subunit β, mitochondrial precursor | 46 | 3 | Mitochondrion-related proteins |
| P10888 | Cox4i1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | 42 | 3 | Mitochondrion-related proteins |
| Q9WV55 | Vapa | Vesicle-associated membrane protein-associated protein A | 41 | 3 | Trafficking proteins |
| P49755 | TMED10 (TMP21) | Transmembrane emp24 domain-containing protein 10 precursor | 36 | 3 | Trafficking proteins |
| Q66HA6 | Arl8b | ADP-ribosylation factor-like protein 8B | 36 | 2 | Trafficking proteins |
| P23978 | Slc6a1 | Sodium- and chloride-dependent GABA transporter 1 | 33 | 2 | Transporter and channel proteins |
| P24942 | Slc1a3 | Excitatory amino acid transporter 1 | 32 | 2 | Transporter and channel proteins |
| P31647 | Slc6a11 | Sodium- and chloride-dependent GABA transporter 3 | 32 | 2 | Transporter and channel proteins |
| P29975 | Aqp1 | Aquaporin-1 | 31 | 2 | Transporter and channel proteins |
| P54311 | Gnb1 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-1 | 30 | 2 | Trafficking proteins |
| Q5FVI6 | Atp6v1c1 | Vacuolar proton pump subunit C1 | 28 | 2 | Transporter and channel proteins |
| P19086 | GNAZ | Guanine nucleotide-binding protein G(z) subunit α | 28 | 2 | Trafficking proteins |
| P11142 | HSPA8 | Heat shock cognate 71-kDa protein | 26 | 2 | Chaperones |
| O75348 | ATP6V1G1 | Vacuolar proton pump subunit G1 | 21 | 2 | Transporter and channel proteins |
TABLE 3.
List of additional DRM-GSAP candidate proteins
An additional five candidates were added based on the spectra mean intensity ratio.
| Additional candidates | |||||
|---|---|---|---|---|---|
| Accession no. | Gene symbol | Protein name | Score | Unique peptide | Main functional category |
| O54991 | Cntnap1 | Contactin-associated protein 1 | 139 | 9 | Cell surface proteins |
| Q8BJI1 | Slc6a17 | Solute carrier family 6, member 17 | 55 | 4 | Transporter |
| Q63198 | Cntn1 | Contactin 1 | 173 | 13 | Cell surface proteins |
| P07825 | Syp | Synaptophysin | 70 | 5 | Trafficking proteins |
| Q6PHU5 | Sort1 | Sortilin 1 | 16 | 1 | Trafficking proteins |
Functional Analysis of Candidate Proteins
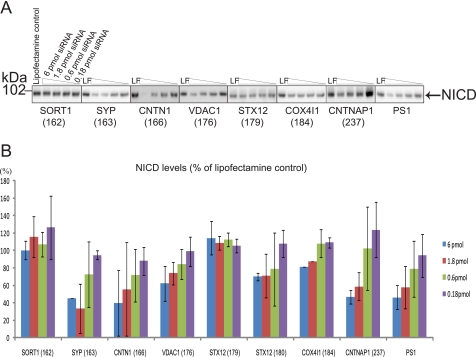
From an AD perspective, the effect of the potential GSAPs on Aβ production is highly interesting. To find out whether the GSAPs affect Aβ production, we silenced the candidate genes in different cell lines by siRNA and measured Aβ production by ELISA. In neuroblastoma BE(2)-C cells, the effect of siRNA directed to PS1 (positive control) on Aβ was poor (supplemental Fig. 1). Hence, we chose to work with HEK-293 APP695 cells, which were efficiently transfected, secreted Aβ at levels that were readily quantified by ELISA, and showed a dose-dependent Aβ reduction upon treatment with siRNA directed to PS1. Because the brain is the most important organ for Alzheimer disease pathology, we selected candidates that are expressed in brain. The mRNA levels and tissue expression levels of each candidate in different organs were checked in the public databases BioGPS and UniProt. Genes which are not expressed in HEK-293 cells according to Affymetrix and Nimblegen were excluded. A schematic view of the siRNA screening process is shown in Fig. 5A. Thirty five siRNAs directed to 11 candidates were transfected into HEK-293 APP695 cells. The knockdown efficiency of each gene on mRNA levels was measured by RT-PCR, and cell viability was measured by Alamar Blue (data not shown). The siRNAs that gave more than 70% knockdown of gene expression and showed more than 70% cell viability were selected for further studies. Next, the effect of siRNA mediated knockdown of the selected GSAPs on Aβ production was measured by a sandwich ELISA. siRNA directed to PS1 was used as a positive control. The expression of excitatory amino acid transporter 1, sodium- and chloride-dependent GABA transporter 3, and solute carrier family 6, member 17, was not silenced by siRNA treatment. siRNAs for guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-1 were toxic. Therefore, those four candidates were excluded from further studies. The rest of the candidates, sortilin1 (SORT1), synaptophysin (SYP), contactin1 (CNTN1), voltage-dependent anion channel 1 (VDAC1), syntaxin12 (STX12), cytochrome c oxidase subunit IV isoform 1 (COX4I1), and contactin-associated protein 1 (CNTNAP1) were analyzed in more detail. First, the knockdown at the protein level was evaluated by Western blot analysis and found to be efficient (supplemental Fig. 2). Then the effect of siRNA treatment on Aβ production was analyzed three times. In brief, the Aβ40 and Aβ42 levels of each sample were adjusted to cell viability data and compared with Lipofectamine control (Fig. 5B). The negative control, a scrambled siRNA, did not affect Aβ production (data not shown). The positive control, siRNA directed to PS1, showed a dose-dependent decrease of Aβ production. Silencing of SORT1, CNTN1, STX12, and COX4I1 did not show any major reduction in Aβ levels (less than 30%). For SYP, Aβ42 and Aβ40 levels were reduced by 44 and 11% at 6 pmol of siRNA, respectively. When siRNA was used to silence VDAC1 and CNTNAP1 at four different concentrations, both Aβ40 and 42 levels were decreased up to 70% in a dose-dependent manner. Thus, the reduction in Aβ levels was even greater than for the control, PS1 (42 and 50% reduction at 6 pmol of siRNA for Aβ40 and Aβ42 levels, respectively). These siRNAs had no significant effect on the levels of γ-secretase components or APP C-terminal fragments as analyzed by Western blot (supplemental Fig. 3). In summary, we suggest that VDAC1 and CNTNAP1 could regulate Aβ production by interacting with γ-secretase.
