Background: Nuclear receptor corepressors form complexes with histone deacetylase 3 (HDAC3).
Results: Interplay of HDAC3 degradation and its complex formation with corepressors maintains free HDAC3 levels, allowing independent formation of nuclear receptor corepressor complexes.
Conclusion: By controlling HDAC3 expression, N-CoR and SMRT corepressors do not interfere with their independent complex formation with HDAC3.
Significance: Independent formation of nuclear receptor corepressor complexes ensures their independent biological functions.
Keywords: Corepressor Transcription, Histone Deacetylase, Nuclear Receptors, Protein Complexes, Protein Degradation, Nuclear Receptor Corepressor
Abstract
An important step in transcriptional regulation by corepressors N-CoR and SMRT is the formation of a stable and active histone deacetylase 3 (HDAC3)-containing complex. Although N-CoR and SMRT are thought to bind HDAC3 competitively, multiple studies have shown that they do not interfere with the function of each other. How this functional independence is sustained under the competitive interaction is unclear. Here, we show that the coupling of corepressor expression with HDAC3 degradation allows cells to maintain a stable level of uncomplexed HDAC3, thereby preventing mutual interference in the assembly of N-CoR and SMRT complexes. The free uncomplexed HDAC3 is highly unstable. Unexpectedly, the rate of HDAC3 degradation is inversely correlated with the expression level of corepressors. Our results indicate that reducing one corepressor accelerates HDAC3 clearance, thus preventing an increase in complex formation between HDAC3 and the other corepressor. In addition, this study also indicates that the formation of a stable and active HDAC3-corepressor complex is a stepwise process in which the C terminus of HDAC3 plays a critical role at late steps of the assembly process.
Introduction
Proper regulation of gene transcription requires the function of histone deacetylases (HDACs),2 which catalyze the removal of acetyl groups from lysine residues of histones as well as non-histone proteins (1–4). The ubiquitously expressed HDACs such as HDAC1 and HDAC3 comprise the class I family of HDACs. A unique feature of HDACs is the requirement of the formation of multisubunit complexes for their function. HDACs can interact with different partners to assemble into distinct multisubunit complexes. These complexes often have different functions, as best exemplified by HDAC1, which is a shared subunit of the functionally distinct NuRD (nucleosome remodeling and deacetylase complex) (5), Sin3A (SIN3 homolog A) (6–8), and CoREST (9–12) corepressor complexes.
HDAC3 also interacts with different proteins, including the two homologous nuclear receptor corepressors N-CoR and SMRT (13–17). N-CoR and SMRT share three conserved repression domains in the N-terminal region and two nuclear receptor interaction regions in the C terminus. In addition, they contain two highly conserved SANT (SWI3/ADA2/N-CoR/TFIIIB-like) domains. The N-terminal SANT domain, along with an upstream N-CoR/SMRT-specific motif, mediates the direct interaction with HDAC3 (13, 18), which is required for the activation of HDAC3 enzymatic activity (13, 18). The conserved HDAC3-interacting regions of N-CoR and SMRT are referred to as deacetylase-activating domains (DAD), also called deacetylase-interacting domains.
The homology between HDAC1 and HDAC3 resides in the conserved histone deacetylase domain, which does not include the unique C-terminal region of HDAC3. Previous studies have shown that both N- and C-terminal regions of HDAC3 are required for SMRT interaction with HDAC3 (18). However, whether these regions form a single binding surface or mediate different but mutually dependent steps in the formation of stable HDAC3 corepressor complexes is unknown.
Similar to the observation that HDAC1 forms heterogeneous complexes, biochemical purification of HDAC3-containing complexes does not support the co-existence of N-CoR and SMRT in the same HDAC3 complex (13–15). The notion that N-CoR and SMRT assemble into different HDAC3 complexes is consistent with their independent biological functions and their differential recruitment to nuclear receptors and other transcription factors (19–23). In mouse studies, depletion of each corepressor caused embryonic lethality, which occurred at different embryonic stages and showed distinct phenotypes (19–21). Knock-in mice carrying a mutated N-CoR DAD domain defective in HDAC3 binding showed defects in functions mediated by thyroid hormone receptor and peroxisome proliferator-activated receptor, which cannot be compensated by SMRT (24, 25), even though SMRT is considered a bona fide corepressor of both receptors (16, 26). In cell culture studies, the lack of N-CoR or SMRT in macrophages impacts specific signaling pathways associated with only N-CoR or SMRT, respectively (22). Similarly, knockdown of N-CoR or SMRT in MCF7 cells causes distinct effects in gene regulation and growth behavior (27), consistent with segregation of N-CoR and SMRT functions.
