Background: The compaction of DNA into nucleosomes interferes with DNA repair.
Results: Monoubiquitination of core histone H2A destabilizes nucleosomes containing UV-damaged DNA.
Conclusion: Destabilized nucleosomes enable the release of the DNA damage-binding complex DDB1-CUL4BDDB2, which assists in histone ubiquitination.
Significance: This mechanism explains how the ubiquitination of histone H2A, in addition to chromatin remodeling, promotes repair and facilitates genome stability.
Keywords: Cancer, Chromatin Histone Modification, DNA-binding Protein, DNA Damage, E3 Ubiquitin Ligase, DDB1-CUL4BDDB2, H2A Lys-119/Lys-120, NER, Nucleosome, Ubiquitination
Abstract
How the nucleotide excision repair (NER) machinery gains access to damaged chromatinized DNA templates and how the chromatin structure is modified to promote efficient repair of the non-transcribed genome remain poorly understood. The UV-damaged DNA-binding protein complex (UV-DDB, consisting of DDB1 and DDB2, the latter of which is mutated in xeroderma pigmentosum group E patients, is a substrate-recruiting module of the cullin 4B-based E3 ligase complex, DDB1-CUL4BDDB2. We previously reported that the deficiency of UV-DDB E3 ligases in ubiquitinating histone H2A at UV-damaged DNA sites in the xeroderma pigmentosum group E cells contributes to the faulty NER in these skin cancer-prone patients. Here, we reveal the mechanism by which monoubiquitination of specific H2A lysine residues alters nucleosomal dynamics and subsequently initiates NER. We show that DDB1-CUL4BDDB2 E3 ligase specifically binds to mononucleosomes assembled with human recombinant histone octamers and nucleosome-positioning DNA containing cyclobutane pyrimidine dimers or 6-4 photoproducts photolesions. We demonstrate functionally that ubiquitination of H2A Lys-119/Lys-120 is necessary for destabilization of nucleosomes and concomitant release of DDB1-CUL4BDDB2 from photolesion-containing DNA. Nucleosomes in which these lysines are replaced with arginines are resistant to such structural changes, and arginine mutants prevent the eviction of H2A and dissociation of polyubiquitinated DDB2 from UV-damaged nucleosomes. The partial eviction of H3 from the nucleosomes is dependent on ubiquitinated H2A Lys-119/Lys-120. Our results provide mechanistic insight into how post-translational modification of H2A at the site of a photolesion initiates the repair process and directly affects the stability of the human genome.
Introduction
Nucleotide excision repair (NER)6 removes a wide range of DNA helix-distorting lesions and is the only pathway that repairs UV-induced cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts (6-4PP) in human cells. Defects in NER give rise to several heritable human disorders characterized by cancer predisposition (e.g. xeroderma pigmentosum) or premature aging (e.g. Cockayne syndrome and trichothiodystrophy) (1). More than 30 proteins cooperate in forming the sequentially assembled NER complex intermediates, which recognize a distorted helix around the lesion, excise the damaged DNA fragment, and re-synthesize the repaired DNA (2, 3). However, we do not yet understand how the cellular machinery detects UV-induced damage in chromatin-embedded DNA substrates, particularly in the non-transcribed DNA that is under surveillance of the global genome repair initiator complexes: UV-damaged DNA-binding protein complex (UV-DDB) and XPC-RAD23 (4).
Chromatin, which comprises assembled histone proteins and DNA (147 bp of DNA is wrapped around an octamer of the core histones), compacts genomic DNA in the nucleus and occludes access to DNA templates (5, 6). Evidence that chromatin rearrangements occur during NER in vivo (7) and that the presence of nucleosomes reduces the repair rate of damaged DNA in vitro (8, 9) led to the hypothesis that the chromatin structure itself must be changed to promote repair initiation in the non-transcribed DNA (4, 10). The specific mechanism of chromatin modulation in response to DNA damage that facilitates access of the NER machinery to the lesions is among the most important and currently poorly understood problems in research on DNA damage and repair.
Nucleosome structure can be altered locally through the covalent modification of histones, incorporation of histone variants, and/or non-covalent remodeling by ATP-dependent enzymes (11). Presumably, repair proteins responsible for recognizing the helix distortion in the nucleosome direct chromatin modulators to the site of the DNA lesion to facilitate the removal of core histones (12). In addition to the post-translational modifications of histones (Ref. 13 and references therein), several chromatin remodeling complexes, including ACF (14), SWI/SNF (15), and INO80 (16) are implicated in initiation of NER. Cells with disrupted remodelers have impaired NER and increased sensitivity to UV irradiation (17). Although INO80 and BRG1 subunits are present at the sites of the UV lesion and promote the recruitment of damage-binding proteins (XPC, DDB2, and XPA) to the lesion, it remains unclear whether they form direct protein-protein interactions with the proteins that bind the DNA lesions (16, 18, 19).
Direct association between UV-induced ubiquitin modification of the core histones and a NER damage sensor, UV-DDB, has been well established (20–22). UV-DDB comprises two proteins: DDB2, mutated in xeroderma pigmentosum group E patients, and DDB1 (23). The DDB1 protein is part of the substrate-recruiting module of cullin E3 ligases, CUL4A and CUL4B, which target proteins for ubiquitination and participate in various aspects of the UV-damage response (24). DDB2 is both a DNA damage binding subunit and a substrate receptor for the DDB1-CUL4-based E3 ligases, DDB1-CUL4ADDB2 and DDB1-CUL4BDDB2; DDB2 recruits the CUL4A and CUL4B E3 ligase complexes to the site of damaged DNA in the chromatin of UV-irradiated cells (22, 25–27).
In previous studies we demonstrated that mutations in DDB2 alter the co-localization of the DDB1-CUL4DDB2 ligases at the site of UV lesions and the monoubiquitination of the surrounding H2A (20), resulting in the partial impairment of global genome repair in the cells of xeroderma pigmentosum group E patients compared with repair-proficient cells. The direct link between DDB1-CUL4DDB2 and the UV-damage-dependent ubiquitination of other core histones has been confirmed in vitro and in vivo (21), suggesting that this type of modification could facilitate NER in the compact chromatin of the non-transcribed DNA.
Indeed, Wang et al. (21) reported that the in vitro UV-damage-dependent ubiquitination of mononucleosomes with a mixture of CUL4A and CUL4B E3 ligases resulted in the release of modified H3 from nucleosomes and assisted with the recruitment of XPC to a lesion. However, Takedachi et al. (22) found that ubiquitination of H3 and H2B by the CUL4A complex was not sufficient to destabilize the nucleosome and proposed that ubiquitination around damaged sites functions as a signal that enhances the recruitment of XPA repair proteins to lesions. As noted above, our in vivo experiments clearly established the connection between the monoubiquitination of H2A and the binding of DDB1-CUL4DDB2 ligases to UV-damaged chromatin (20, 26), which was not examined in either of these studies. To resolve those contradictory data, here we set up an in vitro system to directly test if the DDB1-CUL4BDDB2-mediated ubiquitination of H2A alone (or in concert with H3) would destabilize mononucleosomes with imbedded photolesions.
Functional DDB2 is required for a second global genome repair initiator complex, XPC-RAD23B, that forms an NER preincision complex (28). Upon recognizing the lesion, the DDB1-CUL4ADDB2 E3 ligase ubiquitinates XPC and autoubiquitinates DDB2. Ubiquitination stabilizes XPC, which increases its affinity for damaged DNA, whereas polyubiquitination of DDB2 reduces its affinity for damaged DNA and leads to its degradation by the proteasome (29, 30). Silencing of CUL4A in human cells leads to the retention of DDB2 on DNA and prevents the loading of XPC on damaged sites, consequently reducing repair of CPDs (31). Notably, XPC binds to 6-4PP lesions severalfold less than UV-DDB, and XPC has no significant affinity for CPD lesions (32–34). FRAP studies indicate that the amount of immobilized XPC on lesions has a biphasic character that is dependent on the number of 6-4PP and amount of DDB2 in the cells (35); these results suggest that UV-induced ubiquitination regulates handover of the 6-4PP lesions from DDB2 to XPC (3). Mutated DDB2 has a minimal effect on the excision of 6-4PP, but repair of the prevailing CPD lesion is severely decreased in the cells of xeroderma pigmentosum group E patients (36).
We sought to examine the consequences of damage-dependent ubiquitination by DDB1-CUL4BDDB2 E3 ligase on nucleosome dynamics utilizing mononucleosomes assembled with human recombinant-tagged histone octamers and nucleosome-positioning DNA containing CPD or 6-4PP photolesions. We show for the first time a link between the ubiquitination of H2A lysines 119 and 120, the destabilization of the UV-damaged nucleosome, resulting from the eviction of the H2A, and the dissociation of polyubiquitinated DDB2 from the nucleosome.
EXPERIMENTAL PROCEDURES
Purification of Human Recombinant E3 Ligase Complexes
For expression in Sf9 cells, the human RBX1 clone (Open Biosystems, ID# 3138751) was amplified as N-terminal His-10-tagged RBX and cloned into the Invitrogen™ pBlueBac4.5 plasmid. The plasmids expressing human CUL4A and CUL4B have been previously described (26). Baculoviruses expressing N-terminal His-10-tagged RBX1, CUL4A, and CUL4B were prepared utilizing the Bac-N-Blue transfection kit (Invitrogen) following the manufacturer's protocols. Baculoviruses encoding N-terminal FLAG-tagged DDB2 and untagged DDB1 were previously described (32). The protein components of the DDB1-CUL4ADDB2 and DDB1-CUL4BDDB2 E3 ligases were co-expressed in Sf9 cells and purified through tandem-affinity chromatography with a His tag on RBX1 and a FLAG tag on DDB2 following the method that was developed for the purification of the UV-DDB complex (32).