FIGURE 5.
Functional analysis of candidate proteins. A, schematic view of the screening process. Gene knockdown efficiencies by RT-PCR, cell viability by the Alamar Blue assay, and Aβ production by sandwich ELISA were measured after silencing 11 GSAP candidate genes by siRNA transfection in HEK-293 APP695 cells. More than 70% knockdown and more than 70% cell viability were set as criteria for the siRNAs to be further studied. Candidates showing more than 30% decrease in Aβ production were selected and examined with respect to Notch cleavage. B, levels of Aβ40 and Aβ42 were measured by sandwich Aβ ELISA after the transfection with different amounts of siRNA (6, 1.8, 0.6, and 0.18 pmol) were adjusted to Lipofectamine control and divided by cell viability data (which was also adjusted to Lipofectamine control). The results are presented as means ± S.D. (error bars) and are representative of three independent experiments with exception of PS1 (n = 6).
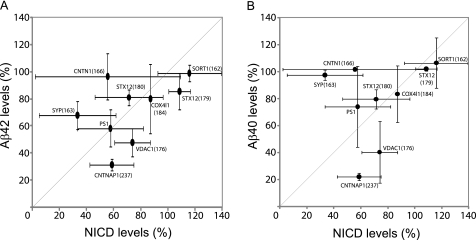
For the purpose of therapeutic inhibition of γ-secretase, ideal GSAPs should affect APP processing but not affect vital pathways such as Notch signaling. To investigate whether these GSAPs are involved in regulating Notch processing, the genes were silenced by siRNAs in HEK-293 cells or in HEK-293 Notch ΔE cells (which overexpress the immediate substrate for γ-secretase). The samples were subjected to gel electrophoresis and Western blotting, and NICD was detected by an antibody specific for cleaved Notch1. No NICD signal was observed in samples from HEK-293 cells (data not shown), whereas there was a significant NICD band in the samples from HEK-293 Notch ΔE cells (Fig. 6A). This difference in expression levels was confirmed by immunohistochemical analysis, using the C20 antibody, of the two cell lines. There was only weak staining in the WT HEK-293 cells, whereas there was a strong nuclear staining in the Notch ΔE-transfected cells. Upon treatment with L-685,458, there was a clear perinuclear staining in both cell lines, indicating that overexpression did not alter the subcellular distribution of the substrate (supplemental Fig. 4). The intensity of the NICD band was quantified by using a CCD camera, and the relative levels were calculated (Fig. 6B). The gene knockdown of SORT1, STX12, and COX4I1 did not change the NICD levels. SYP, CNTN1, and CNTNAP1 affected Notch cleavage to a similar degree as PS1 (around 55%). VDAC1 reduced NICD production to a lower degree (38% at the highest dose (6 pmol) of siRNA). To compare the changes in Aβ42/Aβ40 levels and NICD levels, the data from Figs. 5B and 6B were plotted in Fig. 7. Especially for VDAC1 and CNTNAP1, the effect on Aβ (52–60% and 69–78% reduction, respectively) was greater than the effect on Notch processing (26 and 41%, respectively). Hence, we concluded that VDAC1 and CNTNAP1 affect Aβ and NICD levels differently and decided to further study these GSAPs.
FIGURE 6.
Notch cleavage by candidate proteins. NICD levels were detected by Western blotting. After silencing candidates in HEK-293 Notch ΔE cells, cell lysates were analyzed in triplicate by Western blotting from two to three independent experiments. The representative Western blot is shown in A. Lipofectamine-treated cells served as control. NICD was detected by a cleaved Notch1 (at the cleavage site of Val-1744) antibody. B, levels of NICD were quantified from the Western blot (A) and calculated in the same way as for Aβ levels. The data are presented as means ± S.D. (error bars) and are representative of two (STX12 (179) and CNTNAP1 (237)) or three independent experiments. PS1 data are from six experiments.
FIGURE 7.
Summary of functional analyses. Results of Aβ and Notch analyses at 1.8 pmol siRNA were plotted. The data are presented as means ± S.D. (error bars).
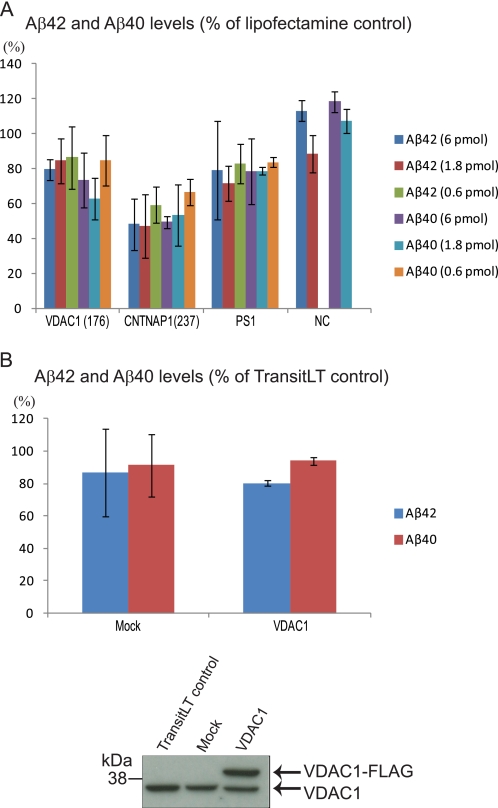
To verify that the effect on Aβ was not depending on overexpression of APP, we studied the effect of siRNA-mediated silencing of VDAC1 and CNTNAP1 in WT HEK-293 cells. Also in this case, the Aβ levels were reduced (Fig. 8A). Finally, we investigated whether overexpression of VDAC1 affects Aβ production. HEK-293 APP695 cells were transfected with VDAC1-FLAG or empty vector. Although the transfection appeared to be efficient as estimated by Western blot analysis, the Aβ levels did not differ from mock-transfected control (Fig. 8B).