These biological studies illustrate an absence of significant functional interference between N-CoR and SMRT corepressors that would be expected to appear as compensating or antagonizing effects arising from their interplay at the level of complex formation. It is not known how the competitive formation of N-CoR and SMRT corepressor complexes permits their functional independency. Here, by studying the regulation of HDAC3 stability, we unexpectedly linked HDAC3 degradation to the stable and independent maintenance of corepressor complex formation. Our results indicate that the free, uncomplexed form of HDAC3 is intrinsically unstable. The rate of its degradation, however, is correlated inversely with the levels of corepressors in cells. This allows the cells to maintain a low and stable level of free HDAC3. Depletion of one corepressor accelerates degradation of free HDAC3, thereby preventing an increase in the assembly of an HDAC3 complex with the other corepressor. In addition, we have also clarified the role of the C-terminal region of HDAC3 in corepressor complex assembly. We found that the C-terminal region is not absolutely required for corepressor binding to HDAC3 but plays an important and direct role in subsequent step(s) in the formation of a stable and active HDAC3-corepressor complex. Given the nonspecific effects of histone deacetylase inhibitors as anti-cancer drugs, understanding how the formation of these complexes is regulated precisely should have important implications for developing new approaches to specifically target HDAC3 in cancers and leukemias.
EXPERIMENTAL PROCEDURES
Plasmids
HDAC3 and N-CoR-derived DAD (amino acid 420–488) and repression domain 1 (RD1; amino acid 1–312) sequences have been reported previously (13). HDAC3 C-terminal deletion mutants HDAC3Δ411 and HDAC3Δ390 were constructed by PCR and confirmed by sequencing.
Cell Culture, Transfection, and Luciferase Assays
All cells were maintained in DMEM supplemented with 10% fetal bovine serum. Standard culture conditions were used. Luciferase assays were performed as described previously (28, 29). In brief, 293T cells grown in 24-well plates were transfected with an SV40 promoter, Gal4-DNA-binding domain, or Gal4-DAD, along with different shRNA constructs, using FuGENE 6 transfection reagent (Roche Applied Science). Luciferase activity was measured 48 h post transfection and normalized to β-galactosidase, which served as the internal control for transfection efficiency. Fold repression was relative to Gal4 DNA-binding domain. All assays were performed in duplicate. Results are reported as the average and S.E. from four independent assays.
Protein Purification and in Vitro HDAC Assay
FLAG-HDAC3 and its truncated mutants were affinity-purified from transfected 293T cells via an in-frame FLAG sequence. The in vitro HDAC assay has been described previously (28). In brief, Xenopus histones were acetylated by p300 and subsequently used as the substrate for the HDAC reaction.
RT Quantitative PCR and Gene Knockdown
Quantitative reverse transcription PCR and shRNA-mediated knockdown were performed as described previously (29).
Immunoprecipitation and Western Blot Analysis
Immunoprecipitation and Western blot analyses were performed as described previously (29). Input lanes show 1% of total. Anti-N-CoR, anti-SMRT, anti-HDAC3, and anti-HDAC1 antibodies were from Thermo Scientific. Anti-FLAG M2-agarose and anti-FLAG antibody were from Sigma. N-19 anti-HDAC3 antibody was from Santa Cruz Biotechnology. Anti-α tubulin antibody was from Active Motif. Anti-GFP antibody was from Antibodies, Inc.. Anti-actin antibody was from Sigma.
