Background: Toxin-antitoxin (TA) systems are broadly conserved in the bacterial kingdom.
Results: Brucella abortus RNase toxin, BrnT, has a RelE-like fold and is neutralized by its antitoxin, BrnA. brnTA transcription is activated by a number of environmental stressors.
Conclusion: BrnTA is a novel, stress-regulated TA system.
Significance: This study structurally and functionally defines a novel member of the RelE toxin family.
Keywords: Bacterial Toxins, RNA, Stress Response, Toxins, X-ray Crystallography, BrnTA, Brucella abortus, Microbiology, RNase, Structure
Abstract
Type II toxin-antitoxin (TA) systems are expressed from two-gene operons that encode a cytoplasmic protein toxin and its cognate protein antitoxin. These gene cassettes are often present in multiple copies on bacterial chromosomes, where they have been reported to regulate stress adaptation and persistence during antimicrobial treatment. We have identified a novel type II TA cassette in the intracellular pathogen Brucella abortus that consists of the toxin gene, brnT, and its antitoxin, brnA. BrnT is coexpressed and forms a 2:2 tetrameric complex with BrnA, which neutralizes BrnT toxicity. The BrnT2-BrnA2 tetramer binds its own promoter via BrnA, and autorepresses its expression; its transcription is strongly induced in B. abortus by various stressors encountered by the bacterial cell during infection of a mammalian host. Although highly divergent at the primary sequence level, an atomic resolution (1.1 Å) crystal structure of BrnT reveals a secondary topology related to the RelE family of type II ribonuclease toxins. However, overall tertiary structural homology to other RelE family toxins is low. A functional characterization of BrnT by site-directed mutagenesis demonstrates a correspondence between its in vitro activity as a ribonuclease and control of bacteriostasis in vivo. We further present an analysis of the conserved and variable features of structure required for RNA scission in BrnT and the RelE toxin family. This structural investigation informs a model of the RelE-fold as an evolutionarily flexible scaffold that has been selected to bind structurally disparate antitoxins, and exhibit distinct toxin activities including RNA scission and DNA gyrase inhibition.
Introduction
The Gram-negative intracellular pathogen, Brucella abortus, naturally infects ruminant hosts where it causes both a chronic and an acute infection. In humans, infection is strictly zoonotic and results in a debilitating chronic disease known as undulant fever. B. abortus primarily resides in professional phagocytic cells and placental trophoblasts in its mammalian host (1, 2). While trafficking through the host system, B. abortus must adapt to a number of stressors including oxidative bursts from macrophages (3), low pH (4), antimicrobial peptides, and nutrient limitation (5). A handful of B. abortus proteins that mediate adaptation to host-generated stress are known (6). However, many putative stress response genes encoded in the genome remain entirely undefined. Among these are type II toxin-antitoxin (TA)4 systems.
Type II TA systems consist of a stable toxin protein and a labile antitoxin protein that form a high-affinity, neutral complex (7). The genes encoding type II TAs were initially identified on plasmids, where they were demonstrated to promote stable plasmid inheritance (8, 9). Several distinct families of this type of TA system have since been identified on bacterial chromosomes (10–12) and are demonstrated to regulate stress survival (13, 14) and persister cell formation (15–17). As antitoxin proteins are generally degraded at a faster rate than toxins by the cellular proteolytic machinery (18–20), the arrest of protein synthesis as a result of antibiotic insult, nutrient deprivation, or other stresses results in accumulation of free toxin in the cytosol. Upon release from their cognate antitoxins, type II toxins are known to act via one of several mechanisms: degradation of mRNA (21, 22), inhibition of the translating ribosome (23), inhibition of DNA gyrase (24–26), degradation of tRNAfMet (27), or inhibition of peptidoglycan synthesis (28, 29). Depending on the system and dosage, toxins may be bacteriostatic or bactericidal (30, 31).
We report a functional, biochemical, and structural characterization of a novel type II toxin-antitoxin system, BrnT-BrnA, from B. abortus. BrnT is a small (81 residues) ribonuclease that is present in bacteria, archaea, bacteriophage, and plasmids. BrnT-BrnA forms a 2:2 tetrameric complex and autoregulates its expression, which is induced by a number of stress insults. Our atomic resolution (1.1 Å) crystal structure of BrnT revealed an overall secondary structural topology that is related to the RelE family of ribonuclease toxins (13). Indeed, the BrnT toxin cleaves RNA in vitro, and site-directed mutagenesis of the BrnT coding sequence has identified a set of polar residues clustered in the α1–β5 region of the structure that are required for RNA scission in vitro and control of bacteriostasis in vivo. We further define the conserved and variable features of structure required for RNA scission in BrnT and the RelE toxin family.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Escherichia coli strains used for growth curves, protein purification, and in vivo protein synthesis assays were E. coli Rosetta pLysS/DE3 with bab1_0993, bab1_0994, or bab1_0993-4 cloned into pET151/D-TOPO (Table 1). E. coli used for bacteriostasis experiments were in Top 10 with bab1_0994 cloned into NdeI/KpnI sites of pSRK-Km (32) and either pBAD 24 (33) as an empty vector control or bab1_0993 cloned into the NdeI/HindIII site of pBAD 24. Cultures were grown with 100 μg/ml of ampicillin and 50 μg/ml of kanamycin. Cultures were induced with 0.5 mm IPTG and plated on LB agar + 100 μg/ml of ampicillin, 50 μg/ml of kanamycin ± 0.2% arabinose. B. abortus 2308 strains were grown in Brucella broth at 37 °C. brnT, brnA, and brnTA expression strains were constructed in a wild-type B. abortus 2308 background by introducing pSRK-Km alone, or with brnT, brnA, and brnTA cloned into NdeI/KpnI sites. Protein expression in B. abortus was induced with 1 mm IPTG. An in-frame, unmarked brnTA deletion strain was made by cloning an upstream fragment of brnT (primers used to amplify the fragments were: forward, 5′-GGATCCGCCACGGATTTCAAAATATC, reverse, 5′-GCGAGCCCAGACGAATAGGGCCGTATGAACGATG), and a downstream fragment of brnA (primers used to amplify the fragments were: forward, 5′-CATCGTTCATACGGCCCTATTCGTCTGGGCTCGC, and reverse, 5′-GGTACCCGTATTGCATGTCTGTCTCC). These two fragments were cloned into pNPTS138, which was subsequently mated into B. abortus 2308 using E. coli S17-1. Strains were first mixed and plated without selection overnight. Bacterial cells were then scraped and plated on Schadelar blood agar with kanamycin and nalidixic acid to select for single B. abortus recombinants. Single colonies were then picked and grown overnight in Brucella broth and then plated the next day on Schadelar blood agar plates with 10% sucrose. Colonies that grew were screened by PCR to confirm that the gene replacement had occurred and that the pNPTS138 backbone had been lost. The PbrnTA-lacZ promoter fusion was made cloning ∼500 nucleotides upstream of the brnT ATG start codon. Primers used were: forward, 5′-AAGCTTGAGCGGGCGATGATCTTCCGC, and reverse, 5′-GGATCCAAAACGTATGTACAATAATTCGTCTGGGC. This fragment was cloned into the HindIII/BamHI sites of pMR15. This transcriptional fusion plasmid was transformed into B. abortus 2308 and B. abortus 2308 ΔbrnTA.
TABLE 1.