FIGURE 8.
Silencing of VDAC1 and CNTNAP1 in WT HEK-293 cells and overexpression of VDAC1 in HEK-293 APP695 cells. A, siRNAs were transfected into WT HEK-293 cells, and Aβ levels were determined by sandwich Aβ ELISA. Aβ levels are expressed as percent of cells treated with Lipofectamine only, and all numbers were adjusted for cell viability. The results are from three independent experiments and presented as mean ± S.D. (error bars). The data of PS1 and negative control (NC) (transfection with scrambled siRNA) are from two experiments. B, HEK-293 APP695 cells were transiently transfected with empty pCMV6 vector (Mock) or pCMV6-VDAC1, and the medium was harvested and analyzed for Aβ levels by sandwich ELISA. Aβ levels are expressed as percent of cells treated with transfection agent alone. All numbers are adjusted for cell viability. The results are from two independent experiments and presented as mean ± S.D. (error bars). The expression levels of endogenous and FLAG-tagged VDAC1 were assayed by Western blotting.
In conclusion, we have identified a number of possible novel GSAPs in DRMs by using the biotinylated γ-secretase inhibitor, GCB, followed by LC-MS/MS analysis. By silencing candidate genes and measuring Aβ and NICD production, we found that voltage-dependent anion channel 1 and contactin-associated protein 1 affect Aβ production and, to a lower degree, NICD levels.
DISCUSSION
Although four proteins (PS, nicastrin, Aph-1, and Pen-2) are sufficient and necessary for the formation of an active γ-secretase complex, GSAPs may be of importance for modulating γ-secretase activity. In an earlier study, we used GCB for affinity pulldown of γ-secretase from microsomal membrane fractions prepared from brain (19). Here, we optimized this approach for DRMs prepared from rat brain and used GCB to purify γ-secretase and GSAPs from DRMs. To our knowledge, this is the first time GSAPs that are associated with brain DRMs are presented.
Characterization of CHAPSO-resistant Membranes
DRMs are not equivalent to lipid rafts, but they reflect to some extent their composition. The protein and lipid compositions of DRMs depend on starting materials as well as the detergents used for their preparation (33, 34). Triton X-100 is most frequently used for preparing DRMs. Regrettably, this detergent disrupts the γ-secretase complex, and we and others have found that CHAPSO is a suitable detergent for preparing DRMs containing active γ-secretase (23, 29). Thus, we used CHAPSO to prepare DRMs from rat brain in this study. Electron microscopy revealed a mixture of mostly vesicular material around 100–200 nm in diameter. Hence, several hundreds of protein molecules could be present in one vesicle. This is in accordance with our previous study where we found γ-secretase in the void volume (>2000 kDa) when a DRM preparation was subjected to size exclusion chromatography (23). To find out which proteins are present in the DRMs prepared using CHAPSO, we subjected them to LC-MS/MS analysis. Because they contain high amounts of lipids and detergents, the proteins were enriched either by Folch extraction (to remove lipids) or by acetone precipitation (to precipitate proteins) prior to analysis. Both approaches gave similar results, and in total 281 proteins were identified. Many of these were previously found in Triton X-100 preparations, which were from cell lines, rodent tissues, or human brain (35–40), as marked in boldface in supplemental Table S1, but many of them have not been reported to localize to DRMs. Hence, as expected, CHAPSO extracts a partly different set of proteins than does Triton X-100. The DRM proteome includes raft proteins, raft-associated proteins, proteins having secondary interactions with raft proteins, and/or false-positive hits (39). We identified classical raft proteins (e.g. flotillin-1, flotillin-2, and Thy-1 membrane glycoprotein) (37), other reported raft proteins (e.g. clathrin heavy chain 1, cofilin-1, and cadherin-13) (39), and raft-associated proteins (e.g. 14-3-3 protein zeta/delta, Ras-related protein Ral-A, contactin-2, and contactin associated protein 1) (36, 39). The identification of calnexin in our DRM proteome might appear unexpectedly because calnexin is frequently used as a non-raft/ER marker protein on Western blot. However, calnexin has previously been identified as a raft or a raft-associated protein from different microdomain proteome studies (37–39, 41–44). In addition, ER lipid raft proteins (Erlin)-1 and Erlin-2 have been identified, which supports the possibility of lipid raft-like domains of the ER (45). Still, some of our identifications might be contaminating proteins, as for example serum albumin.
Even though the known γ-secretase components, PS, nicastrin, Aph-1, and Pen-2, as well as APP, APP-CTFs, and BACE1 were detected by Western blotting (23), they were not identified by mass spectrometry. This may partly be due to the relatively low percentage of these proteins compared with other proteins present in DRMs, how efficiently they are cleaved by trypsin, and the chromatographic and mass spectrometric properties of the resulting peptides. However, two of the previously reported GSAPs, syntaxin1 and CD147, were identified with significant scores (SpectrumMill score 52 and 15, respectively). Although the DRMs were prepared from a microsomal pellet, relatively free from mitochondria, several mitochondrial proteins such as ATP synthase subunits α and β and dihydrolipoyl dehydrogenase were identified. These proteins could in some cases come from mitochondria present in the sample, and in some cases also reside in organelles other than mitochondria. For example, ATP synthase subunits α and β were reported to localize also to the cell surface (38).