Pulse-Chase Analysis
The labeling medium was Met/Cys-free DMEM (Invitrogen, 21013-024) supplemented with 10% dialyzed fetal bovine serum (Invitrogen, 26400) and EXPRE35S35S Protein Labeling Mix (PerkinElmer Life Sciences). After incubating the cells in labeling medium for 30 min (defined as 0-h), the labeling medium was removed, and the cells were incubated for 4 h (4-h time point) in chase medium containing Met/Cys-free DMEM supplemented with 10% dialyzed fetal bovine serum, 500 μg/ml l-cysteine (Sigma), and 100 μg/ml l-methionine (Sigma). Cells were harvested at both 0-h and 4-h time points and lysed in radioimmune precipitation assay buffer. The lysates were immunoprecipitated using anti-FLAG or N-19 antibodies and then subjected to SDS-PAGE. For experiments using MG132, cells were pretreated with MG132 or vehicle for 2 h prior to labeling. The amount of HDAC3 was quantified using Imagequant (Amersham Biosciences) or ImageJ (30) software following phosphorimaging or densitometry scanning, respectively.
shRNA Sequences
All shRNA constructs were purchased from Sigma and were based on the pLKO.1 backbone unless otherwise noted. The control shRNA plasmid (Addgene plasmid 1864) contains a scrambled sequence (CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG) and was used in most experiments except in Fig. 1, in which another nonspecific control shRNA containing the sequence (CCGGTCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG) was used. The human gene-specific shRNA constructs are listed below as sense sequences cloned in pLKO.1 (HDAC3, CCGGCAAGAGTCTTAATGCCTTCAACTCGAGTTGAAGGCATTAAGACTCTTGTTTTTTG; N-CoR, CCGGGCCATCAAACACAATGTCAAACTCGAGTTTGACATTGTGTTTGATGGCTTTTTG; and SMRT, CCGGGCAGCGCATCAAGTTCATCAACTCGAGTTGATGAACTTGATGCGCTGCTTTTTG).
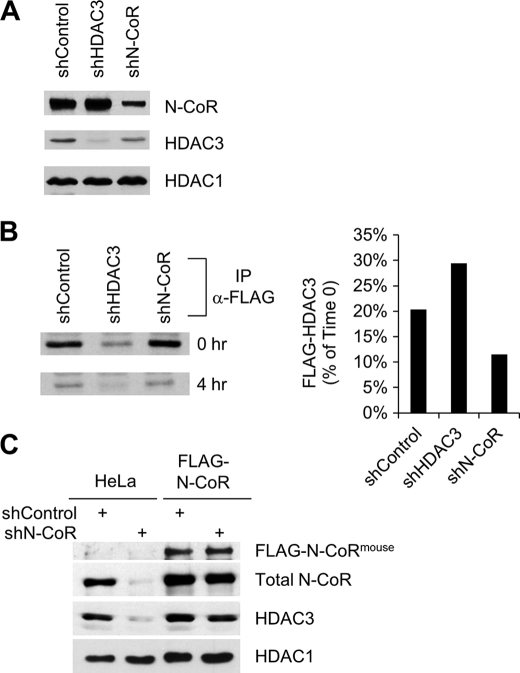
FIGURE 1.
N-CoR regulates HDAC3 stability. A, Western blot analysis of the expression of indicated proteins in FLAG-HDAC3-expressing HeLa cells transduced with the indicated shRNAs. B, pulse-chase assays detecting the turnover rate of FLAG-HDAC3 in FLAG-HDAC3-expressing HeLa cells transduced with the indicated shRNAs. Left panel, autoradiography of immunoprecipitated FLAG-HDAC3 obtained using anti-FLAG M2 agarose at 0-h and 4-h time points. Right panel, quantification of the immunoprecipitated FLAG-HDAC3 shown on the left panel. C, Western blot analysis of the expression of indicated proteins in normal HeLa cells and in HeLa cells stably expressing FLAG-tagged mouse N-CoR protein. These cells were transduced with control shRNA or human N-CoR-specific shRNA.
RESULTS
N-CoR Stabilizes HDAC3 Protein
To facilitate analysis of HDAC3, we used a previously created HeLa cell line that stably expresses FLAG-tagged HDAC3 (13). In these cells, FLAG-HDAC3 behaves similarly to endogenous HDAC3 as shown by its ability to form stable complexes with N-CoR and SMRT (13). We found that knockdown of endogenous N-CoR resulted in a concomitant reduction of total HDAC3 protein (Fig. 1A). In contrast, the level of HDAC1 was not affected. These results, together with our finding that the mRNA level of HDAC3 was unaffected by N-CoR knockdown (see Fig. 4E below), led us to test whether the reduced HDAC3 expression was due to increased protein instability. By using pulse-chase analysis, we immunoprecipitated and quantified nascent FLAG-HDAC3 protein from control, N-CoR and HDAC3 knockdown cells at both 0-h and 4-h time points (Fig. 1B). In control cells, FLAG-HDAC3 was reduced to 20% of its original level 4 h after labeling. Consistent with our hypothesis, knockdown of N-CoR enhanced the turnover rate of HDAC3. In contrast, knockdown of HDAC3 slowed the process. These results are consistent with a protective role for N-CoR in the regulation of FLAG-HDAC3 stability.