Strains
| Strain | Source |
|---|---|
| E. coli strains | |
| E. coli Rosetta (DE3) pLysS | Novagen |
| Rosetta/pET151-brnA | This study |
| Rosetta/pET151-brnT | This study |
| E. coli Top 10 | Invitrogen |
| Top 10/pSRK-km-brnT pBAD 24-brnA | This study |
| Top 10/pSRK-km-brnT pBAD 24 | This study |
| Rosetta/pET151-brnTA | This study |
| Rosetta/pET151-brnT D6A | This study |
| Rosetta/pET151-brnT E7A | This study |
| Rosetta/pET151-brnT K9A | This study |
| Rosetta/pET151-brnT K16A | This study |
| Rosetta/pET151-brnT H17A | This study |
| Rosetta/pET151-brnT H25A | This study |
| Rosetta/pET151-brnT R41A | This study |
| Rosetta/pET151-brnT R72A | This study |
| Rosetta/pET151-brnT K77A | This study |
| Rosetta/pET151-brnT E78A | This study |
| Rosetta/pET151-brnT R79A | This study |
| Brucella abortus strains | |
| B. abortus 2308 | R. M. Roop II |
| 2308ΔbrnTA | This study |
| 2308/pMR15 | This study |
| 2308/pMR15-PbrnTA | This study |
| 2308ΔbrnTA/pMR15 | This study |
| 2308ΔbrnTA/pMR15-PbrnTA | This study |
| 2308/pSRK-km | This study |
| 2308/pSRK-km-brnA | This study |
| 2308/pSRK-km-brnT | This study |
| 2308/pSRK-km-brnTA | This study |
Growth Assays
E. coli growth curves were done in LB + 100 μg/ml of ampicillin at 37 °C. Overnight cultures were diluted 1:100 and grown for 2 h. Cultures were then diluted to an A600 of 0.02. After 1 h of growth, cultures were induced with 0.5 mm IPTG (final concentration). Time points were taken every 30 min after induction. To enumerate viable colony forming units, 10 μl of culture was removed, 1:10 dilutions were prepared in PBS, and cells were plated on LB + 100 μg/ml of ampicillin. For bacteriostasis experiments, the same protocol was followed as above except cultures were plated on media without arabinose or with 0.2% arabinose. Time points were taken at 30 and 60 min after induction and every hour thereafter for 5 h. For B. abortus growth curves, overnight cultures were diluted to an A600 of 0.02 in Brucella broth + 50 μg/ml of kanamycin. Time points were taken every hour. To quantify viable colony forming units, 10 μl of culture was removed, 1:10 dilutions were prepared in PBS, and cells were plated on Schadeler blood agar + 50 μg/ml of kanamycin.
Protein Purification
The same strains used to measure E. coli growth were used for protein expression and purification. Protein expression for solution and mass spectrometry analysis was induced with 1 mm IPTG. Cells were lysed by sonication in 20 mm Tris, pH 7.6, 200 mm NaCl and centrifuged at 15,000 × g for 30 min to remove cell debris. Soluble recombinant proteins were affinity purified by using Ni2+ Chelating Resin (GE Healthcare) using a 50–500 mm imidazole gradient. Further purification was carried out on a Superdex 75 size exclusion column. Purity was confirmed by 14% PAGE. Expression and purification for crystallization was carried out as follows: a 50-ml overnight LB medium culture supplemented with 100 μg/ml of ampicillin was used to inoculate 2 liters of LB + 100 μg/ml of ampicillin; cultures were incubated at 37 °C in a rotary shaker (Infors-HT, Bottmingen, Switzerland) at 220 rpm. Transcription of the complex was induced at an A600 of 0.8 by adding 1 mm IPTG. After 1 h and 30 min of incubation under the same conditions, the cells were harvested by centrifugation at 8,000 × g for 20 min at 4 °C. Cell pellets were resuspended in 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10 mm imidazole with 5 μg/ml of DNase I and 80 μg/ml of phenylmethylsulfonyl fluoride (PMSF) protease inhibitor.
Cells were disrupted by two passages in a French pressure cell, and the cell debris was removed by centrifugation for 20 min at 20,000 × g. The supernatant was loaded onto a Ni2+-Sepharose affinity column (GE Healthcare) pre-equilibrated with the binding buffer. Two washing steps were performed using 10 and 75 mm imidazole followed by the elution step with 200 mm imidazole in the binding buffer. The protein solution was then dialyzed against 10 mm Tris-HCl, pH 7.4, and 150 mm NaCl buffer to remove imidazole. All purification steps were carried out 4 °C.
Size Exclusion Chromatography
All BrnT and BrnA size exclusion chromatography was performed on a 10/300 Superdex 75 column with 20 mm Tris, pH 7.6, 200 mm NaCl buffer. Columns were calibrated with proteins from a Low Molecular Weight Gel Filtration Calibration kit (GE Healthcare). Elution profiles and times of proteins in a concentration range of 25 to 0.3 mg/ml were used to assess molecular size and shape.
Analytical Ultracentrifugation (AUC)
Sedimentation velocity studies were performed in a Beckman Optima XL-A analytical ultracentrifuge equipped with an 60 Ti rotor (Beckman Coulter) using absorption optics at a wavelength of 230 and 280 nm. Experiments were carried out using a 1.2-mm two-channel Epon centerpiece at a rotor speed of 40,000 rpm for 18 h at 20 °C. Absorbance of 0.3–1.0 mg/ml of samples was monitored in a continuous mode time interval of 360–480 s and a step size of 0.003 cm. Multiple scans at different time points were fit to the continuous size distribution, c(S), model and the integrated distribution function by using SEDFIT version 11.3. The partial specific volume of the proteins and buffer density were calculated from standard tables using SEDNTERP.
Nanoelectrospray Ionization (nEIS) and Collision-induced Dissociation (CID) Mass Spectrometry
The protein complex was buffer exchanged into pH ∼7.5, 100 mm ammonium acetate using Micro Biospin columns (Bio-Rad). Solutions of ∼15 μm protein were nanoelectrosprayed into a Synapt G2 (Waters Corporation, Manchester, UK) using a home-built source with capillaries pulled in-house on a P-97 micropipette puller (Sutter Instruments, Hercules, CA). Source conditions were selected for best transmission of the intact protein complex. Typical conditions were: capillary, 1.2 kV; sample cone, 75 V; extractor cone, 2 V; backing pressure ∼5.0 mbar. CID was performed in the trap region of the instrument using argon as the collision target.
Surface Plasmon Resonance
Binding between BrnT and BrnA was measured with a BIAcore 3000 (GE Healthcare) biosensor through surface plasmon resonance. His6-BrnT was bound to the dextran matrix of a sensor chip nitrilotriacetic acid. The His6 tag on the BrnA analyte was removed using TEV protease. All steps in the immobilization process were carried out at a flow rate of 10 μl/min. The control surface was prepared similarly except that running buffer was injected instead of NiCl2 solution. All kinetic experiments were performed at 15 °C in HBS (10 mm HEPES, pH 7.4, 0.15 m NaCl, 0.05% Tween 20) using flow rates of 20 μl/min. These conditions prevented significant leakage of bound target protein from the chip. The analyte (BrnA) at various concentrations (see “Results”) was added in the flow during the binding phase. The amount of protein bound to the sensor chip was monitored by the change in refractive index (given in arbitrary response units). For the measurements of kinetic parameters, the amount of immobilized His6-BrnT was minimized to avoid saturation and generally did not exceed 130 response units. After each binding experiment, the sensor chip was regenerated by sequential washing with 200 μl of 0.3 m imidazole, 0.5 m NaCl followed by 200 μl of 0.35 m Na-EDTA in HBS. Duplicate or triplicate injections of each concentration of BrnA were performed. The data were prepared by the “double referencing“ method. In this method, parallel injections of each BrnA sample over a control dextran surface were performed as well as running buffer injections over both the immobilized BrnT and control dextran surfaces. Subtraction of these sensorgrams yielded the control; this was then subtracted from the experimental sensorgram. Data were simultaneously fit using a 1:1 binding model in BIAevaluation (global fitting algorithm). The early binding phase (<300 s) was used to determine the association constant (ka) between BrnT and BrnA. The dissociation phase (kd) was measured for 600 s using the rate of decline in response units on introduction of free buffer at the end of BrnA injections. The equilibrium dissociation constant (KD) of the complexes was calculated from the ratio kd/ka.