Affinity Purification and Methodological Considerations
In the next step, we purified γ-secretase and GSAPs from DRMs by GCB pulldown. In our previous study on CHAPSO-soluble γ-secretase prepared from microsomal membranes, all the known γ-secretase complex components were detected by mass spectrometry after GCB pulldown (19). In our current study in DRMs, we could not detect the known four γ-secretase components by mass spectrometry even though they were detected by Western blot analysis. It is possible that proteins in DRMs are more stable to tryptic digestion or that the recovery of γ-secretase from DRMs is lower than from microsomal membranes. For instance, the active site of γ-secretase could be less accessible to GCB in DRMs than in preparation of CHAPSO soluble γ-secretase. The DRM preparations gave a higher background than the microsomal preparations, and this problem was reduced by increasing the CHAPSO concentration to 1% in the washing buffer. The amount of competing inhibitor was increased five times, compared with our previous study, to get an efficient competition. One possible explanation for the need for higher concentrations could be that there are several copies of active γ-secretase in one piece of DRM, and all of them must be blocked. To reduce the background arising from proteins that were nonspecifically bound to the beads, we used mild and reducing conditions (10 mm DTT and 0.01% RapiGest) for elution. Thereby the inhibitor part of GCB was released from the beads, leaving most of the nonspecifically bound proteins on the beads. This mild elution is more compatible with mass spectrometry than sample buffer elution. As shown in Fig. 2B, Western blotting showed that the elution of PS1-NTF and nicastrin by 10 mm DTT and 0.01% RapiGest was efficient. Compared with input, the background was substantially decreased (shown by Colloidal gold staining) and γ-secretase components were enriched (shown by Western blotting) after GCB pulldown.
Identification of GSAPs by LC-MS/MS
For the unbiased identification of GSAPs from DRMs, the eluted samples were then subjected to LC-MS/MS analysis. To reduce the risk of finding false-positive GSAPs, we analyzed three independent preparations. We set the criterion for a top candidate to be identified only in the noncompeted sample (−L-685,458) at least in two out of three experiments. Twenty two of the identified proteins fulfilled this criterion and were subjected to further analysis. Based on the spectra mean intensity ratio, we selected five more proteins for further studies. Among the identified proteins were two previously reported GSAPs, TMP21 and syntaxin1 (16, 19). Thus, these two GSAPs may interact with detergent-soluble (19) as well as DRM-associated active γ-secretase.
Syntaxin1 was reported to be a PS1-associated protein (46), and we have previously shown that less than 1% of syntaxin1 was associated with detergent-soluble active γ-secretase (19). The association of syntaxin1 with active γ-secretase in DRMs is also less than 1% (Fig. 2A). TMP21 (also known as p23) is a member of the p24 cargo-protein family (47), and it is involved in protein transport and quality control in the ER and Golgi (48). TMP21 was identified as a GSAP (16, 17, 19) and is a negative modulator for Aβ production (16, 49, 50) via γ-secretase activity (16) and/or altered APP trafficking (49, 50). In contrast to our data, TMP21 was not detected as a GSAP in preparations where the γ-secretase inhibitor Merck C was used for affinity purification (51). However, it was suggested that a small pool of TMP21 might be transiently associated with γ-secretase (16, 51) and that the transmembrane domain of TMP21 is important to interact with the γ-secretase complex (49). It is also likely that different starting materials and methodological approaches could explain the discrepancy (19, 51). Indeed, compared with our previous results from CHAPSO soluble γ-secretase (19), TMP21 was to a larger extent associated with γ-secretase in DRMs. One of our other candidates, VDAC1 was identified from a single distinct peptide as a potential GSAP in the study by Wakabayashi et al. (17). In our study, we found VDAC1 as a significant hit with a high score (SpectrumMill score = 79) and six unique peptides. More importantly, VDAC1 was uniquely identified only in the absence of γ-secretase competing inhibitor from three independent MS experiments. This result strengthens the identification of VDAC1 as a novel GSAP.
Thus, although we have taken a novel approach for purification of GSAPs, some of the candidates we identify have been found in other studies, supporting our notion that the GSAPs we find are relevant. We have not excluded the possibility that some of the proteins we identified, but not met the criteria we set, could be a GSAP. For instance, 14 of the proteins that we identified were found also in the study by Wakabayashi et al. (17). Conversely, some of the novel GSAPs may be false-positive. A clear advantage of using DRMs prepared from brain, compared with cell lines, is that all the relevant proteins are expressed. However, it is likely that some GSAPs, e.g. because of their hydrophobicity or low abundance, will escape detection also by our approach.
Subset of Identified GSAPs Affects Aβ Production
Next, we used siRNA-mediated gene knockdown to find out whether the identified GSAPs could affect Aβ production. For this purpose, we chose to use HEK-293 cells stably transfected with human APP, because they express suitable levels of Aβ and respond well to siRNA treatment. We validated a set of siRNAs, directed to the GSAPs of interest, with respect to knockdown efficacy and effect on cell number/viability. Nine out of 35 siRNAs passed the test, and their effect on Aβ production was tested in triplicate. Interestingly, three of the GSAPs, synaptophysin (SYP), voltage-dependent anion channel 1 (VDAC1), and contactin associated protein 1 (CNTNAP1), showed a reduced Aβ production upon knockdown. SYP and CNTNAP1 have not previously been reported to be GSAPs or to affect Aβ production, while VDAC1 was reported to associate with γ-secretase but no functional data were presented (17). Interestingly, the reduced Aβ levels were not accompanied by increased CTF levels as evaluated by Western blot. Possible explanations for this could be that Western blot is a poor technique for quantification of small changes in protein levels or that the CTFs could be degraded by enzymes other than γ-secretase. It should also be noted that Aβ was collected in the medium for 24 h, while the CTFs were analyzed at end point.