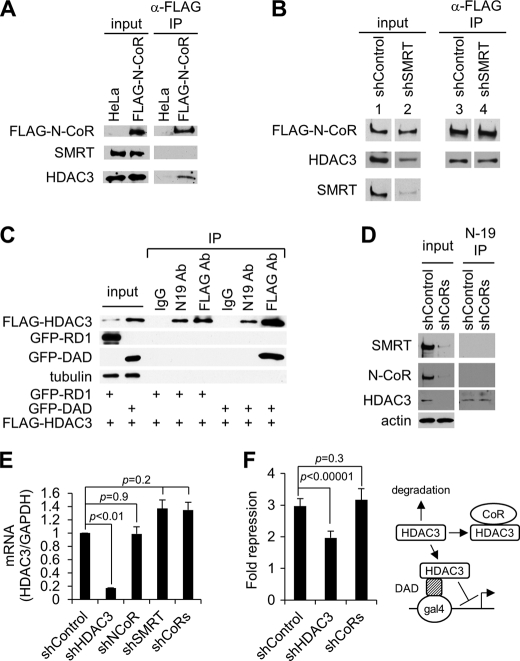
FIGURE 4.
Effect of SMRT knockdown on the formation of N-CoR-HDAC3 complex and effect of corepressor depletion on the expression of free HDAC3. A, Western blot analysis of indicated proteins in the input and anti-FLAG M2-agarose immunoprecipitates obtained from nuclear extract of HeLa cells stably expressing FLAG-N-CoR. B, Western blot analysis of indicated proteins in the input and anti-FLAG M2-agarose immunoprecipitates obtained from cell extracts of the FLAG-N-CoR cell line transduced with either control or SMRT shRNA. Lanes 1–4 were from non-adjacent lanes on the same gel. C, Western blot analysis of indicated proteins in the input and immunoprecipitates from cells co-transfected with FLAG-HDAC3 along with GFP-RD1 or GFP-DAD. Immunoprecipitation was carried out using either control IgG, N-19 anti-HDAC3, or anti-FLAG antibodies (Ab). FLAG-HDAC3 was detected by using the anti-FLAG antibody. D, Western blot analysis of indicated proteins in the input as well as the N19 anti-HDAC3 immunoprecipitates obtained from HeLa cells depleted of both N-CoR and SMRT by shRNAs (shCoRs). E, quantitative real-time PCR showing that knockdown of N-CoR and SMRT corepressors does not significantly affect the mRNA level of HDAC3 in HeLa cells. HDAC3 levels were normalized to the level of GAPDH under each treatment condition, and the relative expression of HDAC3 was set as 1 in control shRNA-transduced cells. F, luciferase reporter assays performed in 293T cells co-transfected with a Gal4-UAS-SV40 luciferase reporter along with Gal4-DAD fusion protein as well as the different shRNA constructs, as indicated. The fold repression is relative to the luciferase activities observed in cells transfected with Gal4 DNA-binding domain along with each shRNA construct. Right panel, schematic representation showing the different HDAC3 pathways in this assay system. Only free HDAC3 can be recruited by Gal4-DAD to repress transcription from the transfected reporter.
To explore whether N-CoR plays a similar role in the regulation of endogenous HDAC3, and to ascertain whether the effect observed with the N-CoR shRNA was indeed N-CoR-specific, we knocked down N-CoR in normal HeLa cells as well as in an engineered HeLa cell line that stably expresses a FLAG-tagged mouse N-CoR cDNA, which is not targeted by the human-specific shRNA used. In HeLa cells, depletion of N-CoR protein resulted in a similarly strong reduction of endogenous HDAC3, but the level of HDAC1 was unaffected (Fig. 1C). In the cells expressing mouse FLAG-N-CoR, as expected, the total level of N-CoR was only slightly reduced by the human N-CoR shRNA. Accordingly, the level of HDAC3 was much less affected (Fig. 1C). Thus, both the depletion and the rescue experiments consistently demonstrated that N-CoR is an important stabilizing factor of HDAC3.