Purification of Human Recombinant Histones and Histone Octamers
Human core histones were purified from a bacterial expression system (from J. Parvin, Ohio State University) as described (37). A specific tag sequence on the histone N terminus (Myc tag on H2A and HA tag on H3) was generated by PCR and cloned in a pET vector. The mutated histone Myc-H2A in which lysine 119 or/and lysine 120 is replaced with arginine (H2A K119R; H2A K120R, and H2A K119R/K120R) was produced by PCR using specific primers (supplemental Table 1). Bacteria from the Rosetta strain (Novagen) were transformed with the appropriate pET vector-based histone clone and induced with isopropyl 1-thio-β-d-galactopyranoside. The expressed untagged and tagged histone proteins were extracted from inclusion bodies, dialyzed against salt and urea buffers (SAU buffers), purified on an HiTrap SP HP cartridge (GE Healthcare), and re-natured as histone octamers in the appropriate refolding buffers. Histone octamers were purified through gel filtration in a Superdex 200 HR16/60 column (ACTA Purifier-10 FPLC, GE Healthcare) as a mass of 110 kDa. We purified histone octamers in three combinations of untagged and tagged histone proteins: Myc-H2A (Myc-tag on H2A histone and the other three untagged core histones); Myc-H2A K119R/K120R (Myc-tag on H2A K119R/K120R histone and the other three untagged core histones), and HA-H3 (HA-tag on H3 histone and the other three untagged core histones).
Purification of CPD and 6-4PP Photolyases
Photolyases are enzymes that utilize the visible light to directly repair (photoreactivate) UVC-induced photoproducts. Although photolyases are present in all bacteria, prokaryotes and eukaryotes, placental mammals lost these enzymes during evolution (38). The Potorous tridactylus CPD photolyase gene (39) or Arabidopsis thaliana 6-4PP photolyase gene (40) was cloned into a pCold-IMPACT vector, which combines a His tag at the N terminus from the pCold vector (Takara) and an intein tag at the C terminus from the pTYB1 expression vector (New England Biolabs). The fusion proteins were expressed in Escherichia coli. The CPD photolyases (CPD phr) and 6-4 photolyases (6-4PP phr) were purified with chitin beads (following the protocol of the IMPACT System of New England Biolabs), and the eluted fractions were further purified with Q-Sepharose.
Photoreactivation of 6-4PP and CPD
The 177- or 147-bp nucleosome-positioning 601 DNA sequence (41, 42) with biotin at the 5′ end was amplified by PCR. After purification, the 177-bp DNA (1 μg/μl) was spotted on a plate and UVC-irradiated from above with 5 kJ/m2 to generate the UV-damaged DNA (UV-DNA: with CPD and 6-4PP). To selectively repair the 6-4PP, CPD, or both photoproducts, the UV-DNA (500 ng/μl) was mixed with 6-4PP, CPD, or both photolyases (0.5 μg/reaction), respectively, and illuminated with fluorescent light at room temperature for 1 h (43). The efficiency of the photoreactivation (photo-repair) was monitored with an immunoblot assay as previously described (44). Briefly, after purification using a column (MERmaid SPIN Kit; Fisher), the indicated amounts of DNAs (±UV-DNA and +UV-DNA after photo-repair) were spotted and cross-linked to a nitrocellulose membrane with a MINIFOLD II Slot Blot System (Schleicher & Schuell), then probed with antibody to CPD or 6-4PP and detected by the Chemi-Doc System (Bio-Rad). Utilizing the above treatment (in addition to control, un-damaged DNA, and UV-DNA), we produced three substrates of the nucleosome positioning DNA: CPD-DNA (with CPD only), 6-4PP-DNA (with 6-4PP only), and CPD+6-4PP phr treated-DNA (without CPD and 6-4PP).
Nucleosome Assembly
We reconstituted the nucleosomes with one of the 177- or 147-bp nucleosome-positioning 601 DNA sequence (32pmol) and one of the histone octamer combinations: Myc-H2A, Myc-H2A K119R/K120R, or HA-H3 (described above). The DNA was assembled into mononucleosomes using the salt-jump method in which DNA was incubated with histone octamers (DNA:octamers at 1:1.1 molar ratio) in a stepwise dilution of sodium chloride (2 to 0.25 m NaCl concentration). The quality of assembled nucleosomes was monitored by co-migration of the protein and DNA on a 0.9% agarose/1× Tris borate EDTA gel stained with ethidium bromide. We assembled the mononucleosomes in six combinations: Myc-H2A nucleosomes (with DNA, UV-DNA, CPD-DNA, or 6-4PP-DNA), Myc-H2A K119R/K120R nucleosomes (with UV-DNA), and HA-H3 nucleosomes (with UV-DNA).
Nucleosome Binding Assay
Reconstituted nucleosomes (−UV, +UV, CPD, or 6-4PP nucleosomes) or the same 177-bp DNA template devoid of the histone octamers was incubated with the various concentrations of E3 ligase at room temperature for 30 min in 100 μl of binding buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 0.2 mm EDTA, 1.0 mm MgCl2, and 5% glycerol). A 5′-biotinylated DNA or mononucleosome was pulled down with streptavidin-bound magnetic beads (Dynabeads, Invitrogen) at room temperature for 15 min. The formed complex was eluted with 2× SDS sample buffer and examined by Coomassie staining or immunoblotting for the presence of the E3 ligase proteins (DDB1, DDB2, CUL4B, and RBX1) and core histones.
Ubiquitination Assay
The ubiquitin ligase activity of recombinant DDB1-CUL4DDB2 complexes (indicated concentration) was assayed with a slight modification from previously published methods (26). Human recombinant tagged free histone (70 pmol) or mononucleosomes (32 pmol) were used as substrates. Reactions were performed in a reaction mixture (30 μl) that contained 50 mm Tris, pH 7.4, 1 mm DTT, 10 mm MgCl2, 0.2 mm CaCl2, 4 mm ATP, 0.1 mg/ml bovine serum albumin, 2 mm ubiquitin aldehyde, 100 ng of E1 (Boston Biochem), 400 ng of E2 (UbcH5a or a panel of E2 proteins; Boston Biochem), and 1 mg of human recombinant ubiquitin (Boston Biochem): FLAG-ubiquitin (FLAG-Ub), His6-ubiquitin Lys-48 only (ubiquitin mutant contains only a single lysine residue, Lys-48; other lysines mutated to arginine), or His6-ubiquitinated lysines (ubiquitin mutant contains no lysine residues; all lysines are mutated to arginine). The reactions were incubated for 4 h on a shaking platform (1050 rpm) at 34 °C and were terminated by suspending in SDS sample buffer and analyzed by immunoblotting for the presence of unmodified and modified histones.
Nucleosome Disassembly Assay
A complex of immobilized UV-damaged nucleosomes (48 pmol) ± recombinant DDB1-CUL4BDDB2 ligase was subjected to the ubiquitination assay as described above. Where indicated, ATP was omitted from the reaction. After the ubiquitination assay, the supernatant and pulldown fraction (beads with immobilized UV-damaged nucleosomes) were divided. The beads were subsequently washed with binding buffer (30 μl) supplemented with increased salt concentration (0.1, 0.3, 0.6, 0.9, 1.2, 1.5, and 2 m NaCl). The supernatants (0.1–1.2 m released fractions) and the pulldown beads were suspended in SDS sample buffer and analyzed by immunoblotting for the presence of unmodified and modified histones and/or DDB1 and DDB2 proteins.
Stable Cell Line Expressing FLAG-His-H2A Wild Type or Mutants
The human histone H2A type 3 (NM_033445.2) open reading frame (4–391) was amplified by PCR using the primers, which generated a XhoI site at the 5′ terminus and a NotI site at the 3′ terminus of the amplified fragment. The fragment was cloned into pBluescript II vector, sequenced, and subcloned into a pcDNA5/FRT/TO expression vector (modified with an in-frame XhoI site at the start and a NotI site at the stop codons), which harbors an N-terminal FLAG-His tag (Invitrogen). The recombinant histone FLAG-His-H2A mutants (H2A K119R; H2A K120R, and H2A K119R/K120R) were cloned by amplifying wild type H2A with specific PCR primers (supplemental Table 1). The Flp-In T-REx-293 (FT293) cell line, a derivative of human 293 embryonic kidney fibroblasts (Invitrogen), was used as a host to generate a tetracycline-inducible expression vector of the FLAG-His-H2A by cotransfecting the pcDNA5/FRT/TO expression vector containing wild type or mutated H2A and the Flp recombinase expression plasmid, pOG44. The cells expressing the recombinant histones were selected by hygromycin (150 mm) and maintained in Dulbecco's modified minimum Eagle's medium supplemented with 10% FCS, glutamax, and tetracycline (1 μg/ml). The expression of recombinant histones H2A was analyzed in the chromatin soluble fraction by immunoblotting.