Crystallization of Toxin-Antitoxin Complex
To produce selenomethionine toxin-antitoxin complex for experimental phase determination, we expressed toxin-antitoxin complex from pET151/D-TOPO in defined medium as previously described (34). Purified toxin-antitoxin complex was concentrated using a centrifugal filter unit (3-kDa membrane, Amicon-Millipore, Billerica, MA). Protein purity, estimated to be 90%, was assessed by 14% SDS-PAGE and stained with Coomassie Brilliant Blue. Before crystallization screening, 0.17 μg/μl of subtilisin was added to the protein solution. Initial crystallization screening was carried out using the hanging-drop, vapor-diffusion technique in 96-well microplates. Trays were set using a Mosquito robot (TTP LabTech, Cambridge, MA) and commercial crystallization kits (Nextal-Qiagen, Valencia, CA). The drops were set up by mixing equal volumes (0.1 μl) of the protein and the precipitant solutions equilibrated against 75 μl of the precipitant solution. In all trials, the protein concentration was 38 mg/ml. The best crystals were obtained at 14 °C by manual refinement with the following crystallization solution: 100 mm Tris buffer, pH 8, 8% PEG 6000, 150 mm NaCl. Crystals grew to their final size in 2 weeks. All manual crystallization attempts were carried using the sitting-drop, vapor-diffusion technique in 24-well plates. The drops were set up by mixing equal volumes of the protein and the precipitant solutions equilibrated against 500 μl of the precipitant solution. After 1 min of soaking in crystallization buffer supplemented with 20% of glycerol and 5 mm β-mercaptoethanol, crystals were flash-frozen directly at 100 K in liquid nitrogen. Incorporation of selenomethionine in the crystal was confirmed by measuring x-ray fluorescence.
Crystallographic Data Collection and Processing
Crystal diffraction data were collected at a temperature of 100 K on beamline 21-ID-D (LS-CAT, Advanced Photon Source, Argonne, IL) using a MAR Mosaic 300 detector and an oscillation range of 1 degree. Diffraction images were reduced using the HKL 2000 suite (35).
Phasing and Refinement
Diffraction from a single selenomethionine-BrnT protein crystal was measured at an energy of 12.66 keV (0.979 Å), and the structure of BrnT was phased from the resulting 1.4-Å dataset by selenium single-wavelength anomalous dispersion (36). Three selenium sites were located within the asymmetric unit using the Autosol single-wavelength anomalous dispersion routine in Phenix (37). An initial structure was built into these maps and refined to an Rfree of 21%. This structure was then used as a molecular replacement search model to phase a 1.1-Å native BrnT dataset.
BrnT was further refined against the 1.1-Å data to an Rwork of 14.0% and an Rfree of 15.8% using phenix.refine in the Phenix software suite. Manual model building, solvent addition, and refinement of this structure was conducted iteratively using Coot (38) and phenix.refine (see Table 3).
TABLE 3.
Crystallographic data and refinement statistics
a Rmerge = ΣhklΣi|Ii-<I>|/ΣhklΣiIi, for all data > −3.
b Experimental phases were determined by SAD using the anomalous signal from selenium. Total figure of merit values are based on experimental phase information (prior) for all reflections.
c Rcryst = Σhkl ‖Fobs| − |Fcalc‖/Σhkl|Fobs|.
d Rfree uses 1906 total reflections for cross-validation.
e rmsd, root mean square deviation.
Sequence Alignment and Protein Visualization Methods
Protein sequence alignments were carried out in ClustalW2. BrnT ribbon structure rendering, electrostatic potential surfaces calculation (by using the generate-vacuum electrostatics function), and visualization of the hydrophobic surfaces (according to the Eisenberg hydrophobicity scale) (39) were performed with PyMOL. Visualization of the conserved residues in the structure was performed by using the Consurf server (40). Predicted residues involved in RNA binding have been predicted with three different programs: RNABindR (41), BindN (42), and PiRaNhA (43).
Protein Synthesis Assay
E. coli Rosetta pLysS/DE3 with pET151/D+bab1_0993, pET151/D+bab1_0994, or pET151/D+bab1_0993-4 were grown to an A600 0.5 and then induced with IPTG to a final concentration of 0.5 mm for 30 min. Cells were washed twice by centrifuging at 7,000 × g for 1 min and resuspended in 1 ml of M9 medium + all amino acids but methionine and cysteine. [35S]Methionine and cysteine were added to media for 1 min and then protein synthesis was stopped by adding 75 μl of ice-cold 100% TCA followed by a standard TCA precipitation. Samples were then resuspended in sample buffer (0.5 m Tris, 4% SDS, pH 8.0), boiled for 10 min, and 5 μl were added to scintillation fluid and scintillation counted.
In Vitro RNase Assay
In vitro lacZ RNA transcript was made with pET151/D + lacZ and the MEGAscript high yield transcription kit (Ambion) according to the manufacturer's protocol. RNA was incubated with water, buffer, 8, 4, 2, 1, or 0.5 μm BrnT, 16 μm BrnA, or 4 μm BrnT + 16 μm BrnA for 20 min at room temperature. All proteins were purified with a Ni2+ affinity column and a Superdex 75 size exclusion column. Samples were loaded in 1% TAE-agarose gel. In the RNase assay with mutant proteins, 0.6 μm BrnT was incubated with RNA for 30 min at 37 °C.
Electrophoretic Mobility Shift Assay
Fifty nucleotide oligos were purchased from IDT (Coralville, IA) (bab1_0994, −50 forward, 5′-GTACGATTATTATTGCGAGCCCAGACGAATTATTGTACATACGTTTTATG, bab1_0994, −50 reverse, 5′-CATAAAACGTATGTACAATAATTCGTCTGGGCTCGCAATAATAATCGTAC). Oligos were labeled with 32P using T4 polynucleotide kinase (Fermentas) by incubating the reaction at 37 °C for 20 min. Unincorporated nucleotides were removed using Micro Bio-spin 6 chromatography columns (Bio-Rad) and oligos were annealed by mixing and incubating at 80 °C for 20 min, then letting them cool to room temperature overnight. Binding reactions consisted of 1 μl of 20 nm 32P-annealed oligo, 20 ng of salmon sperm DNA, 4 μl of 5× binding buffer (100 mm Tris, pH 8.0, 250 mm NaCl, 25% glycerol, 250 ng/μl of BSA), 13 μl of H2O, and 1 μl of protein at concentration of interest. The binding reaction took place for 30 min at room temperature and then 4 μl of 80% glycerol was added to each sample. Samples were loaded in a 12% polyacrylamide native gel and run at 140 V for 2 h. The gel was placed on a phosphorscreen overnight and imaged with a Typhoon scanner.
β-Galactosidase Transcription Reporter Assay
Transcriptional activity of the brnTA promoter was assayed using a PbrnTA-lacZ promoter fusion plasmid incorporated in wild-type B. abortus or ΔbrnTA. Overnight cultures of both strains were diluted to A600 0.05. Cultures were grown for 3 h and then the A600 was measured. β-Galactosidase activity was determined as previously described (44).