Overexpression of VDAC1 did not result in increased Aβ levels. The lack of effect upon overexpression of proteins has been observed previously, for instance for TMP21, which upon silencing reduced Aβ levels (16). Possibly the endogenous levels could be sufficiently high for maximal interaction with γ-secretase.
These candidates were further studied with respect to their effect on Notch processing. NICD production from Notch ΔE, which is a direct substrate for γ-secretase, was measured after transfection with siRNAs in HEK-293 Notch ΔE cells. Knockdown of SYP reduced the NICD levels more than the Aβ levels. In contrast, knockdown of VDAC1 as well as CNTNAP1 reduced NICD to a lower degree than Aβ. Hence, these GSAPs regulate APP and Notch processing differently.
Voltage-dependent anion channel protein 1 and contactin-associated protein 1 are involved in many different cellular functions. Voltage-dependent anion channel protein (VDAC) is a major component of the outer mitochondrial membrane where it regulates fluxes of ions and metabolites. To find this protein to be a GSAP may seem unexpected. However, γ-secretase is found also in mitochondria, and accumulation of Aβ in the mitochondria is common in AD (52, 53). Alternatively, the VDAC that we found could come from the neuronal plasma membrane (VDAC), where it can form channels and be involved in redox homeostasis and apoptosis (54, 55). There are several reports indicating that VDAC could be involved in AD. For instance, VDAC accumulates in amyloid plaques in APP/PS1 transgenic mice (54), and nitrated VDAC1 has been shown to be increased in AD (56). Furthermore, as shown by immunoprecipitation, the plasma membrane VDAC/caveolin-1 (a marker protein for a lipid raft structure called caveolae) complex is highly expressed in AD human brains compared with healthy controls (55). This might indicate that VDAC1 is indeed associated with DRMs. Contactin-associated protein 1 (CNTNAP1 as gene symbol, Caspr, Neurexin IV, or Paranodin as protein name) is a transmembrane protein that forms a cis-interaction with the glycosylphosphatidylinositol-anchored protein contactin (57). These proteins form a complex in the paranodal regions, where glial cells form junctions with axons in myelinated fibers (including the nodes of Ranvier) (58). Caspr regulates the transport and processing of contactin (59), a protein previously shown to interact with APP and possibly Notch (60, 61). A homozygous mutation of CNTN1 in human causes a reduced expression level of CNTN1 at the neuromuscular junction, which leads to a defect in neuromuscular transmission (62). Experiments in mice lacking Caspr showed that it is required in the formation of paranodal junctions that are important for saltatory conduction (63). Caspr is newly reported to be involved in inhibition of neurite outgrowth in CNS neurons (64). Regarding the potential association of Caspr to AD, the CSF and plasma levels of CNTNAP1 and/or CNTN (CNTN1 and/or -2) are altered in AD individuals, and it is possible that these changes could be used for diagnosis of AD (65–68).
In conclusion, we have found by affinity purification several novel γ-secretase-associated proteins in DRMs prepared from rat brain. Two of these, by siRNA-mediated knockdown, were shown to have a clear effect on Aβ production. Because they were found to affect Notch cleavage to a lower extent, we suggest that inhibiting their interactions with γ-secretase may be a way to specifically decrease Aβ levels in Alzheimer disease.
Supplementary Material
Acknowledgments
We thank Dr. Eirikur Benedikz (Karolinska Institutet) for the HEK-293 APP695 cells and Dr. Helena Karlström (Karolinska Institutet) for the HEK-293 Notch ΔE cells. We thank Dr. Maria Ankarcrona (Karolinska Institutet) for the anti-cytochrome oxidase subunit IV antibody. We thank Dr. Jesper Brohede (Karolinska Institutet) for the analysis of gene expression patterns in HEK-293 cells and Dr. Kjell Hultenby (Karolinska Institutet) for electron microscopy. We also thank Birgitta Wiehager (Karolinska Institutet) for the excellent technical assistance.
This work was supported by Gamla Tjänarinnor (to J.-Y. H.), Knut and Alice Wallenberg Foundation, and Dainippon Sumitomo Pharma.

This article contains supplemental Table 1 and Figs. 1–4.
- AD
- Alzheimer disease
- Aβ
- amyloid β-peptide
- Ambic
- ammonium bicarbonate
- APP
- amyloid precursor protein
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- DRM
- detergent-resistant membranes
- GCB
- γ-secretase inhibitor with a cleavable biotin group
- GSAP
- γ-secretase-associated protein
- NICD
- Notch intracellular domain
- NTF
- N-terminal fragment
- NICD
- Notch intracellular domain
- CTF
- C-terminal fragment.