Cellular Fractionations
The FT293 cells were treated with ±UVC radiation (40 J/m2), incubated in a fresh medium for 30 min, and lysed with Nonidet P-40 buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% Nonidet P-40, 5 mm MgCl2, protease inhibitor mixture) for 30 min at 4 °C with rotation. The pellet, recovered after centrifugation, was treated with chromatin lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% Nonidet P-40, 5 mm MgCl2, protease inhibitor mixture, 1 mm CaCl2, 0.6 unit/μl of micrococcal nuclease) for 30 min at 37 °C with vortexing each 5 min. Samples were centrifuged, and the supernatant used as the solubilized chromatin fraction.
Immunoblotting
Proteins were separated on a 15% or 4–20% gradient SDS-polyacrylamide electrophoresis gel and (unless otherwise indicated) transferred onto an Immobilon-P membrane (Novel Experimental Technology) (upper part, including the components of the E3 complex >30 kDa) or a nitrocellulose membrane (Whatman) (lower part, including the core histone proteins between 10 and 30 kDa). The membranes were incubated overnight with the appropriate primary antibodies (supplemental Table 2) at 4 °C and for 1 h with the corresponding horseradish peroxidase (HRP) or alkaline phosphatase-conjugated secondary antibodies at room temperature. Proteins were detected by a chemiluminescent signal produced by HRP/ alkaline phosphatase substrate (Sigma) and captured by BioMax-Light film (Eastman Kodak Co.) and/or a Chemi-Doc System (Bio-Rad).
RESULTS
DDB1-CUL4BDDB2 E3 Ligase Specifically Binds to CPD and 6-4PP Lesions in Mononucleosomes
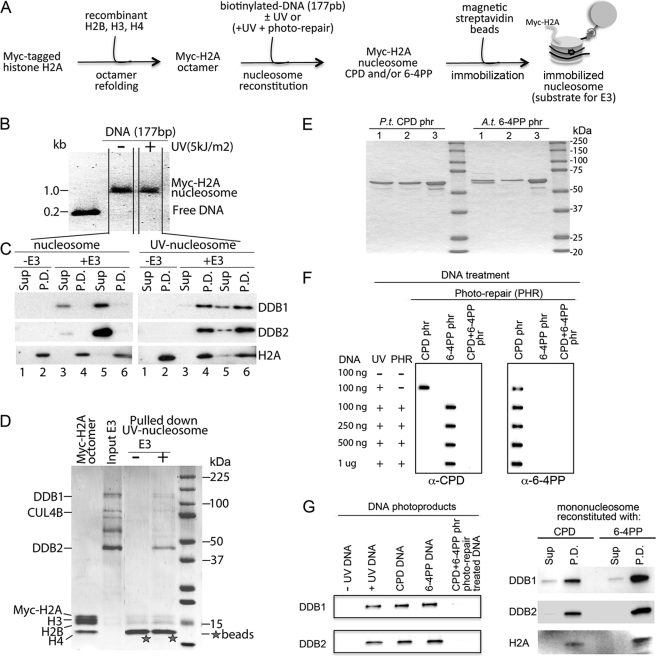
To define the role of human recombinant DDB1-CUL4BDDB2 E3 ligase in the ubiquitination of core histones, we developed an in vitro system with the photoproducts incorporated into a reconstituted mononucleosome in which one of the human core histones is replaced with tagged proteins (Fig. 1A). Importantly, the presence of UV-induced photolesions in a 177-bp nucleosome-positioning 601 DNA sequence and Myc-H2A octamers did not affect efficient reconstitution of the nucleosomes (Fig. 1B).
FIGURE 1.
DDB1-CUL4BDDB2 E3 ligase binds mononucleosomes reconstituted with UV-damaged DNA. A, shown is the experimental strategy for the assembly of nucleosomes used as a binding substrate for the DDB1-CUL4BDDB2 E3 ligase in pulldown studies. B, mononucleosomes were reconstituted in vitro from the mock or UV-irradiated nucleosome-positioning DNA sequence and human recombinant octamers and analyzed on ethidium bromide-stained agarose gel. C, the reconstituted nucleosomes with undamaged DNA (nucleosomes) or UV-damaged DNA (UV nucleosomes) were incubated ± DDB1-CUL4BDDB2, and the DNA was pulled down with streptavidin-bound Dynabeads. The supernatants (20%) and pulled-down beads were boiled and run on a gradient SDS-PAGE. The separated proteins were probed with the DDB1, DDB2, and H2A antibodies. Samples: supernatant (Sup) and pulled-down material (P.D.) containing 4 pmol of mononucleosomes without E3 (lanes 1 and 2), 4 pmol of mononucleosomes and 2 pmol of E3 (lanes 3 and 4), and 4 pmol of mononucleosomes and 4 pmol of E3 (lanes 5 and 6). D, shown is the presence of core histones in a complex ±DDB1-CUL4BDDB2 after pulling down the UV-damaged nucleosomes, confirmed by Coomassie staining of the gel. The input of the binding assay (16 pmol of the E3 and 36 pmol of histone octamers) and pulled-down UV-damaged nucleosomes (32 pmol) were separated as in C. The strong staining overlapping with histone H4 is a nonspecific staining of the beads (marked with stars). E, recombinant proteins, P. tridactylis CPD photolyase (P.t. CPD phr) and A. thaliana 6-4 photolyase (A.t. 6-4 phr) expressed in E. coli, were purified with chitin beads and Q-Sepharose and visualized with Coomassie staining. Samples: chitin bead eluted fraction (lane 1), Q-Sepharose flow-through (lane 2), and concentrated Q-Sepharose elution (5 μg) (lane 3). F, UV-DNA was photoreactivated (PHR: photo-repaired) with 6-4PP or CPD photolyase to generate the nucleosome-positioning DNA sequences with CPD or 6-4PP photoproducts, respectively. The indicated amounts of DNA (1 μg of 177-bp nucleosome-positioning DNA sequence is equivalent to 8 pmol) photoreactivated with one or both photolyases were spotted on nitrocellulose membranes and probed with CPD or 6-4PP antibodies. Samples of ±UV-irradiated DNA were included as controls. G, shown is binding of DDB1-CUL4BDDB2 (2 pmol for the left panel or 4 pmol for the right panel) to ±UV-irradiated and ±photoreactivated DNA (4 pmol) (left panel) and mononucleosomes assembled with CPD or 6-4PP-DNA (8 pmol) (right panel), tested as in C.
We first examined whether the DDB1-CUL4BDDB2 E3 ligase complex directly binds to a UV-induced lesion and, thus, discriminates between damaged and undamaged DNA within the nucleosome structure, as a prerequisite for UV-damage-dependent ubiquitination. When the immobilized nucleosomes with DNA or UV-damaged DNA were incubated with the ligase complex at a molar ratio of 1 to 0.5, the presence of DDB1 and DDB2 proteins was detected only in the unbound supernatant or the pulldown bound fractions, respectively (Fig. 1C, lanes 3 and 4). Increasing the amount of E3 complex in the reactions to a molar ratio of 1 to 1 (Fig. 1C, lanes 5 and 6) did not result in additional binding, indicating that less than one molecule of E3 was specifically bound to a damaged nucleosome. We detected the same efficiency of E3 binding to a damaged nucleosome, reconstituted with a 147-bp nucleosome-positioning 601 DNA sequence (supplemental Fig. S1), which rules out the possibility of predominant binding to the lesion in linker DNA. Thereafter, we used a molar ratio of 1 to 0.5 in all binding assays. The dose of UV irradiation applied here (5 kJ/m2) randomly produces about 11 CPDs per molecule of the positioning DNA sequence (45). We detected less than one molecule of E3 bound to a damaged nucleosome; assembly of DNA into a nucleosome may make it less accessible for DDB2 binding. In preliminary experiments, we did not detect binding of E3 to nucleosomes reconstituted with DNA irradiated with 0.5 kJ/m2 (not shown). However, we do not know if the structure and spatial requirements for DDB1-CUL4BDDB2 could affect the number of simultaneously bound complexes on a nucleosome.
We further demonstrated the presence of the four core histone proteins in a complex with the E3 ligase by Coomassie staining of the gel (Fig. 1D) after pulling down the UV-damaged nucleosomes, confirming that the binding of the CUL4B complex occurs at the mononucleosomal level and does not affect the stoichiometry of the core histones, as shown for its counterpart CUL4A E3 ligase (22).
In human cells, detection of CPD in non-transcribed DNA by the damage-sensor protein XPC is inefficient (28), indicating that the UV-DDB complex, although binding strongly to DNA fragments containing 6-4PP (32), plays a crucial role in the detection and repair of CPD in the context of chromatin (35). To explore how DDB1-CUL4BDDB2 E3 ligase contributes to the repair of each type of UVC photolesion, we used recombinant P. tridactylis CPD photolyase and A. thaliana 6-4 photolyase, purified almost to homogeneity (Fig. 1E), to produce a mononucleosome with a single type of photolesion. The antibodies specific to each photoproduct were used to monitor the efficiency of the CPD or 6-4PP photolyase to photoreactivate the UV-DNA sequences and generate DNA with a 6-4PP or CPD photoproduct, respectively. The immunoblots confirm the substrate specificity of each enzyme to repair CPD or 6-4PP photoproducts in the UV-DNA sequence (Fig. 1F).