Stress Assays and Quantification of Transcription by RT-qPCR
B. abortus 2308 (1 × 108 cfu/ml) was stressed with either 200 μg/ml of chloramphenicol versus an equal volume of 95% ethanol as a control, 44 versus 37 °C, 5 mm H2O2 versus an equal volume of H2O, or grown in Brucella broth and exposed to pH 4.0 versus 7.0. All stress assays were carried out for 30 min with the exception of low pH, which was done for 3 h. RNA was harvested using TRIzol. Custom TaqMan primers were used to measure brnT transcript: forward, 5′-TCATCGCCGTCATTTTCAAG, reverse, 5′-TGCTGAACGCATGGAGATCA, and probe, 5′-CGGTTGGTTCGGAAGCCCTCTCC. RNA was reverse transcribed for 30 min at 50 °C followed by an inactivation step, 95 °C for 6 min. The cDNA was then amplified for 50 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 15 s. B. abortus RNA levels were normalized to 16 S ribosomal RNA: forward, 5′-CCTTACGGGCTGGGCTAC, reverse: 5′-TGCTCGCTGCCCACTGT, probe, 5′-ACGTGCTACAATGGTGG. All assays were performed on an ABI 7300 system and analyzed with SDS 1.3 software (Applied Biosystems).
RESULTS
BrnTA Is a Novel Toxin-Antitoxin System
Using a combination of bioinformatic metrics, Makarova and colleagues (45) predicted four type II TA systems in the B. abortus 2308 genome. Of these bab1_0993 and bab1_0994, encoding a putative antitoxin and toxin, respectively, have an unusual gene arrangement and physicochemical properties as compared with typical type II systems (31). Atypical features include: 1) the putative toxin gene, bab1_0994, precedes the antitoxin, bab1_0993, in the operon (Fig. 1A); generally the antitoxin gene is positioned upstream of the toxin, 2) the predicted toxin and antitoxin proteins have equivalent theoretical isoelectric points of 9.5; toxins are typically basic and antitoxins acidic, and 3) the predicted antitoxin is larger than the toxin, and has a C-terminal (rather than N-terminal) ribbon-helix-helix DNA binding domain.
FIGURE 1.
The brnTA operon. A, the brnT and brnA genes overlap by 1 bp. B, clustal multiple sequence alignment of B. abortus BrnT toxin and homologous toxin sequences from 10 other Gram-negative species including several human pathogens. Ba, B. abortus (GI: 82615932); Ec, E. coli (GI: 309704811); Yp, Yersinia pestis (GI: 22126312); Ypt, Yersinia pseudotuberculosis (GI: 170024520); Bpp, Bordatella parapertussis (GI: 33596571); Nm, Neisseria meningitidis (GI: 161870004); Hi, Haemophilus influenzae (GI: 270670382); Bp, Burkholderia pseudomallei (GI: 217424545); Vc, Vibrio cholerae (GI: 15601097); Cb, Coxiella burnetii (GI: 154705785); Cc, Caulobacter crescentus (GI: 16124744). Boxes shaded black are amino acids identical in 40% of sequences shown. Boxes shaded gray are similar in 40% of sequences.
To test if bab1_0993–0994 encodes a bona fide TA system, we expressed the predicted toxin, antitoxin, and toxin-antitoxin operon heterologously in E. coli from an inducible promoter. In strains expressing the toxin alone, bacterial growth as measured by visible optical density ceased within 1 h of induction (Fig. 2A). Expression of antitoxin alone did not significantly attenuate growth relative to wild-type. Coordinate expression of the antitoxin significantly reduced toxicity of the toxin (Fig. 2A). When viable colony forming units were plated at time points after induction of toxin, antitoxin, or toxin-antitoxin operon (Fig. 2B), expression of the toxin, but not the antitoxin or full TA operon, resulted in an apparent ∼3 log loss of viable cfu after 1 h, and an over 4 log loss after 2 h.
FIGURE 2.
B. abortus BrnTA is a toxin-antitoxin system. A, antitoxin (brnA, blue), toxin (brnT, red), or brnTA operon (green) were expressed in E. coli and optical density at 600 nm (A600) was measured at 30-min intervals. B, starting at the time of induction (marked with black arrows) colony forming units were quantified on LB agar. C, growth curves of B. abortus 2308 + pSRK-Km (empty vector control, black), pSRK-brnT (red), pSRK-brnA (blue), or pSRK-brnTA (green). A660 was measured every hour; expression of genes from pSRK was induced after 4 h (black arrow). D, B. abortus viable colony forming units after induction of gene expression were enumerated by plating on Schaedler blood agar. E, E. coli Plac-brnT + Para-brnA or Plac-brnT + Para empty vector control (EVC) were grown in the presence or absence of 0.5 mm IPTG for increasing amounts of time. Cells were then plated on LB agar containing ampicillin and kanamycin ± 0.2% arabinose. Solid lines indicate the presence of arabinose in agar; dashed lines indicate agar without arabinose. + or − indicates induction of specific protein. Error bars represent S.D.
An E. coli strain that has toxin under the control of the lac promoter and antitoxin under control of the ara promoter, exhibits the same apparent decrease in “viability” in the presence of IPTG and absence of arabinose (Fig. 2E). However, if we induce toxin and plate on solid medium containing 0.2% arabinose at time points up to 5 h after induction, it is evident that expression of toxin does not result in a decrease in viability, but rather a decrease in culturability (in this time period). Induction of bab1_0994 thus leads to a bacteriostatic state, and subsequent induction of the antitoxin is able to neutralize the toxin and rescue cells from stasis.
To confirm bab1_0993–0994 acts in the same manner in B. abortus 2308, we performed similar experiments in which we ectopically expressed toxin, antitoxin, or TA operon from an inducible promoter. The results of these experiments in B. abortus were consistent with what we observed in E. coli: expression of the toxin resulted in arrest of bacterial growth and a decrease in culturability, and co-expression of the antitoxin neutralized these effects (Fig. 2, C and D). Together these data indicate that BAB1_0993–0994 functions as a toxin-antitoxin system and that expression of the toxin, BAB1_0994, is bacteriostatic.
A search of the BAB1_0994 primary sequence against the Pfam 25.0 data base shows that this toxin is not limited to B. abortus. BAB1_0994 is described by the profile hidden Markov model of domain family DUF497 in Pfam 25.0 (E < 2e-11) (46). This hidden Markov model apprehends 499 sequences from 317 species spanning archaea, plasmids, bacteriophage, and bacteria, several of which are human pathogens (Fig. 1B). Our study, for the first time, ascribes a function to this domain family. Based on data that we will present and discuss further below, we have named this conserved toxin, BrnT and its cognate antitoxin, BrnA.
BrnT-BrnA Toxin-Antitoxin System Forms a High Affinity 2:2 Tetrameric Complex
To test if there is a physical association between BrnT and BrnA, we co-expressed His6-BrnT and BrnA from a single plasmid. His6-BrnT and BrnA were co-purified from Ni2+-chelating Sepharose resin. Resolution of this protein complex by SDS-PAGE revealed a ladder of protein bands at weights larger than monomeric toxin or antitoxin (Fig. 3A). Analysis of each of the five bands on the gel by mass spectrometry showed that four were composed solely of BrnT and one was BrnA (supplemental Table S1). BrnT has no cysteines and formed these unusual oligomers under denaturing conditions (as assessed by SDS-PAGE) in both the presence or absence of the reductant β-mercaptoethanol, suggesting it is not a result of oxidative bond formation. We do not currently understand why BrnT forms higher order oligomers under denaturing conditions.
FIGURE 3.