REFERENCES
- 1. Selkoe D. J. (2001) Presenilin, Notch, and the genesis and treatment of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 98, 11039–11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarrett J. T., Berger E. P., Lansbury P. T., Jr. (1993) The carboxyl terminus of the β-amyloid protein is critical for the seeding of amyloid formation. Implications for the pathogenesis of Alzheimer disease. Biochemistry 32, 4693–4697 [DOI] [PubMed] [Google Scholar]
- 3. Welander H., Frånberg J., Graff C., Sundström E., Winblad B., Tjernberg L. O. (2009) Aβ43 is more frequent than Aβ40 in amyloid plaque cores from Alzheimer disease brains. J. Neurochem. 110, 697–706 [DOI] [PubMed] [Google Scholar]
- 4. Imbimbo B. P., Panza F., Frisardi V., Solfrizzi V., D'Onofrio G., Logroscino G., Seripa D., Pilotto A. (2011) Therapeutic intervention for Alzheimer disease with γ-secretase inhibitors. Still a viable option? Expert Opin. Investig. Drugs 20, 325–341 [DOI] [PubMed] [Google Scholar]
- 5. Dickerson B. C., Stoub T. R., Shah R. C., Sperling R. A., Killiany R. J., Albert M. S., Hyman B. T., Blacker D., Detoledo-Morrell L. (2011) Alzheimer-signature MRI biomarker predicts AD dementia in cognitively normal adults. Neurology 76, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckholtz N. S. (2011) Perspective. In search of biomarkers. Nature 475, S8. [DOI] [PubMed] [Google Scholar]
- 7. Jan A., Gokce O., Luthi-Carter R., Lashuel H. A. (2008) The ratio of monomeric to aggregated forms of Aβ40 and Aβ42 is an important determinant of amyloid-β aggregation, fibrillogenesis, and toxicity. J. Biol. Chem. 283, 28176–28189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aoki M., Volkmann I., Tjernberg L. O., Winblad B., Bogdanovic N. (2008) Amyloid β-peptide levels in laser capture microdissected cornu ammonis 1 pyramidal neurons of Alzheimer brain. Neuroreport 19, 1085–1089 [DOI] [PubMed] [Google Scholar]
- 9. Wolfe M. S. (2007) When loss is gain. Reduced presenilin proteolytic function leads to increased Aβ42/Aβ40. Talking point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 8, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Strooper B. (2007) Loss-of-function presenilin mutations in Alzheimer disease. Talking point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 8, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Selkoe D. J. (2001) Alzheimer disease. Genes, proteins, and therapy. Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 12. Kimberly W. T., Wolfe M. S. (2003) Identity and function of γ-secretase. J. Neurosci. Res. 74, 353–360 [DOI] [PubMed] [Google Scholar]
- 13. Edbauer D., Winkler E., Regula J. T., Pesold B., Steiner H., Haass C. (2003) Reconstitution of γ-secretase activity. Nat. Cell Biol. 5, 486–488 [DOI] [PubMed] [Google Scholar]
- 14. Siemers E. R., Quinn J. F., Kaye J., Farlow M. R., Porsteinsson A., Tariot P., Zoulnouni P., Galvin J. E., Holtzman D. M., Knopman D. S., Satterwhite J., Gonzales C., Dean R. A., May P. C. (2006) Effects of a γ-secretase inhibitor in a randomized study of patients with Alzheimer disease. Neurology 66, 602–604 [DOI] [PubMed] [Google Scholar]
- 15. Bray S. J. (2006) Notch signaling. A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 16. Chen F., Hasegawa H., Schmitt-Ulms G., Kawarai T., Bohm C., Katayama T., Gu Y., Sanjo N., Glista M., Rogaeva E., Wakutani Y., Pardossi-Piquard R., Ruan X., Tandon A., Checler F., Marambaud P., Hansen K., Westaway D., St George-Hyslop P., Fraser P. (2006) TMP21 is a presenilin complex component that modulates γ-secretase but not ϵ-secretase activity. Nature 440, 1208–1212 [DOI] [PubMed] [Google Scholar]
- 17. Wakabayashi T., Craessaerts K., Bammens L., Bentahir M., Borgions F., Herdewijn P., Staes A., Timmerman E., Vandekerckhove J., Rubinstein E., Boucheix C., Gevaert K., De Strooper B. (2009) Analysis of the γ-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat. Cell Biol. 11, 1340–1346 [DOI] [PubMed] [Google Scholar]
- 18. Zhou S., Zhou H., Walian P. J., Jap B. K. (2005) CD147 is a regulatory subunit of the γ-secretase complex in Alzheimer disease amyloid β-peptide production. Proc. Natl. Acad. Sci. U.S.A. 102, 7499–7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teranishi Y., Hur J. Y., Welander H., Frånberg J., Aoki M., Winblad B., Frykman S., Tjernberg L. O. (2010) Affinity pulldown of γ-secretase and associated proteins from human and rat brain. J Cell Mol. Med. 14, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He G., Luo W., Li P., Remmers C., Netzer W. J., Hendrick J., Bettayeb K., Flajolet M., Gorelick F., Wennogle L. P., Greengard P. (2010) γ-Secretase-activating protein is a therapeutic target for Alzheimer disease. Nature 467, 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuda S., Giliberto L., Matsuda Y., McGowan E. M., D'Adamio L. (2008) BRI2 inhibits amyloid β-peptide precursor protein processing by interfering with the docking of secretases to the substrate. J. Neurosci. 28, 8668–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simons K., Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 23. Hur J. Y., Welander H., Behbahani H., Aoki M., Frånberg J., Winblad B., Frykman S., Tjernberg L. O. (2008) Active γ-secretase is localized to detergent-resistant membranes in human brain. FEBS J 275, 1174–1187 [DOI] [PubMed] [Google Scholar]
- 24. Farmery M. R., Tjernberg L. O., Pursglove S. E., Bergman A., Winblad B., Näslund J. (2003) Partial purification and characterization of γ-secretase from postmortem human brain. J. Biol. Chem. 278, 24277–24284 [DOI] [PubMed] [Google Scholar]