The affinity of the DDB1-CUL4BDDB2 E3 ligase to bind to a mononucleosome with CPD or 6-4PP photolesions (Fig. 1G, right panel) or the same DNA template devoid of the histone octamer is demonstrated in Fig. 1G, left panel. When UV-DNA was treated with both photolyases, as in unirradiated control DNA, no binding could be detected (Fig. 1G, left panel). Even though 6-4PPs are induced at one-third the frequency of CPD (45), we detected about 1.3-fold more DDB1 and DDB2 proteins in the pulldown fraction of 6-4PP-DNA and nucleosomes compared with CPD-DNA and nucleosomes. Collectively, these results demonstrate that DDB1-CUL4BDDB2 E3 ligase discriminates between damaged and undamaged DNA within the mononucleosomes and directly binds to mononucleosomes containing CPD or 6-4PP photolesions.
DDB1-CUL4BDDB2 E3 Ligase Targets Histones H2A and H3 Organized in Mononucleosomes with Damaged DNA
We previously demonstrated that free H2A is a substrate for DDB1-CUL4BDDB2 (26), and it was shown that an E3 complex that contained CUL4A and CUL4B could specifically target H3 in vitro and in vivo (21). Therefore, we assessed the capacity of the E3 ligase formed by CUL4B to ubiquitinate histones H2A and H3 organized in nucleosomes. To overcome the lack of suitable antibodies for ubiquitinated core histones, we overproduced H2A and H3 as fusion proteins with a specific epitope tag, Myc and HA, respectively. We reconstituted 2 types of nucleosomes by replacing histone H2A or H3 with the corresponding tagged proteins. Such an approach enabled us to immunodetect unmodified and ubiquitinated histones with an antibody against each tag. Because DDB1-CUL4BDDB2 E3 ligase requires other components of the ubiquitination reaction that are not well characterized, we tested a panel of E2s in a ubiquitination assay with Myc-H2A as a substrate. The UbcH5a was the most efficient type of E2 for the E3 ligase formed by CUL4B (supplemental Fig. S2), as previously demonstrated for its CUL4A counterpart (46). Furthermore, we tested and confirmed that human recombinant DDB1-CUL4BDDB2 purified from Sf9 cells was more efficient than DDB1-CUL4ADDB2 in ubiquitinating free histones H2A and H3 in vitro (supplemental Fig. S3), as we previously had demonstrated for H2A when the CUL4 complexes were purified from human cells (26).
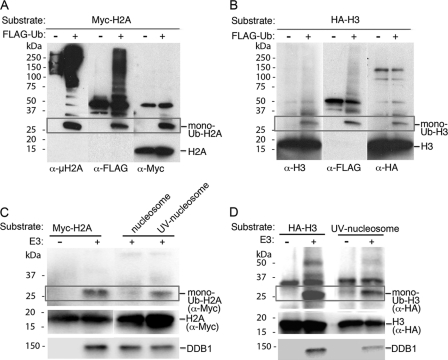
To define the conditions for the reliable detection of modified H2A and H3, we prepared the ubiquitination assay (±FLAG-Ub) in triplicate using FLAG antibody, which detects any FLAG moiety (i.e. unmodified and modified FLAG-tagged DDB2 around position 50 kDa and above, respectively, were detected in Fig. 2, A and B) and antibodies for the individual histones (Myc, HA, H3) and for modified H2A (uH2A). Extensive use of the uH2A antibody revealed recognition of the ubiquitin moiety on various substrates in addition to H2A (not shown), which probably accounts for the detection of bands above 75 kDa in Fig. 2A. Results obtained with three different antibodies indicate that DDB1-CUL4BDDB2 primarily monoubiquitinates free H2A and H3 in vitro (Fig. 2, A and B).
FIGURE 2.
DDB1-CUL4BDDB2 E3 ligase targets tagged nucleosomal H2A and H3 for ubiquitination in a photolesion binding-dependent manner. Free Myc-H2A (A) or HA-H3 (B) was used as a substrate for DDB1-CUL4BDDB2 E3 ligase (4 pmol). Each ubiquitination assay was performed ±FLAG-ubiquitin (FLAG-Ub) in triplicate. The total reaction was separated on 15% SDS-PAGE. Modified histone was detected with uH2A, FLAG, and Myc in A and with H3, FLAG, and HA antibodies in B. C, free Myc-H2A and nucleosomal Myc-H2A (nucleosomes or UV-nucleosomes) in C or free HA-H3 and nucleosomal HA-H3 (UV- nucleosomes) in D were used as a substrate for DDB1-CUL4BDDB2 E3 ligase (8 pmol). Membrane strips were probed with DDB1 and Myc (C) or HA (D) antibodies.
We next measured the ubiquitination of histones H2A and H3 organized in nucleosomes. We immunodetected substrates and the modified products in the total assay reaction without pulling down the nucleosomes to compare the pattern of ubiquitination between free and nucleosomal histones (Figs. 2, C and D). The ubiquitination of core histone H2A preferentially occurs in mononucleosomes with UV-damaged DNA, with the pattern similar to that of free histones. Note that histone H2A in undamaged nucleosomes is not a target for ubiquitination by DDB1-CUL4BDDB2 (Fig. 2C), suggesting that nucleosomal organization makes H2A less accessible to the E3. Although histone H3 in the form of an H3-H4 tetramer is sandwiched between two H2A-H2B dimers in the core particle (5), the pattern of modification of core H3 is similar to that observed for free H3, which suggests that one of the lysine residues on the protruding histone tails is a substrate for DDB1-CUL4BDDB2 (Fig. 2D). Taken together, these results show that DDB1-CUL4BDDB2 E3 ligase has to recognize and bind to a photolesion in the nucleosomal DNA before targeting the core histones H2A and H3 for monoubiquitination. Presumably, the binding enables positioning of the E3 and ubiquitination machinery (i.e. E2 with activated ubiquitin) on UV-damaged mononucleosomes.
DDB1-CUL4BDDB2 E3 Ligase Monoubiquitinates H2A at Multiple Lysine Residues
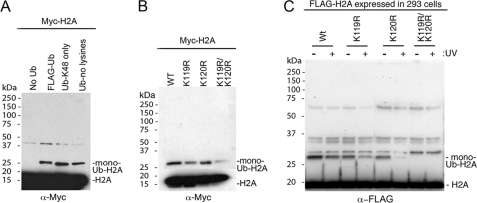
Detecting more than one shifted band of ubiquitinated Myc-H2A in some of the tested conditions in Fig. 2A implied that the E3 ligase could attach several ubiquitin molecules to the substrate. To elucidate this further and discriminate between poly- and multiubiquitination (47, 48), we replaced unmodified FLAG-ubiquitin with a ubiquitin-Lys-48-only (which can support extension of the ubiquitin chain only through Lys-48) or a ubiquitin-no-lysine mutant (which can produce only mono-ubiquitinated substrate) and found that a single ubiquitin molecule is attached to free Myc-H2A with each mutant, suggesting that DDB1-CUL4BDDB2 targets a single lysine residue (Fig. 3A).
FIGURE 3.
DDB1-CUL4BDDB2 ligase activity in targeting H2A mutants. A, ubiquitination of a free Myc-H2A histone was assayed in the presence of unmodified ubiquitin (FLAG-Ub) or mutated ubiquitin: Ub-K48 only (single lysine residue, Lys-48) or Ub-no lysine (all lysine residues mutated). Unmodified and modified H2A was detected with Myc antibody. B, wild type or H2A mutants (H2A K119R, H2A K120R, and H2A K119R/K120R) were used as a substrate for DDB1-CUL4BDDB2 E3 ligase, and products were detected as in A. C, shown is expression of FLAG-His-tagged wild type (WT) or H2A mutants (H2A K119R, H2A K120R, and H2A K119R/K120R) in the chromatin soluble fraction of the stable isogenic 293 cell lines 30 min after ±UV irradiation of 40 J/m2. Unmodified and modified histones were detected with FLAG antibody at the position of 20 and 27 kDa, respectively. The other bands, detected in the chromatin fractions above monoubiquitinated-H2A, presumably represent a different type of modification that occurred endogenously or after damage.
We next sought to identify the target site for DDB1-CUL4BDDB2 in H2A. The identified lysine residue targeted for monoubiquitination in vivo reported in the literature was Lys-119 (49, 50). H2A contains five lysines in its C terminus, and Lys-120 is adjacent to Lys-119. After a closer look at prior publications that described a site of ubiquitination in histone H2A either by purified E3 ligase in vitro (50) or in various cells in vivo (51, 52), we found that in most of the analyzed histone H2A, a ubiquitin was attached to the second lysine, i.e. Lys-120. To clarify this question, we cloned two Myc-H2A single mutants by replacing lysine 119 or 120 with arginine (H2A K119R or H2A K120R) or a double mutant, H2A K119R/K120R. We used wild type and each H2A mutant as substrates for DDB1-CUL4BDDB2 with unmodified FLAG-ubiquitin. After immunoblotting with Myc antibody, a signal corresponding to monoubiquitinated H2A was detected in the three mutants used, with some variation in the intensity of the shifted signals (Fig. 3B). However, the relative level of monoubiquitinated H2A remained unchanged when compared with the corresponding level of unmodified H2A (Fig. 3B). These results suggest that the E3 ligase targets only one lysine in each H2A molecule in vitro and that H2A Lys-119 or Lys-120 is not the only target of DDB1-CUL4BDDB2 as ubiquitination could occur at multiple lysine residues of H2A. This could have resulted from our use of the purified complex in which CUL4B does not contain NEDD8 modification, required for E3 ligase activity in vivo (53).