BrnTA forms a tetramer consisting of two antitoxins and two toxins. A, c(S) distribution calculated from sedimentation velocity measurements of the purified BrnTA complex. BrnT-BrnA sediments as a single 2.9 ± 0.3 S species (root mean square deviation = 0.006). Resolution of this purified complex by 14% SDS-PAGE reveals multiple Coomassie-stained bands; mass spectrometry of tryptic peptides from these excised bands demonstrates that they contain either BrnT or BrnA, labeled as A or T to the right of each band. B, top, nESI-MS of purified BrnA-BrnT identifies a major complex with a molecular weight that matches a tetramer with 2:2 stoichiometry. B, bottom, 70-V CID spectrum of the 15+ tetramer ion species reveals monomeric and trimeric product ions, supporting a model in which the precursor is a 2:2 tetramer. C, size exclusion chromatography of BrnT (red) and BrnA (blue) and SDS-PAGE of major peaks. D, c(S) distribution from AUC of BrnT (red) and BrnA (blue), BrnT is 13.2 kDa, root mean square deviation = 0.022; frictional ratio = 1.2. BrnA is 29.3 kDa, root mean square deviation = 0.006, frictional ratio = 1.8.
To characterize the molecular weight, oligomeric state, and structure of the BrnT-BrnA complex under native conditions, we used AUC and nEIS-MS. Sedimentation velocity analysis of the purified BrnT-BrnA complex revealed one major 2.9 ± 0.3 S particle (Fig. 3A). Although a molecular weight distribution associated with this particle can be calculated from a distribution of Lamm equation solutions, it requires knowledge of the shape and other parameters that affect its hydrodynamic properties (47). Given a molecular weight, one can thus draw conclusions about the shape and folded state of a protein complex from the sedimentation velocity data. As such, we directly determined the molecular weight of the BrnT-BrnA complex by nEIS-MS. The data clearly show that the purified protein complex exists primarily as a species of mass 48,906 Da (Fig. 3B, top); this is the exact predicted molecular mass of a 2:2 antitoxin to toxin tetramer. The 15+ tetramer species (m/z 3261) was isolated and dissociated via CID, resulting in complementary monomer and trimer product ions (A and AT2, T and A2T; Fig. 3B, bottom) that confirm the precursor ion was an A2T2 heterotetramer. Further analysis of the CID data revealed a low level of BrnA-BrnT heterodimer and BrnA2 homodimer. BrnA and BrnT monomers are approximately equally abundant (at low levels) in these spectra. There is no evidence of BrnT2 dimer by CID. There is a significantly greater abundance of the T/A2T pair than the A/AT2 pair, suggesting that it is more difficult to release the antitoxin than the toxin from the complex. Based on these mass spectrometry data, we propose a model in which dimers of antitoxin recruit two individual toxin monomers to form a tetrameric complex consisting of two BrnA proteins and two BrnT proteins (Fig. 3B, inset). Based on the presence of a small amount of AT2 trimer after CID, we think toxins may interact after formation of the tetrameric complex.
We additionally analyzed BrnT and BrnA individually by size exclusion chromatography and AUC. We observed that BrnT is monomeric across a range of concentrations and that BrnA sediments as a likely dimer of high frictional ratio (Fig. 3, C and D). These data support our nEIS-MS results where we observe monomers of both toxin and antitoxin, and dimers of antitoxin, but no toxin dimers. Given a complex molecular mass of 48,906 Da, we reanalyzed our sedimentation velocity data to assess the hydrodynamic properties of the TA complex. From a fit of these data we determined that a frictional ratio of 1.6 is consistent with a 48.9-kDa 2.9 S particle. This frictional ratio is higher than typical globular proteins (1.1–1.3) and suggests that a portion of the BrnT2-BrnA2 complex is loosely structured or random coil (48). Based on the fit frictional ratio of BrnA2, we propose that the BrnA antitoxin has disordered, unstructured regions that contribute to the high apparent frictional ratio. This is consistent with the high relative susceptibility of BrnA to proteolysis in vitro (see our description of BrnT crystallization below), as has been reported for other antitoxins.
We further measured the affinity of the BrnT-BrnA complex by surface plasmon resonance. His6-BrnT was immobilized on a nickel-nitrilotriacetic acid chip, and BrnA flowed at concentrations ranging from 17 to 500 nm. BrnA binds BrnT with association and dissociation rate constants of 2.44 × 105 m−1 s−1 and 1.63 × 10−4 s−1, respectively. The ratio of the dissociation to association rate constant yields an equilibrium affinity of 670 pm (Table 2).
TABLE 2.
BrnT-BrnA binding measured by surface plasmon resonance
| Analyte | ka | Kd | KD | χ2 |
|---|---|---|---|---|
| m−1s−1 | s−1 | pm | ||
| BrnA | 2.44 × 105 | 1.63 × 10−4 | 670 | 0.109 |
Crystal Structure of BrnT Reveals a Fold Topologically Related to RelE Toxin Family
Having defined BrnTA as a toxin-antitoxin system that forms a direct physical association, we next sought to characterize the molecular structure of this novel TA system. Although purified the BrnT-BrnA complex failed to yield crystals, incubation of BrnT-BrnA with a low concentration of subtilisin in the crystallization drop (see “Experimental Procedures”) produced orthorhombic crystals of space group I4, with cell dimensions a and b = 74.4 Å and c = 29.6 Å. The structure was solved from a single selenomethionine crystal phased by single-wavelength anomalous dispersion. This crystal contained one molecule of BrnT per asymmetric unit; the protein structure was refined to a final Rwork of 0.140 and an Rfree of 0.158 (Table 3).
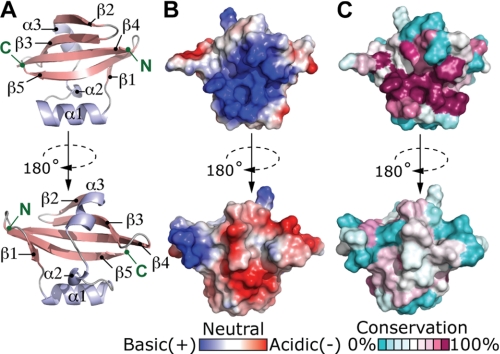
Full-length BrnT (PDB ID 3U97) contains 81 residues; electron density for the five C-terminal residues is not visible, which is likely due to disorder or to proteolysis of this segment of the structure. BrnT is topologically related to the RelE family of bacterial RNase toxins at the level of secondary structure, although overall the tertiary structural homology to RelE and its structural homologs (e.g. YoeB and MqsR) is low (DALI Z-scores <5.3). BrnT is composed of a 4-stranded antiparallel β-scaffold (β2-β3-β4-β5); β5 is flanked by a single parallel β strand (β1) (Fig. 4). Two short α-helices (α2 and α3) are docked on a hydrophobic face of the β-scaffold, and helix α1 interacts directly with β5. The most conserved region of the BrnT primary sequence corresponds to the solvent-exposed face of the β-scaffold and surface of helix α1 that interacts with the β-scaffold. This surface contains a highly positive basic patch that resembles a functional site for RNA interaction and cleavage (Fig. 4). Experiments in which we test this hypothesis are discussed below.
FIGURE 4.
Structure of BrnT. A, ribbon structure of BrnT. β-Strands, pink; α-helices, light blue. B, electrostatic surface map of BrnT; basic, blue; acidic, red. C, conservation surface map of BrnT reveals the structural position of residues that are conserved among BrnT homologs (DUF497 in the Conserved Domain Database). Coordinates of B. abortus BrnT have been deposited in the Protein Data Bank (PDB ID 3U97).