- 25. Abad-Rodriguez J., Ledesma M. D., Craessaerts K., Perga S., Medina M., Delacourte A., Dingwall C., De Strooper B., Dotti C. G. (2004) Neuronal membrane cholesterol loss enhances amyloid peptide generation. J. Cell Biol. 167, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hattori C., Asai M., Onishi H., Sasagawa N., Hashimoto Y., Saido T. C., Maruyama K., Mizutani S., Ishiura S. (2006) BACE1 interacts with lipid raft proteins. J. Neurosci. Res. 84, 912–917 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. J., Liyanage U., Bickel P. E., Xia W., Lansbury P. T., Jr., Kosik K. S. (1998) A detergent-insoluble membrane compartment contains Aβ in vivo. Nat. Med. 4, 730–734 [DOI] [PubMed] [Google Scholar]
- 29. Urano Y., Hayashi I., Isoo N., Reid P. C., Shibasaki Y., Noguchi N., Tomita T., Iwatsubo T., Hamakubo T., Kodama T. (2005) Association of active γ-secretase complex with lipid rafts. J. Lipid Res. 46, 904–912 [DOI] [PubMed] [Google Scholar]
- 30. Wahrle S., Das P., Nyborg A. C., McLendon C., Shoji M., Kawarabayashi T., Younkin L. H., Younkin S. G., Golde T. E. (2002) Cholesterol-dependent γ-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol. Dis. 9, 11–23 [DOI] [PubMed] [Google Scholar]
- 31. Vetrivel K. S., Cheng H., Kim S. H., Chen Y., Barnes N. Y., Parent A. T., Sisodia S. S., Thinakaran G. (2005) Spatial segregation of γ-secretase and substrates in distinct membrane domains. J. Biol. Chem. 280, 25892–25900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rogaeva E., Meng Y., Lee J. H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C. T., Cheng R., Hasegawa H., Chen F., Shibata N., Lunetta K. L., Pardossi-Piquard R., Bohm C., Wakutani Y., Cupples L. A., Cuenco K. T., Green R. C., Pinessi L., Rainero I., Sorbi S., Bruni A., Duara R., Friedland R. P., Inzelberg R., Hampe W., Bujo H., Song Y. Q., Andersen O. M., Willnow T. E., Graff-Radford N., Petersen R. C., Dickson D., Der S. D., Fraser P. E., Schmitt-Ulms G., Younkin S., Mayeux R., Farrer L. A., St George-Hyslop P. (2007) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. (2003) Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U.S.A. 100, 5795–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Williamson R., Thompson A. J., Abu M., Hye A., Usardi A., Lynham S., Anderton B. H., Hanger D. P. (2010) Isolation of detergent-resistant microdomains from cultured neurons. Detergent-dependent alterations in protein composition. BMC Neurosci. 11, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adam R. M., Yang W., Di Vizio D., Mukhopadhyay N. K., Steen H. (2008) Rapid preparation of nuclei-depleted detergent-resistant membrane fractions suitable for proteomics analysis. BMC Cell Biol. 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martosella J., Zolotarjova N., Liu H., Moyer S. C., Perkins P. D., Boyes B. E. (2006) High recovery HPLC separation of lipid rafts for membrane proteome analysis. J. Proteome Res. 5, 1301–1312 [DOI] [PubMed] [Google Scholar]
- 37. Raimondo F., Ceppi P., Guidi K., Masserini M., Foletti C., Pitto M. (2005) Proteomics of plasma membrane microdomains. Expert Rev. Proteomics 2, 793–807 [DOI] [PubMed] [Google Scholar]
- 38. Bae T. J., Kim M. S., Kim J. W., Kim B. W., Choo H. J., Lee J. W., Kim K. B., Lee C. S., Kim J. H., Chang S. Y., Kang C. Y., Lee S. W., Ko Y. G. (2004) Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics 4, 3536–3548 [DOI] [PubMed] [Google Scholar]
- 39. Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim K. B., Lee J. W., Lee C. S., Kim B. W., Choo H. J., Jung S. Y., Chi S. G., Yoon Y. S., Yoon G., Ko Y. G. (2006) Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics 6, 2444–2453 [DOI] [PubMed] [Google Scholar]
- 41. Blonder J., Hale M. L., Chan K. C., Yu L. R., Lucas D. A., Conrads T. P., Zhou M., Popoff M. R., Issaq H. J., Stiles B. G., Veenstra T. D. (2005) Quantitative profiling of the detergent-resistant membrane proteome of ι-b toxin-induced Vero cells. J Proteome Res. 4, 523–531 [DOI] [PubMed] [Google Scholar]
- 42. Li N., Mak A., Richards D. P., Naber C., Keller B. O., Li L., Shaw A. R. (2003) Monocyte lipid rafts contain proteins implicated in vesicular trafficking and phagosome formation. Proteomics 3, 536–548 [DOI] [PubMed] [Google Scholar]
- 43. Li N., Shaw A. R., Zhang N., Mak A., Li L. (2004) Lipid raft proteomics.+ Analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics 4, 3156–3166 [DOI] [PubMed] [Google Scholar]
- 44. Man P., Novák P., Cebecauer M., Horváth O., Fiserová A., Havlícek V., Bezouska K. (2005) Mass spectrometric analysis of the glycosphingolipid-enriched microdomains of rat natural killer cells. Proteomics 5, 113–122 [DOI] [PubMed] [Google Scholar]
- 45. Browman D. T., Resek M. E., Zajchowski L. D., Robbins S. M. (2006) Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid raft-like domains of the ER. J. Cell Sci. 119, 3149–3160 [DOI] [PubMed] [Google Scholar]
- 46. Smith S. K., Anderson H. A., Yu G., Robertson A. G., Allen S. J., Tyler S. J., Naylor R. L., Mason G., Wilcock G. W., Roche P. A., Fraser P. E., Dawbarn D. (2000) Identification of syntaxin 1A as a novel binding protein for presenilin-1. Brain Res. Mol. Brain Res. 78, 100–107 [DOI] [PubMed] [Google Scholar]
- 47. Blum R., Feick P., Puype M., Vandekerckhove J., Klengel R., Nastainczyk W., Schulz I. (1996) Tmp21 and p24A, two type I proteins enriched in pancreatic microsomal membranes, are members of a protein family involved in vesicular trafficking. J. Biol. Chem. 271, 17183–17189 [DOI] [PubMed] [Google Scholar]