To determine whether histones H2A K119R and/or K120R are ubiquitinated under more physiological conditions, we generated the stable isogenic 293 cell lines expressing H2A and its mutants tagged with a FLAG-His epitope. Analysis of chromatin fractions from the 293 cell lines showed that all FLAG-H2A proteins are expressed at a similar level (compare the bands detected with FLAG antibody at 20 kDa in Fig. 3C). Importantly, in non-irradiated cells (−UV lanes in Fig. 3C), a mutation in FLAG-H2A Lys-119 or Lys-120 does not affect the level of monoubiquitinated H2A when compared with wild type FLAG-H2A in the chromatin fraction. Furthermore, ubiquitination was completely eliminated in the double mutant H2A K119R/K120R. These results indicate that either Lys-119 or Lys-120 is a target for monoubiquitination in vivo.
Several E3 ligases are implicated in targeting histone H2A upon UV irradiation (22, 49, 54). We previously observed that the presence of functional DDB1-CUL4DDB2 E3 restores the level of monoubiquitinated H2A in the cells after initial loss of this modification after UV irradiation (20). Therefore, we next compared the level of ubiquitination of the FLAG-H2A wild type and mutants in the chromatin fraction of non-irradiated 293 cells with the fractions collected 30 min after UV treatment of the cells. A decreased signal for monoubiquitinated wild type H2A and FLAG-H2A Lys-119 or Lys-120 was detected; the monoubiquitination of the FLAG-H2A K119R/K120R was not detected even after UV irradiation (compare the −UV with the +UV lanes in Fig. 3C). These data confirm that E3 ligase(s) involved in the cellular response to UV-damaged DNA target the same H2A lysine residues that serve as the endogenous substrate for ubiquitination in vivo.
Monoubiquitinated H2A Lys-119 and Lys-120 Are Required for Destabilization of UV-damaged Nucleosomes
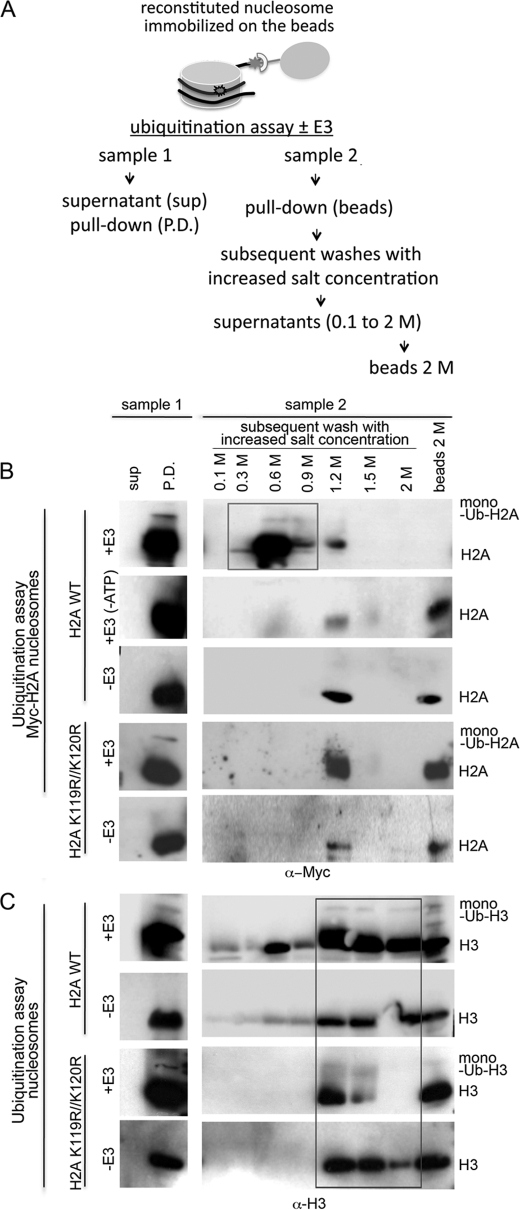
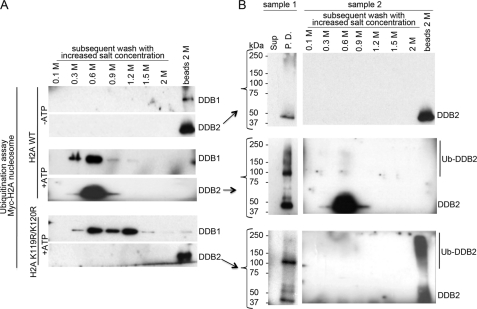
We tested the hypothesis that UV-damaged mononucleosomes are more easily disassembled as a result of weakening the H2A-DNA interaction due to the monoubiquitination of the H2A Lys-119 and Lys-120. We combined a ubiquitination pulldown assay with subsequent washes of immobilized nucleosomes with increased salt (NaCl) concentrations (Fig. 4A), which is a routine method to induce nucleosome disassembly in vitro (i.e. conversely to the order of nucleosome assembly in vitro, as the salt concentrations are increased, the H2A-H2B dimers release from the DNA first and then the (H3-H4)2 tetramer) (55).
FIGURE 4.
Monoubiquitinated H2A Lys-119 and Lys-120 facilitate disassembly of a UV-damaged nucleosome. A, shown is the experimental design of the nucleosomal disassembly assay. Immobilized UV-damaged nucleosomes (wild type H2A or H2A K119R/K120R) were used as a substrate for the ubiquitination assay ± DDB1-CUL4BDDB2 E3. Unless indicated the ubiquitination reaction was supplemented with ATP. After the reaction was terminated, the supernatant and beads (DNA-bound fraction) were divided. Fractions from sample 1 were loaded on the gel as a total yield of unmodified and modified histone per ubiquitin reaction in vitro. Beads from the sample, run in a parallel reaction (sample 2), were washed with buffer using increasing amounts of NaCl and boiled with sample buffer. Samples were run on 4–20% gradient SDS-PAGE, and the membrane was probed with Myc antibody to detect modified and unmodified H2A (B) and H3 antibody to detect modified and unmodified H3 (C).
As shown in Figs. 4, A and B, we performed a ubiquitination assay (±E3) using UV-damaged nucleosomes reconstituted with Myc-H2A as a substrate. Immunoblotting of the pulldown beads with ubiquitinated nucleosomes and the corresponding supernatant (Fig. 4B, sample 1, +E3) did not reveal the presence of dissociated H2A in the supernatant fraction (containing 100 mm salt); this observation prompted us to treat the pulldown nucleosomes from the sample run in a parallel reaction with increased salt concentrations. We reasoned that the appearance of H2A in the fractions of lower salt concentration extracted from the ubiquitinated nucleosomes compared with the unubiquitinated control would indicate the contribution of modified H2A to the disassembly of mononucleosomes in vitro. Indeed, the majority of modified and unmodified Myc-H2A was extracted from the ubiquitinated UV-damaged nucleosomes at 0.3–0.9 m salt concentrations, and a smaller fraction was extracted with a 1.2 m salt concentration. In control nucleosomes with unmodified histones (Fig. 4B, sample 2, −E3), a large portion of H2A was extracted with a 1.2 m salt concentration, and some H2A remained bound to the beads with UV-DNA. To rule out the possibility that destabilization of the nucleosome is the result of DDB1-CUL4BDDB2 binding to UV-DNA rather than the consequence of H2A ubiquitination, we performed a ubiquitination reaction supplemented with the E3 and depleted for ATP on the Myc-H2A WT nucleosome (Fig. 4B, sample 2; +E3, −ATP). The binding of E3 to the nucleosome is documented in Fig. 5A. The pattern of unmodified H2A extraction from this sample was similar to the pattern for the nucleosome incubated without E3 in the ubiquitination reaction (compare Fig. 2B, sample 2; +E3, −ATP, with Fig. 4B, sample 2; −E3).
FIGURE 5.
Histone mutant H2A K119R/K120R prevents dissociation of polyubiquitinated DDB2 from UV-damaged nucleosomes. Samples were prepared as outlined in Fig. 4A with a modified ubiquitination assay; E3 was a component in all reactions, and ATP was supplemented as indicated (±ATP). Samples were run on a 4–20% gradient SDS-PAGE gel after which the membrane strips were probed with DDB1 and DDB2 antibodies (A) and the whole membrane was probed with DDB2 antibody (B).
We next assessed the effect of using a double mutant H2A K119R/K120R on the ubiquitination and consequent disassembly of UV-damaged nucleosomes. As with wild type nucleosomes, monoubiquitinated H2A was detected in pulldown beads with mutated nucleosomes (Fig. 4B, sample 1, +E3), which corroborates the result obtained with a free H2A K119R/K120R (Fig. 3D). In marked contrast, the mutated lysine residues affected the pattern of the H2A distribution in the salt extracts from ubiquitinated mutated nucleosomes when compared with wild type nucleosomes (Fig. 4B). The modified and unmodified H2A K119R/K120R were not extracted with lower salt washes but were detected in a 1.2 m salt fraction, and unmodified mutant was bound to the beads with UV-damaged DNA (Fig. 4B, sample 2, +E3). Furthermore, except for the absence of the modified form, the pattern of H2A K119R/K120R extraction in the control, non-ubiquitinated mutant nucleosomes was similar to the pattern for the sample with ubiquitinated nucleosomes with mutant histones. Collectively, these results demonstrate that DDB1-CUL4BDDB2 can target another lysine residue(s) in nucleosomal H2A; nevertheless, the ubiquitination of H2A Lys-119 and Lys-120 profoundly affects the dissociation of H2A and the destabilization of UV-damaged mononucleosomes in vitro.