BrnT Is a Ribonuclease
As discussed above, type II toxins commonly function to inhibit the expression of new proteins. The conservation of secondary structure topology between BrnT and RNase toxins RelE, YoeB, and MqsR suggested BrnT toxicity may result from the inhibition of protein synthesis via its activity as a ribonuclease. To test this hypothesis, we first investigated whether BrnT expression affected total protein expression in vivo in a heterologous system. E. coli expressing brnT (bab1_0994), brnA (bab1_0993), or brnTA under control of an inducible promoter was grown to mid-log phase, switched to M9 medium lacking methionine and cysteine, and starved for 1 min. [35S]Methionine and [35S]cysteine were pulsed in for 1 min, the reaction was quenched with cold TCA, and the amount of methionine and cysteine incorporated into total cellular protein was quantified after brnT, brnA, or brnTA induction. Within 30 min of induction, the rate of protein synthesis in the strain expressing brnT was 3-fold lower (p < 0.01) than strains expressing brnA or brnTA (Fig. 5A). From this, we conclude that BrnT does inhibit de novo protein expression at some step.
FIGURE 5.
BrnT is a ribonuclease toxin. A, quantification of in vivo protein synthesis in E. coli 30 min after induction of brnA, brnT, or the brnTA operon; expression shown in counts per minute (cpm). Error bars represent S.D; * = p < 0.01 (one-way analysis of variance, Tukey post test). B, degradation of lacZ RNA incubated with water or buffer controls or purified BrnT, BrnA, or BrnT + BrnA (see “Experimental Procedures”). Stars indicate equal concentration of BrnT.
To test if the inhibition of protein expression is due to the degradation of RNA, we incubated purified BrnT toxin with an in vitro transcribed lacZ RNA template. RNA incubated in the presence of BrnT is degraded in a concentration-dependent manner (Fig. 5B). Addition of antitoxin to the reaction neutralizes toxicity and prevents RNA degradation; binding of the neutral BrnT-BrnA complex to the RNA template is evident in the observed bandshift in the gel and is likely mediated by the ribbon-helix-helix DNA binding domain of the antitoxin. At the concentrations used in this reaction, BrnT degrades RNA independent of the ribosome indicating that it may be a ribosome-independent RNase, in contrast to RelE and YoeB, which require the ribosome for their RNase activity. We have named the toxin BrnT for bacterial ribonuclease toxin and the antitoxin BrnA for bacterial ribonuclease antitoxin.
To identify residues required for RNA cleavage we mutated charged amino acids conserved in DUF497 at an identity level higher than 45%, and which are located in regions of structure computationally predicted to bind RNA (41–43) (Fig. 6A). Additionally, we also mutated the two histidine residues in BrnT to alanine as histidines have been shown to act as the catalytic acid in the case of YoeB and the endogenous RNases, RNase T1, RNase Sa, and RNase Sa2 (49–53). Using the heterologous E. coli expression system we measured growth after induction of the 11 mutant toxins or wild-type brnT. Amino acids fell into two general categories, those that did not change toxicity (Lys-77, His-25, Arg-79), and those that render the toxin non-toxic (Arg-41, Arg-72, His-17, Glu-78, Lys-16, Asp-6, Glu-7, and Lys-9) (Fig. 6C). We confirmed that each mutant protein was stable by Western blot (supplemental Fig. S1). Point mutations that completely ablated BrnT toxicity in vivo largely mapped to the α1–β5 region of BrnT, which is part of a larger basic patch on the surface of the protein.
FIGURE 6.
Structure/function analysis of BrnT. A, amino acid sequence of B. abortus BrnT. Mutated residues are colored in red and green depending on their effect on BrnT toxicity (no effect or strong effect, respectively). Sequence position of secondary structure elements are labeled above the alignment. Conserved residues are highlighted with black (identical residues) and gray (similar residues) squares; shading is thresholded at 50% (based on alignment with 97 homologous sequences). Residues predicted to be involved in RNA binding as assessed by three different algorithms are highlighted with purple (RNABindR), pink (BindN), and orange (PiRaNhA) squares. B, ribbon structure of BrnT. β-Strands, pink; α-helices, light blue. Side chains of mutated residues are shown as sticks and colored in red and green depending on their effect on BrnT toxicity (no effect or strong effect, respectively). C, site-directed mutagenesis of conserved charged amino acids in BrnT reveals residues required for full toxicity when compared with wild-type toxin. Red bars indicate toxicity similar to wild-type, green bars indicate diminished toxicity, and white bars indicate uninduced controls. D, RNase activity of purified wild-type BrnT or BrnT point mutants was assessed by monitoring uncleaved RNA on an agarose gel. Error bars represent S.D.
To test the correspondence between in vivo toxicity and in vitro ribonuclease activity, we purified each of the toxic point mutants and compared their ability to degrade RNA in the in vitro RNase assay. Although most non-toxic mutant BrnT proteins (R41A, E78A, D6A, E7A, and K9A) could no longer degrade RNA, BrnT R72A, H17A, and K16A retained RNase activity (Fig. 6D). Interestingly, when compared to other RelE family members, the structural position of residues reqquired for RNase activity is divergent (Fig. 7).
FIGURE 7.
Structural comparison between BrnT and members of the RelE toxin family. For each toxin (BrnT, RelE, YoeB, and MqsR) a secondary structure diagram and a surface rendered model (white) are shown. Residues that are known to be required for RNA cleavage in each of these structures are highlighted in red. PDB codes of each structure are in parentheses.
brnTA Transcription Is Autoregulated and Controlled by Multiple Environmental Stressors
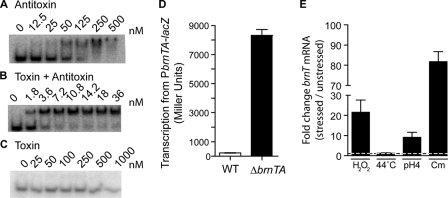
BrnA is predicted by the Conserved Domain Database (54) to contain a ribbon-helix-helix domain at its C terminus, which suggested that it may bind DNA. To test this, we first purified BrnA, BrnT, and the BrnTA complex and incubated each of these proteins with double-stranded DNA corresponding to the ATG translational start of brnT plus 47 base pairs 5′ of the start codon. By electrophoretic mobility shift (EMSA) we showed that both BrnA and BrnTA bind this putative promoter DNA with high affinity, whereas the BrnT protein alone does not bind at concentrations up to 1 μm (Fig. 8, A–C). The BrnTA complex binds the putative promoter region of the brnTA operon with a higher equilibrium affinity than BrnA alone (BrnA, KD = 47.9 nm and BrnT + A, KD = 2.0 nm). This may be due to an allosteric effect of BrnT binding on the conformation of the BrnA ribbon-helix-helix domain or an effect of BrnT binding on general BrnA stability.
FIGURE 8.
BrnA negatively autoregulates the brnTA operon; brnTA transcription is activated by multiple stressors. Purified antitoxin (A) and TA complex (B), but not toxin alone (C) bind to 32P-labeled probe corresponding to the 50 nucleotides upstream of the brnT start codon, and cause a gel shift in EMSA. D, β-galactosidase assay of wild-type B. abortus or B. abortus ΔbrnTA carrying a PbrnTA-lacZ promoter fusion plasmid. E, TaqMan assay quantifying brnT transcript in wild-type B. abortus in various stress conditions including 5 mm H2O2 in Gerhardt's minimal media, heat shock (44 °C), pH 4.0, and 200 μg/ml of chloramphenicol. Each sample is normalized to 16 S RNA and then compared with a relevant non-stressed control. Error bars represent S.D.
To assess the functional relevance of BrnTA binding of the region upstream of brnTA we constructed a transcriptional fusion of lacZ to a DNA fragment that contains the putative brnTA promoter (see “Experimental Procedures”). Wild-type B. abortus 2308 and a B. abortus strain in which the chromosomal copy of brnTA was deleted (B. abortus ΔbrnTA) were transformed with the PbrnTA-lacZ transcriptional reporter plasmid. In a wild-type genetic background (i.e. when BrnTA is expressed in the cell), transcription from the brnTA promoter is strongly repressed (Fig. 8D). In B. abortus ΔbrnTA, transcription from PbrnTA is derepressed by a factor of ∼50. Based on these data we conclude that DNA binding by the BrnA component of the BrnTA complex functions to negatively autoregulate transcription of brnTA.