- 48. Jenne N., Frey K., Brugger B., Wieland F. T. (2002) Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 277, 46504–46511 [DOI] [PubMed] [Google Scholar]
- 49. Pardossi-Piquard R., Böhm C., Chen F., Kanemoto S., Checler F., Schmitt-Ulms G., St George-Hyslop P., Fraser P. E. (2009) TMP21 transmembrane domain regulates γ-secretase cleavage. J. Biol. Chem. 284, 28634–28641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vetrivel K. S., Gong P., Bowen J. W., Cheng H., Chen Y., Carter M., Nguyen P. D., Placanica L., Wieland F. T., Li Y. M., Kounnas M. Z., Thinakaran G. (2007) Dual roles of the transmembrane protein p23/TMP21 in the modulation of amyloid precursor protein metabolism. Mol. Neurodegener. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winkler E., Hobson S., Fukumori A., Dümpelfeld B., Luebbers T., Baumann K., Haass C., Hopf C., Steiner H. (2009) Purification, pharmacological modulation, and biochemical characterization of interactors of endogenous human γ-secretase. Biochemistry 48, 1183–1197 [DOI] [PubMed] [Google Scholar]
- 52. Hansson C. A., Frykman S., Farmery M. R., Tjernberg L. O., Nilsberth C., Pursglove S. E., Ito A., Winblad B., Cowburn R. F., Thyberg J., Ankarcrona M. (2004) Nicastrin, presenilin, APH-1, and PEN-2 form active γ-secretase complexes in mitochondria. J. Biol. Chem. 279, 51654–51660 [DOI] [PubMed] [Google Scholar]
- 53. Pavlov P. F., Hansson Petersen C., Glaser E., Ankarcrona M. (2009) Mitochondrial accumulation of APP and Aβ. Significance for Alzheimer disease pathogenesis. J. Cell Mol. Med. 13, 4137–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ferrer I. (2009) Altered mitochondria, energy metabolism, voltage-dependent anion channel, and lipid rafts converge to exhaust neurons in Alzheimer disease. J. Bioenerg. Biomembr. 41, 425–431 [DOI] [PubMed] [Google Scholar]
- 55. Ramírez C. M., González M., Díaz M., Alonso R., Ferrer I., Santpere G., Puig B., Meyer G., Marin R. (2009) VDAC and ERα interaction in caveolae from human cortex is altered in Alzheimer disease. Mol. Cell Neurosci. 42, 172–183 [DOI] [PubMed] [Google Scholar]
- 56. Sultana R., Poon H. F., Cai J., Pierce W. M., Merchant M., Klein J. B., Markesbery W. R., Butterfield D. A. (2006) Identification of nitrated proteins in Alzheimer disease brain using a redox proteomics approach. Neurobiol. Dis. 22, 76–87 [DOI] [PubMed] [Google Scholar]
- 57. Peles E., Nativ M., Lustig M., Grumet M., Schilling J., Martinez R., Plowman G. D., Schlessinger J. (1997) Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 16, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rios J. C., Melendez-Vasquez C. V., Einheber S., Lustig M., Grumet M., Hemperly J., Peles E., Salzer J. L. (2000) Contactin-associated protein (Caspr) and contactin form a complex that is targeted to the paranodal junctions during myelination. J. Neurosci. 20, 8354–8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gollan L., Salomon D., Salzer J. L., Peles E. (2003) Caspr regulates the processing of contactin and inhibits its binding to neurofascin. J. Cell Biol. 163, 1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bai Y., Markham K., Chen F., Weerasekera R., Watts J., Horne P., Wakutani Y., Bagshaw R., Mathews P. M., Fraser P. E., Westaway D., St George-Hyslop P., Schmitt-Ulms G. (2008) The in vivo brain interactome of the amyloid precursor protein. Mol. Cell Proteomics 7, 15–34 [DOI] [PubMed] [Google Scholar]
- 61. Perreau V. M., Orchard S., Adlard P. A., Bellingham S. A., Cappai R., Ciccotosto G. D., Cowie T. F., Crouch P. J., Duce J. A., Evin G., Faux N. G., Hill A. F., Hung Y. H., James S. A., Li Q. X., Mok S. S., Tew D. J., White A. R., Bush A. I., Hermjakob H., Masters C. L. (2010) A domain level interaction network of amyloid precursor protein and Aβ of Alzheimer disease. Proteomics 10, 2377–2395 [DOI] [PubMed] [Google Scholar]
- 62. Compton A. G., Albrecht D. E., Seto J. T., Cooper S. T., Ilkovski B., Jones K. J., Challis D., Mowat D., Ranscht B., Bahlo M., Froehner S. C., North K. N. (2008) Mutations in contactin-1, a neural adhesion and neuromuscular junction protein, cause a familial form of lethal congenital myopathy. Am. J. Hum. Genet. 83, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bhat M. A., Rios J. C., Lu Y., Garcia-Fresco G. P., Ching W., St Martin M., Li J., Einheber S., Chesler M., Rosenbluth J., Salzer J. L., Bellen H. J. (2001) Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron 30, 369–383 [DOI] [PubMed] [Google Scholar]
- 64. Devanathan V., Jakovcevski I., Santuccione A., Li S., Lee H. J., Peles E., Leshchyns'ka I., Sytnyk V., Schachner M. (2010) Cellular form of prion protein inhibits Reelin-mediated shedding of Caspr from the neuronal cell surface to potentiate Caspr-mediated inhibition of neurite outgrowth. J. Neurosci. 30, 9292–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee K. H., Relkin N. R., Finehout E. (October 19, 2006) World Intellectual Property Organization Patent WO/2006/110621
- 66. Berntenis N., Bohrmann B., Guentert A., Mueller B. (December 13, 2007) World Intellectual Property Organization Patent WO/2007/140971
- 67. Luider T. M., Sillevis Smitt P. A. E. (July 17, 2008) World Intellectual Property Organization Patent WO/2008/085035
- 68. Rees R. C., Ball G. R., Matharoo-Ball B., Morgan K., Kalshekar N. A. (December 30, 2009) World Intellectual Property Organization Patent WO/2009/156747
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.