We also assessed whether ubiquitinated H3 contributes to the destabilization of mononucleosomes by comparing the appearance of H3 in the salt washes of wild type H2A or H2A K119R/K120R UV-damaged nucleosomes after ubiquitination (±E3). Dissociation of H3 from nucleosomes should require higher salt concentrations than H2A in the chromatin soluble fraction (55, 56). Thus, the majority of H3 was extracted with 1.2–2 m salt concentration and some H3 that had been retained bound to the beads in all samples (Fig. 4C). Variable amounts of H3 were detected in 0.1–0.6 m salt fractions extracted from wild type H2A nucleosomes independent of ubiquitination. We cannot rule out that the appearance of H3 in these fractions could have resulted from our use of the tagged HA-H3 to reconstitute wild type H2A nucleosomes. Importantly, neither modified nor unmodified H3 was present in lower salt fractions extracted from the nucleosome with mutated H2A K119R/K120R that contained untagged H3 (Fig. 4C). These results suggest that DDB1-CUL4BDDB2-mediated ubiquitination of H3 does not significantly affect the stability of UV-damaged mononucleosomes in vitro when H2A K119R/K120R are not available for ubiquitination.
Release of Polyubiquitinated DDB2 Requires Disassembled UV-damaged Nucleosomes
After demonstrating an association between the ubiquitination of H2A and structural changes in nucleosomes, we set out to determine whether the release of auto-polyubiquitinated DDB2 is affected by nucleosomal structure. The release of DDB2 from the UV-nucleosomes was assessed in H2A wild type and H2A K119R/K120R nucleosomes as outlined in Fig. 4A. To monitor the binding and release of DDB2 and DDB1, presumably as the E3 ligase complex, with either activated (+ATP) or inhibited ubiquitination (−ATP), we modified the ubiquitination assay accordingly. The unmodified DDB2 remained bound to UV-damaged DNA in the beads fraction even after washed with increasing salt concentrations for the H2A wild type nucleosomes under the conditions of inhibited ubiquitination (−ATP); DDB1 co-localized with DDB2 (Fig. 5, A and B). Conversely, the majority of DDB2 was extracted in the 0.6 m salt fraction under the condition of active ubiquitination; a small fraction of DBB2 was detected in the 0.9 m salt extract. Distribution of the extracted DDB1 resembled the distribution of DDB2; DDB1 was mostly detected in the 0.6 m fraction, and a small amount was extracted with 0.3 and 0.9 m salt washes. Unexpectedly, when H2A K119R/K120R nucleosomes were ubiquitinated and subjected to increased monovalent salt washes, polyubiquitinated DDB2 remained bound to UV-DNA in the beads fraction but dissociated from DDB1, which was extracted at 0.6–1.2 m salt concentrations. Overall, the distribution of modified DDB2 correlates with the distribution of monoubiquitinated H2A in wild type and mutant UV-nucleosomes (compare Fig. 5B and Fig. 4B). Results shown in Figs. 5 and 4B indicate that nucleosomes must be disassembled to release polyubiquitinated DDB2 from UV-damaged DNA. Notably, polyubiquitination of DDB2 seems to weaken the DDB1-DDB2 protein interaction in addition to affecting the binding to damaged DNA.
DISCUSSION
With regard to the impact of monoubiquitination on nucleosome stability, prior reports are inconclusive as to whether DDB2-mediated ubiquitination of the core histones destabilizes the nucleosomes, thereby leading to histone eviction and assembly of the NER complex at a lesion (21, 22). Furthermore, it has not been reported whether ubiquitination of only one core histone can affect interaction with the DNA and, therefore, destabilize the UV-damaged nucleosome or if coordinated modification of several core histones is required. Here, we provide convincing data that monoubiquitination of H2A Lys-119 and Lys-120 by DDB1-CUL4BDDB2 is indispensible for destabilization of the photolesion-containing nucleosomes, leading to eviction of H2A from the nucleosome, and that the partial eviction of H3 from the nucleosomes also depends on ubiquitinated H2A Lys-119/Lys-120. Furthermore, nucleosomal structure has consequences for the bound E3 ligase complex; polyubiquitinated DDB2 is only released from the destabilized nucleosome, presumably freeing space around the lesion to load the NER preincision complex and proceed with repair.
We used photolyases to generate the CPD and 6-4PP lesions, the latter of which is not commercially available, to demonstrate that the E3 ligase can recognize both CPD and 6-4PP efficiently. We found that the affinity of DDB1-CUL4BDDB2 for nucleosomes with 6-4PP lesions is 1.2–1.5-fold greater than for those with CPD lesions, consistent with previous findings of UV-DDB binding to naked DNA fragments in vitro (32). While this manuscript was under review, it was published that EMSA analysis of UV-DDB binding to nucleosomes, containing a chemically synthesized 6-4PP or CPD lesion, confirmed the weaker binding to CPD nucleosomes than to 6-4PP-nucleosomes (57). However, these findings alone cannot account for the significant difference in NER capacity for these lesions. Repair of the CPD and 6-4PP differs both in vitro and in vivo. The half-life of 6-4PP and CPD lesions is about 2 and 24 h, respectively (1). In cells a 6-4PP is repaired by the cooperative actions of XPC and DDB2, whereas the repair of CPD depends on DDB2 recognition (35, 36, 58). Together, the low amount of DDB2 protein (∼1 × 105 molecules per cell) and the UV dose-dependent degradation within 2 h post-treatment implicate DDB2 as a limiting factor in repair of CPD lesions (58). Enhanced expression of DDB2 improved repair of both CPD and 6-4PP lesions in dermal fibroblasts (59). Thus, the amount of endogenous DDB2 protein in addition to the binding affinity for photolesions defines the outcome of repair of CPD and 6-4PP lesions.
Although we showed that CPD or 6-4PP lesions are present in DNA fragments after photolyase treatment, we did not determine the exact number of lesions in the damaged DNA nor their location within the 177-bp nucleosome-positioning 601 DNA sequence. However, our data reflect DDB1-CUL4BDDB2 E3 ligase binding to DNA damage located in the nucleosome core, because the same binding efficiency of the E3 was obtained when the UV nucleosome was reconstituted with a 147-bp DNA sequence that does not contain the 30-bp of linker DNA sequence. Whether the number or location of photolesions affects ubiquitination and destabilization of the nucleosome must be determined in experiments using a nucleosome containing a single chemically synthesized lesion. Nonetheless, a single UV lesion at a specific site can promote the transient, spontaneous unwinding of nucleosomal DNA. This “breathing” activity is limited to DNA at the terminal edges of the nucleosomal template, as detected by FRET, and might facilitate the detection of photolesions (60).
Monoubiquitinated H2A is an abundant histone modification affecting 5–15% of nucleosomal H2A (61) that is mediated by various E3 ligases. H2A is endogenously monoubiquitinated at Lys-120, predominantly by the Ring1B (Ring2) E3 ligase (50); the same lysine residue is targeted upon UV treatment (49). Importantly, the Ring1B-Mel-18 E3 complex has specificity for lysines 119 and 120 of nucleosomal histone H2A; in in vitro experiments, nucleosomes in which these lysines are replaced with arginines remain largely unmodified (62), which corroborates our finding that either Lys-119 or Lys-120 is monoubiquitinated in vivo. After UV damage, which E3 ligase is involved and when it does so appears to determine whether the ubiquitinated histone aids NER or is involved in another aspect of the cellular response to UV-damaged DNA (63). The persistence (24 h post-UV irradiation) of H2A ubiquitinated by the MDC1-RNF8 complex suggests a role for modified-H2A in the response to amplification of the DNA damage signal rather than actual damage recognition (54). Furthermore, ubiquitination of H2A, which depends on the functional endonucleases XPF and XPG and the histone chaperone CAF1, suggests a role for modified-H2A in chromatin restoration after the completion of NER (64). The Ring2 E3 ligase is associated with UV damage-induced monoubiquitinated H2A in an ATR-dependent manner (49), but the most recent data do not support its role in the initiation of NER (64), emphasizing the involvement of DDB1-CUL4DDB2 E3 ligases in H2A modification immediately upon UV insult. Notably, DDB2 is the first XP factor that co-localizes with a photolesion, loading the E3 activity around damaged sites in chromatin independently of the functional status of other NER proteins (26, 65).
The structural impact of ubiquitinated H2A on the bulk configuration of the undamaged nucleosome remains inconclusive, with published reports confirming nucleosomal assembly (66) and the binding of linker histone H1 (67). Conversely, ubiquitinated-H2A:ubiquitinated-H2B.1 dimers dissociate at lower salt concentrations than H2A:ubiquitinated-H2B.1 dimers in nucleosomes reconstituted with the histones purified from chicken cells (68). Moreover, ubiquitination may serve as a signal for recognition or operate synergistically with other post-translational modifications of H2A in an ATM-driven DNA damage response. After DNA double strand breaks, H2A Lys-120 is ubiquitinated through Lys-63 linkage of ubiquitin moieties by the E3 ligases RNF8 and RNF168, leading to the recruitment of repair proteins to the sites of DNA damage (69). However, our findings demonstrate that monoubiquitinated-H2A Lys-119/120, in response to CPD and 6-4PP photolesions, destabilizes the recombinant nucleosome core particle, allowing easier removal of H2A and suggesting altered or disrupted histone-DNA interactions. Our studies with recombinant nucleosomes allowed us to concentrate specifically on the posttranslational modification of ubiquitination, unlike previous studies, whereas nucleosomes purified from cells contain multiple types of posttranslational modifications (66–68). Despite the fact that DDB1-CUL4BDDB2 ligase can target lysine residues in H2A other than Lys-119 and Lys-120 as well as residues in H3, loss of nucleosomal stability occurs only when the lysines on the distal part of the H2A C termini are ubiquitinated. Thus, although ubiquitinated-H3 has been implicated in the destabilization of a UV-damaged nucleosome (21), neither our data nor those obtained with the DDB-CUL4ADDB2 E3 (22) support such a role.