The expression of type II toxin-antitoxin systems has been shown to be induced under specific stress conditions (7, 21, 55–57), often as a result of degradation of the antitoxin repressor by cellular proteases (18, 19, 57–59). B. abortus encounters many different stressors throughout its life cycle in the host, including low pH, oxidative stress, and nutrient deprivation. To test whether these and other stressors have an effect on transcription of brnTA we applied heat shock (44 °C), chloramphenicol, 5 mm H2O2, and acidic pH, pH 4.0, to wild-type B. abortus and quantified the brnTA transcript levels by RT-qPCR. Chloramphenicol, H2O2 stress, and low pH stress all result in a 9–80-fold increase in transcription of brnTA (Fig. 8E). We presume these stressors lead to the degradation of BrnA through an unknown protease and this degradation results in derepression of the brnTA operon. However, we cannot rule out other mechanisms of brnTA transcriptional activation.
DISCUSSION
Homeostatic Regulation and Stoichiometry
We have presented evidence that B. abortus BrnTA constitutes a novel, conserved ribonuclease toxin-antitoxin system. brnT expression results in cessation of bacterial growth that can be rescued by subsequent expression of brnA, even 5 h after BrnT induction. This effect on bacterial cell physiology is similar to what has been reported for the RelE and MazF toxins (60). BrnTA is a heterotetramer consisting of two molecules of BrnA and two molecules of BrnT. brnTA is organized with the brnT toxin gene preceding brnA antitoxin on B. abortus chromosome 1. It has been postulated that the canonical genetic organization of type II TA systems (i.e. antitoxin preceding toxin in the operon) ensures that there is an excess of antitoxin protein, as the gene expression level is often proportional to the gene order on a polycistronic message (10, 61, 62). However, there are a handful of systems, such as BrnTA, in which toxin precedes antitoxin (e.g. mqsRA, higBA, and hicAB) (61, 63–65). This raises the question of whether there are additional regulatory mechanisms that ensure proper toxin:antitoxin ratios in the cell. In the case of BrnTA, regulation may occur at the level of translation, as there is an apparent Shine-Dalgarno sequence upstream of the BrnA translation start, but no clear Shine-Dalgarno upstream of BrnT. Thus, BrnT and BrnA expression may be affected by mRNA-ribosome pairing efficiency near the toxin and antitoxin translation start sites. Such a mechanism could ensure an excess of antitoxin relative to toxin under normal growth conditions.
Structure and RNA Cleavage
BrnT is related to the RelE family of RNase toxins in terms of its secondary topology. However, the overall structure of BrnT differs from known RelE family members in several respects. BrnT lacks the C-terminal helix that spans the solvent-exposed β-sheet of RelE. Additionally, α2–α3 that are docked across the hydrophobic face of the β-sheet are much smaller than the two corresponding helices found in RelE and YoeB. Moreover, the β-sheet of BrnT has 5 strands in contrast to the 4 of RelE and the 6 of MqsR (Fig. 7). Despite these structural differences, the function of BrnT, similar to other RelE family members, is conserved as it rapidly attenuates protein synthesis in vivo. Similar to the toxins RelE, YoeB, and MqsR, BrnT most likely attenuates protein synthesis via its activity as a ribonuclease. Our functional analysis of BrnT by site-directed mutagenesis identified several amino acids that are required for RNA degradation. Surprisingly, not all of the amino acids required for toxicity in vivo are required for BrnT RNase activity in vitro under our assay conditions. Mutation of residues Arg-72, His-17, and Lys-16 to alanine blocks BrnT toxicity in cells, but does not completely abrogate RNA cleavage. It is not known whether mutation of these residues affects Km or Vmax of the RNA scission reaction; clearly under the conditions we have assayed in vitro RNA cleavage can still occur.
Scission of RNA by RelE family toxins requires specific general acid and general base residues, as well as residues required for orientation of nucleotides and stabilization of the transition state (51). The structural position of putative general acid and base residues that we have identified in BrnT by site-directed mutagenesis do not correspond to those described for other RelE family ribonucleases (Fig. 7). Our structural analysis is thus consistent with the emerging picture of the RelE fold as a scaffold on which particular residues required for RNA scission are plastic; the position and identity of these residues may confer different properties/specificities to this family of ribonucleases (13).
Environmental Regulation and Function
B. abortus brnA is a member of COG3514 in the Conserved Domain Database. Based on secondary structure prediction, BrnA consists of 3 α-helices and a C-terminal ribbon-helix-helix DNA binding domain. Like other toxin-antitoxin systems (66, 67), BrnA negatively autoregulates the brnTA operon and has higher affinity for the DNA operator when complexed with BrnT. In response to various environmental stressors, such as low pH and oxidative stress, we have shown that transcription of the brnTA operon is up-regulated in B. abortus. B. abortus encounters these stressors throughout the course of an infection, and toxin-antitoxin systems such as BrnTA may facilitate persistence of this bacterium within hostile host niches. We do not currently know if regulated proteolysis of BrnA provides a mechanism by which expression of brnTA is up-regulated during stress.
An intriguing hypothesis is that BrnTA and the three other TA systems in B. abortus underpin persistence and recurrent infection in those individuals that contract Brucellosis. Indeed, TA systems such as HigBA and MqsRA can contribute to the establishment of persister cells (17) and confer the ability of bacteria to enter a dormant state during antibiotic treatment (16). Persistence is particularly problematic in Brucella spp. infection; even after rigorous antibiotic treatment, relapse rates range from 3 to 30% (2). Future studies on the functional role of this system and how it contributes to the fitness of B. abortus during infection may inform new strategies for the treatment of Brucellosis.
Supplementary Material
Acknowledgments
We thank Elena Solomaha for help with AUC and surface plasmon resonance experiments and Dick Winant at the Stanford PAN facility for help with mass spectrometry protein fingerprinting. The Advanced Photon Source is supported by the Department of Energy Office of Basic Energy Sciences (contract DE-AC02-06CH11357). LS-CAT is supported by the Michigan Economic Development Corp. and Michigan Technology Tri-Corridor Grant 085P1000817. We also thank Aretha Fiebig and Nicholas Heaton for critical discussions and comments on the manuscript and Marty Roop at East Carolina University for plasmids and B. abortus 2308.
This work was supported, in whole or in part, by National Institutes of Health Grant 1-U54-AI-057153 to the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE) from the NIAID (to S. C.), by a National Science Foundation MRI grant for development of the custom tandem mass spectrometer, and the Pacific Southwest Regional Center for Excellence Grant 1U54-AI-065359 (to V. H. W.).

This article contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (code 3U97) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TA
- type II toxin-antitoxin
- AUC
- analytical ultracentrifugation
- CID
- collision-induced dissociation
- nEIS
- nanoelectrospray ionization
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. Anderson T. D., Meador V. P., Cheville N. F. (1986) Pathogenesis of placentitis in the goat inoculated with Brucella abortus. I. Gross and histologic lesions. Vet. Pathol. 23, 219–226 [DOI] [PubMed] [Google Scholar]
- 2. Moreno E., Moriyon I. (2006) in The Prokaryotes: A Handbook on the Biology of Bacteria (Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E., eds) 3rd Ed., pp. 315–456, Springer, New York [Google Scholar]
- 3. Jiang X., Leonard B., Benson R., Baldwin C. L. (1993) Macrophage control of Brucella abortus. Role of reactive oxygen intermediates and nitric oxide. Cell. Immunol. 151, 309–319 [DOI] [PubMed] [Google Scholar]
- 4. Porte F., Liautard J. P., Köhler S. (1999) Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun 67, 4041–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Köhler S., Michaux-Charachon S., Porte F., Ramuz M., Liautard J. P. (2003) What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 11, 215–219 [DOI] [PubMed] [Google Scholar]
- 6. Roop R. M., 2nd, Gaines J. M., Anderson E. S., Caswell C. C., Martin D. W. (2009) Survival of the fittest. How Brucella strains adapt to their intracellular niche in the host. Med. Microbiol Immunol. 198, 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 8. Gerdes K., Larsen J. E., Molin S. (1985) Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol 161, 292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaffé A., Ogura T., Hiraga S. (1985) Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163, 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pandey D. P., Gerdes K. (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Melderen L., Saavedra De Bast M. (2009) Bacterial toxin-antitoxin systems. More than selfish entities? PLoS Genet. 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moyed H. S., Bertrand K. P. (1983) hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blower T. R., Salmond G. P., Luisi B. F. (2011) Balancing at survival's edge. The structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr. Opin. Struct. Biol. 21, 109–118 [DOI] [PubMed] [Google Scholar]
- 14. Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 15. Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. (2004) Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol 186, 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maisonneuve E., Shakespeare L. J., Jørgensen M. G., Gerdes K. (2011) Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 108, 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Wang X., Wood T. K. (2011) Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsuchimoto S., Nishimura Y., Ohtsubo E. (1992) The stable maintenance system pem of plasmid R100. Degradation of PemI protein may allow PemK protein to inhibit cell growth. J. Bacteriol. 174, 4205–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Melderen L., Bernard P., Couturier M. (1994) Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11, 1151–1157 [DOI] [PubMed] [Google Scholar]
- 20. Lehnherr H., Yarmolinsky M. B. (1995) Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 3274–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christensen S. K., Pedersen K., Hansen F. G., Gerdes K. (2003) Toxin-antitoxin loci as stress-response elements. ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 22. Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 23. Schumacher M. A., Piro K. M., Xu W., Hansen S., Lewis K., Brennan R. G. (2009) Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang Y., Pogliano J., Helinski D. R., Konieczny I. (2002) ParE toxin encoded by the broad-host range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979 [DOI] [PubMed] [Google Scholar]
- 25. Maki S., Takiguchi S., Miki T., Horiuchi T. (1992) Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. Antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J. Biol. Chem. 267, 12244–12251 [PubMed] [Google Scholar]
- 26. Yuan J., Yamaichi Y., Waldor M. K. (2011) The three vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J. Bacteriol. 193, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winther K. S., Gerdes K. (2011) Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mutschler H., Gebhardt M., Shoeman R. L., Meinhart A. (2011) A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9, e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lioy V. S., Martín M. T., Camacho A. G., Lurz R., Antelmann H., Hecker M., Hitchin E., Ridge Y., Wells J. M., Alonso J. C. (2006) pSM19035-encoded ζ toxin induces stasis followed by death in a subpopulation of cells. Microbiology 152, 2365–2379 [DOI] [PubMed] [Google Scholar]
- 30. Van Melderen L. (2010) Toxin-antitoxin systems. Why so many, what for? Curr. Opin. Microbiol. 13, 781–785 [DOI] [PubMed] [Google Scholar]
- 31. Hayes F., Van Melderen L. (2011) Toxins-antitoxins: Diversity, evolution, and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408 [DOI] [PubMed] [Google Scholar]
- 32. Khan S. R., Gaines J., Roop R. M., 2nd, Farrand S. K. (2008) Broad-host range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74, 5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W. (1997)Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36. Dauter Z. (2002) One-and-a-half wavelength approach. Acta Crystallogr. D 58, 1958–1967 [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX, a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eisenberg D., Schwarz E., Komaromy M., Wall R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 [DOI] [PubMed] [Google Scholar]
- 40. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010, calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Terribilini M., Sander J. D., Lee J. H., Zaback P., Jernigan R. L., Honavar V., Dobbs D. (2007) RNABindR, a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 35, W578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L., Brown S. J. (2006) BindN, a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34, W243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murakami Y., Spriggs R. V., Nakamura H., Jones S. (2010) PiRaNhA, a server for the computational prediction of RNA-binding residues in protein sequences. Nucleic Acids Res. 38, W412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 45. Makarova K. S., Wolf Y. I., Koonin E. V. (2009) Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tanford C., Kawahara K., Lapanje S., Hooker T. M., Jr., Zarlengo M. H., Salahuddin A., Aune K. C., Takagi T. (1967) Proteins as random coils. 3. Optical rotatory dispersion in 6 m guanidine hydrochloride. J. Am. Chem. Soc. 89, 5023–5029 [DOI] [PubMed] [Google Scholar]
- 49. Kamada K., Hanaoka F. (2005) Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19, 497–509 [DOI] [PubMed] [Google Scholar]
- 50. Bauerová-Hlinková V., Dvorský R., Perecko D., Povazanec F., Sevcík J. (2009) Structure of RNase Sa2 complexes with mononucleotides. New aspects of catalytic reaction and substrate recognition. FEBS J. 276, 4156–4168 [DOI] [PubMed] [Google Scholar]
- 51. Heinemann U., Saenger W. (1983) Crystallographic study of mechanism of ribonuclease T1-catalyzed specific RNA hydrolysis. J. Biomol. Struct. Dyn. 1, 523–538 [DOI] [PubMed] [Google Scholar]
- 52. Yakovlev G. I., Mitkevich V. A., Shaw K. L., Trevino S., Newsom S., Pace C. N., Makarov A. A. (2003) Contribution of active site residues to the activity and thermal stability of ribonuclease Sa. Protein Sci. 12, 2367–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sevcik J., Zegers I., Wyns L., Dauter Z., Wilson K. S. (1993) Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate. Eur. J. Biochem. 216, 301–305 [DOI] [PubMed] [Google Scholar]
- 54. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD, a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fiebig A., Castro Rojas C. M., Siegal-Gaskins D., Crosson S. (2010) Interaction specificity, toxicity, and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol. Microbiol. 77, 236–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hazan R., Sat B., Engelberg-Kulka H. (2004) Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186, 3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K. (2001) RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U.S.A. 98, 14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aizenman E., Engelberg-Kulka H., Glaser G. (1996) An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate. A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Donegan N. P., Thompson E. T., Fu Z., Cheung A. L. (2010) Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol 192, 1416–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pedersen K., Christensen S. K., Gerdes K. (2002) Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 61. Leplae R., Geeraerts D., Hallez R., Guglielmini J., Drèze P., Van Melderen L. (2011) Diversity of bacterial type II toxin-antitoxin systems. A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamaguchi Y., Inouye M. (2011) Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790 [DOI] [PubMed] [Google Scholar]
- 63. Brown B. L., Grigoriu S., Kim Y., Arruda J. M., Davenport A., Wood T. K., Peti W., Page R. (2009) Three-dimensional structure of the MqsR·MqsA complex. A novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5, e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jørgensen M. G., Pandey D. P., Jaskolska M., Gerdes K. (2009) HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191, 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tian Q. B., Hayashi T., Murata T., Terawaki Y. (1996) Gene product identification and promoter analysis of high locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 225, 679–684 [DOI] [PubMed] [Google Scholar]
- 66. Kedzierska B., Lian L. Y., Hayes F. (2007) Toxin-antitoxin regulation. Bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res. 35, 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bailey S. E., Hayes F. (2009) Influence of operator site geometry on transcriptional control by the YefM-YoeB toxin-antitoxin complex. J. Bacteriol. 191, 762–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.