H2A is the only one of the four core histones with a significant C-terminal tail that is not a part of the histone-fold domain (70), and on the nucleosome core particle electron-density map (PDB code 1EQZ) the tail is visible only to Leu-115, leaving the rest of the C-terminal residues disordered (71). Thus, the disordered tail precludes speculation as to how the modified H2A Lys-119/120 could affect interaction with DNA. Nevertheless, the ubiquitinated Lys-119/120 are localized within the docking domain of H2A that has been shown to affect histone octamer and nucleosome stability (72, 73).
Unexpectedly, we also observed that nucleosomal structure, specifically the eviction of ubiquitinated H2A Lys-119/Lys-120, affects the release of polyubiquitinated DDB2 from the damaged DNA. This release corroborates results reported for DDB1-CUL4ADDB2 ligase purified from HeLa cells in which a significant amount of unmodified or moderately ubiquitinated DDB2 was retained in the nucleosomal fraction after ubiquitination (22). Interestingly, the polyubiquitinated DDB2 is efficiently released from UV-damaged DNA, whereas naked DNA is a binding substrate for DDB1-CUL4ADDB2 ligase (29). This raises the question of whether a different conformation of the DDB1-CUL4DDB2 ligase is required to target substrates for mono- (i.e. nucleosomal histone H2A) versus polyubiquitination (i.e. autopolyubiquitination of DDB2). Indeed, we recently reported that DDB2 dimerizes upon binding to damaged DNA, with consequences for loading the E3 ligase on the damaged nucleosome (74). The finding corroborates the notion that dimerization of cullin-based E3 enables interaction with various protein targets and increases the efficiency of ubiquitination (75).
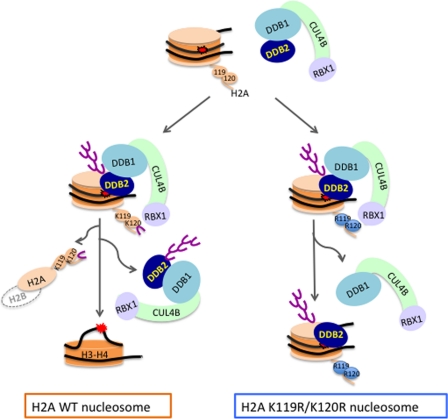
Here, we propose a model for the role of monoubiquitinated-H2A mediated by DDB1-CUL4BDDB2 E3 ligase in the initiation of NER by regulating release of the damage-binding protein DDB2 from the UV-damaged nucleosome (Fig. 6). The E3 ligase ubiquitinates H2A at lysine 119 or 120 in nucleosomes with a CPD or 6-4PP lesion, concomitant with the dissociation of the H2A and the release of DDB1-CUL4BDDB2. Replacement of these two lysines with arginines generates nucleosomes that resist ubiquitination, thus preventing H2A removal and leaving polyubiquitinated-DDB2 bound to the lesion, which presumably obstructs the loading of XPC that is required for the assembly of the NER preincision complex. Our results reveal how post-translational modification of H2A at the site of a photolesion initiates the repair process, which affects the stability of the human genome.
FIGURE 6.
Proposed model for the role of ubiquitinated H2A Lys-119 and Lys-120 in UV-damaged nucleosomes in the initiation of NER. DDB2 binds to a 6-4PP or CPD photolesion and positions the DDB1-CUL4BDDB2 E3 ligase over the damaged nucleosome. The E3 ligase facilitates the transfer of activated ubiquitin from UbcH5 (E2) to the core histone H2A Lys-119/Lys-120 and to DDB2 itself. Monoubiquitination of the H2A Lys-119/Lys-120 leads to dissociation of H2A, presumably as (H2A-H2B) dimers, and destabilization of the nucleosome. This destabilization facilitates release of polyubiquitinated DDB2, freeing space around the lesion for loading of the NER preincision complex. Nucleosomes in which these lysines are replaced with arginines (i.e. H2A K119R/K120R) are resistant to such structural changes; thus, the polyubiquitinated DDB2 is not disassociated from the damaged nucleosome, leading instead to the release of the DDB1-CUL4-RBX1 E3 subcomplex. Note that Andrews et al. (76) described an equilibrium constant for the transition between 2(H2A/H2B)-(H3-H4)2-DNA and 2(H2A/H2B) + DNA-(H3-H4)2 nucleosomal structural states, confirming a function of tetrasomes ((H3-H4)2-DNA), after eviction of the 2(H2A/H2B).
Supplementary Material
Acknowledgments
We thank Alana Cheeks-Lomax for helping with the cloning and expression of CUL4B, Jelena Grahovac for assisting with the cloning and purifying the core histones, Dr. Jennifer Guerrero-Santoro for E3 ligase expertise, and Dr. Guillermo Calero for protein purification expertise. We appreciate the helpful discussion with our colleagues at the University of Pittsburgh Cancer Institute and members of the Program of Molecular and Cellular Cancer Biology. University of Pittsburgh Cancer Institute shared resources were supported in part by National Institutes of Health Award P30CA047904.
This work was supported, in whole or in part, by National Institutes of Health Grants GM07787 (to S. H. L.) and CA122177 (to P. R.-C.). This work was also supported by a University of Pittsburgh School of Medicine start-up fund to V. R.-O.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- NER
- nucleotide excision repair
- CPD
- cyclobutane pyrimidine dimer
- 6-4PP
- 6-4 photoproducts
- UV-DDB
- UV-damaged DNA-binding protein complex
- phr
- photolyase
- Ub
- ubiquitin
- XPC
- xero derma pigmentosum group C protein.
REFERENCES
- 1. Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2005) DNA Repair and Mutagenesis, 2nd Edition, American Society for Microbiology, Washington, D. C [Google Scholar]
- 2. Gillet L. C., Schärer O. D. (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106, 253–276 [DOI] [PubMed] [Google Scholar]
- 3. Naegeli H., Sugasawa K. (2011) The xeroderma pigmentosum pathway. Decision tree analysis of DNA quality. DNA Repair 10, 673–683 [DOI] [PubMed] [Google Scholar]
- 4. Reed S. H. (2011) Nucleotide excision repair in chromatin. Damage removal at the drop of a HAT. DNA Repair 10, 734–742 [DOI] [PubMed] [Google Scholar]
- 5. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 6. van Holde K. E. (1988) Chromatin, Springer-Verlag New York, LLC, New York [Google Scholar]
- 7. Smerdon M. J., Lieberman M. W. (1978) Nucleosome rearrangement in human chromatin during UV-induced DNA- reapir synthesis. Proc. Natl. Acad. Sci. U.S.A. 75, 4238–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Araki M., Masutani C., Maekawa T., Watanabe Y., Yamada A., Kusumoto R., Sakai D., Sugasawa K., Ohkuma Y., Hanaoka F. (2000) Reconstitution of damage DNA excision reaction from SV40 minichromosomes with purified nucleotide excision repair proteins. Mutat Res. 459, 147–160 [DOI] [PubMed] [Google Scholar]
- 9. Hara R., Mo J., Sancar A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20, 9173–9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gong F., Kwon Y., Smerdon M. J. (2005) Nucleotide excision repair in chromatin and the right of entry. DNA Repair 4, 884–896 [DOI] [PubMed] [Google Scholar]
- 11. Andrews A. J., Luger K. (2011) Nucleosome structure(s) and stability. Variations on a theme. Annu. Rev. Biophys. 40, 99–117 [DOI] [PubMed] [Google Scholar]
- 12. Green C. M., Almouzni G. (2002) When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 3, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palomera-Sanchez Z., Zurita M. (2011) Open, repair, and close again. Chromatin dynamics and the response to UV-induced DNA damage. DNA Repair 10, 119–125 [DOI] [PubMed] [Google Scholar]
- 14. Ura K., Araki M., Saeki H., Masutani C., Ito T., Iwai S., Mizukoshi T., Kaneda Y., Hanaoka F. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 20, 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hara R., Sancar A. (2002) The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 22, 6779–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang Y., Wang X., Bao S., Guo R., Johnson D. G., Shen X., Li L. (2010) INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 17274–17279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong F., Fahy D., Liu H., Wang W., Smerdon M. J. (2008) Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang L., Zhang Q., Jones K., Patel M., Gong F. (2009) The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 8, 3953–3959 [DOI] [PubMed] [Google Scholar]
- 19. Zhao Q., Wang Q. E., Ray A., Wani G., Han C., Milum K., Wani A. A. (2009) Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem. 284, 30424–30432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kapetanaki M. G., Guerrero-Santoro J., Bisi D. C., Hsieh C. L., Rapi-Otrin V., Levine A. S. (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U.S.A. 103, 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H., Zhai L., Xu J., Joo H. Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006) Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 [DOI] [PubMed] [Google Scholar]
- 22. Takedachi A., Saijo M., Tanaka K. (2010) DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku and contributes to the recruitment of XPA. Mol. Cell. Biol. 30, 2708–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rapic-Otrin V., Navazza V., Nardo T., Botta E., McLenigan M., Bisi D. C., Levine A. S., Stefanini M. (2003) True XP group E patients have a defective UV-damaged DNA-binding protein complex and mutations in DDB2 that reveal the functional domains of its p48 product. Hum. Mol. Genet. 12, 1507–1522 [DOI] [PubMed] [Google Scholar]
- 24. Higa L. A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006) CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 11, 1277–1283 [DOI] [PubMed] [Google Scholar]
- 25. Scrima A., Konícková R., Czyzewski B. K., Kawasaki Y., Jeffrey P. D., Groisman R., Nakatani Y., Iwai S., Pavletich N. P., Thomä N. H. (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guerrero-Santoro J., Kapetanaki M. G., Hsieh C. L., Gorbachinsky I., Levine A. S., Rapi-Otrin V. (2008) The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68, 5014–5022 [DOI] [PubMed] [Google Scholar]
- 27. Luijsterburg M. S., Goedhart J., Moser J., Kool H., Geverts B., Houtsmuller A. B., Mullenders L. H., Vermeulen W., van Driel R. (2007) Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J. Cell Sci. 120, 2706–2716 [DOI] [PubMed] [Google Scholar]
- 28. Fitch M. E., Nakajima S., Yasui A., Ford J. M. (2003) In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 278, 46906–46910 [DOI] [PubMed] [Google Scholar]
- 29. Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. (2005) UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 [DOI] [PubMed] [Google Scholar]
- 30. Rapic-Otrin V., McLenigan M. P., Bisi D. C., Gonzalez M., Levine A. S. (2002) Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 30, 2588–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. El-Mahdy M. A., Zhu Q., Wang Q. E., Wani G., Praetorius-Ibba M., Wani A. A. (2006) Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 281, 13404–13411 [DOI] [PubMed] [Google Scholar]
- 32. Wittschieben B. Ø., Iwai S., Wood R. D. (2005) DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 280, 39982–39989 [DOI] [PubMed] [Google Scholar]
- 33. Kusumoto R., Masutani C., Sugasawa K., Iwai S., Araki M., Uchida A., Mizukoshi T., Hanaoka F. (2001) Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat Res. 485, 219–227 [DOI] [PubMed] [Google Scholar]
- 34. Batty D., Rapic'-Otrin V., Levine A. S., Wood R. D. (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 300, 275–290 [DOI] [PubMed] [Google Scholar]
- 35. Nishi R., Alekseev S., Dinant C., Hoogstraten D., Houtsmuller A. B., Hoeijmakers J. H., Vermeulen W., Hanaoka F., Sugasawa K. (2009) UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair 8, 767–776 [DOI] [PubMed] [Google Scholar]
- 36. Hwang B. J., Ford J. M., Hanawalt P. C., Chu G. (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. U.S.A. 96, 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luger K., Rechsteiner T. J., Richmond T. J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 38. Essen L. O., Klar T. (2006) Light-driven DNA repair by photolyases. Cell. Mol. Life Sci. 63, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yasui A., Eker A. P., Yasuhira S., Yajima H., Kobayashi T., Takao M., Oikawa A. (1994) A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 13, 6143–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakajima S., Sugiyama M., Iwai S., Hitomi K., Otoshi E., Kim S. T., Jiang C. Z., Todo T., Britt A. B., Yamamoto K. (1998) Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 26, 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomschik M., Zheng H., van Holde K., Zlatanova J., Leuba S. H. (2005) Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 102, 3278–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lowary P. T., Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 43. Nakajima S., Lan L., Kanno S., Takao M., Yamamoto K., Eker A. P., Yasui A. (2004) UV light-induced DNA damage and tolerance for the survival of nucleotide excision repair-deficient human cells. J. Biol. Chem. 279, 46674–46677 [DOI] [PubMed] [Google Scholar]
- 44. Köberle B., Roginskaya V., Wood R. D. (2006) XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair 5, 641–648 [DOI] [PubMed] [Google Scholar]
- 45. You Y. H., Lee D. H., Yoon J. H., Nakajima S., Yasui A., Pfeifer G. P. (2001) Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 276, 44688–44694 [DOI] [PubMed] [Google Scholar]
- 46. Matsuda N., Azuma K., Saijo M., Iemura S., Hioki Y., Natsume T., Chiba T., Tanaka K., Tanaka K. (2005) DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair 4, 537–545 [DOI] [PubMed] [Google Scholar]
- 47. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 48. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 49. Bergink S., Salomons F. A., Hoogstraten D., Groothuis T. A., de Waard H., Wu J., Yuan L., Citterio E., Houtsmuller A. B., Neefjes J., Hoeijmakers J. H., Vermeulen W., Dantuma N. P. (2006) DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 20, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 51. Nickel B. E., Davie J. R. (1989) Structure of polyubiquitinated histone H2A. Biochemistry 28, 964–968 [DOI] [PubMed] [Google Scholar]
- 52. Böhm L., Crane-Robinson C., Sautière P. (1980) Proteolytic digestion studies of chromatin core-histone structure. Identification of a limit peptide of histone H2A. Eur. J. Biochem. 106, 525–530 [DOI] [PubMed] [Google Scholar]
- 53. Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A. F., Tanaka K., Nakatani Y. (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367 [DOI] [PubMed] [Google Scholar]
- 54. Marteijn J. A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A. M., Dantuma N. P., Lukas J., Vermeulen W. (2009) Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Böhm V., Hieb A. R., Andrews A. J., Gansen A., Rocker A., Tóth K., Luger K., Langowski J. (2011) Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 39, 3093–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rodriguez-Collazo P., Leuba S. H., Zlatanova J. (2009) Robust methods for purification of histones from cultured mammalian cells with the preservation of their native modifications. Nucleic Acids Res. 37, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fischer E. S., Scrima A., Böhm K., Matsumoto S., Lingaraju G. M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F., Iwai S., Gut H., Sugasawa K., Thomä N. H. (2011) The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 [DOI] [PubMed] [Google Scholar]
- 58. Moser J., Volker M., Kool H., Alekseev S., Vrieling H., Yasui A., van Zeeland A. A., Mullenders L. H. (2005) The UV-damaged DNA-binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair 4, 571–582 [DOI] [PubMed] [Google Scholar]
- 59. Alekseev S., Kool H., Rebel H., Fousteri M., Moser J., Backendorf C., de Gruijl F. R., Vrieling H., Mullenders L. H. (2005) Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 65, 10298–10306 [DOI] [PubMed] [Google Scholar]
- 60. Duan M. R., Smerdon M. J. (2010) UV damage in DNA promotes nucleosome unwrapping. J. Biol. Chem. 285, 26295–26303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. West M. H., Bonner W. M. (1980) Histone 2A, a heteromorphous family of eight protein species. Biochemistry 19, 3238–3245 [DOI] [PubMed] [Google Scholar]
- 62. Elderkin S., Maertens G. N., Endoh M., Mallery D. L., Morrice N., Koseki H., Peters G., Brockdorff N., Hiom K. (2007) A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell 28, 107–120 [DOI] [PubMed] [Google Scholar]
- 63. Zhu Q., Wani A. A. (2010) Histone modifications. Crucial elements for damage response and chromatin restoration. J. Cell Physiol. 223, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu Q., Wani G., Arab H. H., El-Mahdy M. A., Ray A., Wani A. A. (2009) Chromatin restoration after nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair 8, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wakasugi M., Kawashima A., Morioka H., Linn S., Sancar A., Mori T., Nikaido O., Matsunaga T. (2002) DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277, 1637–1640 [DOI] [PubMed] [Google Scholar]
- 66. Davies N., Lindsey G. G. (1994) Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim. Biophys. Acta 1218, 187–193 [DOI] [PubMed] [Google Scholar]
- 67. Jason L. J., Finn R. M., Lindsey G., Ausió J. (2005) Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J. Biol. Chem. 280, 4975–4982 [DOI] [PubMed] [Google Scholar]
- 68. Li W., Nagaraja S., Delcuve G. P., Hendzel M. J., Davie J. R. (1993) Effects of histone acetylation, ubiquitination, and variants on nucleosome stability. Biochem. J. 296, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 70. Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. (1991) The nucleosomal core histone octamer at 3.1 A resolution. A tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. U.S.A. 88, 10148–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Harp J. M., Hanson B. L., Timm D. E., Bunick G. J. (2000) Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 56, 1513–1534 [DOI] [PubMed] [Google Scholar]
- 72. Eickbush T. H., Godfrey J. E., Elia M. C., Moudrianakis E. N. (1988) H2a-specific proteolysis as a unique probe in the analysis of the histone octamer. J. Biol. Chem. 263, 18972–18978 [PubMed] [Google Scholar]
- 73. Shukla M. S., Syed S. H., Goutte-Gattat D., Richard J. L., Montel F., Hamiche A., Travers A., Faivre-Moskalenko C., Bednar J., Hayes J. J., Angelov D., Dimitrov S. (2011) The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 39, 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yeh J. I., Levine A. S., Du S., Chinte U., Ghodke H., Wang H., Shi H., Hsieh C. L., Conway J. F., Van Houten B., Rapi-Otrin V. (2012) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bosu D. R., Kipreos E. T. (2008) Cullin-RING ubiquitin ligases. Global regulation and activation cycles. Cell Div. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andrews A. J., Chen X., Zevin A., Stargell L. A., Luger K. (2010) The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.