Free Uncomplexed HDAC3 Is Unstable and Is Protected by Binding to Corepressors
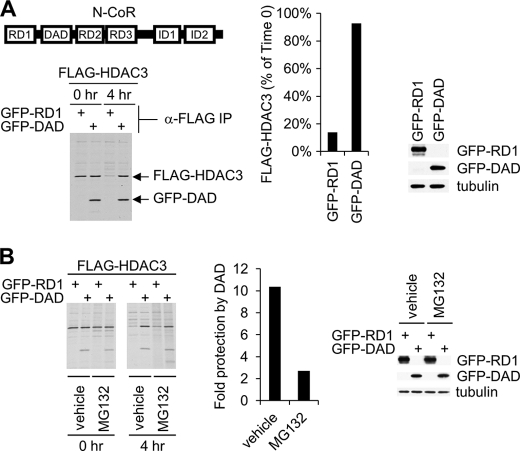
Because N-CoR binds HDAC3 but not HDAC1, we hypothesized that the stabilizing effect of N-CoR is attributed to its binding to HDAC3. To test this, we asked whether the short DAD region of N-CoR (amino acids 420 to 488, Fig. 2A) was sufficient to stabilize HDAC3. The repression domain 1 of N-CoR (RD1), which does not bind to HDAC3, was used as a negative control. Both polypeptides were expressed as fusion proteins to GFP. These fusion protein constructs were co-transfected with FLAG-HDAC3 into 293T cells followed by pulse-chase experiments. In cells expressing GFP-RD1, newly synthesized FLAG-HDAC3 was degraded to 10% of its original level in 4 h (Fig. 2A). Remarkably, expression of GFP-DAD resulted in ∼90% protection of HDAC3 from degradation (Fig. 2A). GFP-RD1 and GFP-DAD were expressed at similar levels in transfected cells (Fig. 2A, right panel). We further studied the possible involvement of proteasomal pathways in the degradation of HDAC3. Treatment of the cells with the proteasome inhibitor, MG132, resulted in modest protection of HDAC3 from degradation in cells coexpressing GFP-RD1 (Fig. 2B). This result shows that the proteasomal pathway is partially responsible for the degradation of HDAC3 in these cells. In cells co-expressing GFP-DAD, treatment with MG132 significantly reduced but did not abolish the ability of DAD to stabilize HDAC3 (Fig. 2B). The levels of GFP-RD1 or GFP-DAD were similar in vehicle- and MG132-treated cells (Fig. 2B, right panel). Taken together, these results show that the corepressor binding step is crucial for the protection of HDAC3 from degradation.
FIGURE 2.
Role of the corepressor DAD domain in the regulation of HDAC3 stability. A, pulse-chase analysis of FLAG-HDAC3 turnover in 293T cells transfected with FLAG-HDAC3 along with GFP-RD1 or GFP-DAD. The middle panel shows the quantification of the remaining HDAC3 at the 4-h time point as a percentage of the initial HDAC3 level at the 0-h time point. The right panel is a Western blot analysis of whole cell extracts showing the expression of GFP-RD1 and GFP-DAD in 293T cells transfected with FLAG-HDAC3 along with GFP-RD1 or GFP-DAD. GFP-RD1 and GFP-DAD were detected using the anti-GFP antibody. B, pulse-chase analysis performed as described in A in the presence of MG132 (20 μm) or dimethyl sulfoxide (DMSO; vehicle control). The middle panel shows the ratio of remaining HDAC3 percentage at 4-h time point in DAD-transfected to RD1-transfected cells in the presence of MG132 or dimethyl sulfoxide. The right panel is a Western blot analysis of cell extracts showing the expression of GFP-RD1 and GFP-DAD in vehicle (dimethyl sulfoxide)-treated and MG132-treated 293T cells transfected with FLAG-HDAC3 along with GFP-RD1 or GFP-DAD. GFP-RD1 and GFP-DAD were detected as described in A. Cells were treated with dimethyl sulfoxide or MG132 for 6 h before lysis.
Deletion of C Terminus of HDAC3 Uncouples DAD Binding from Its Ability to Stabilize and Activate HDAC3
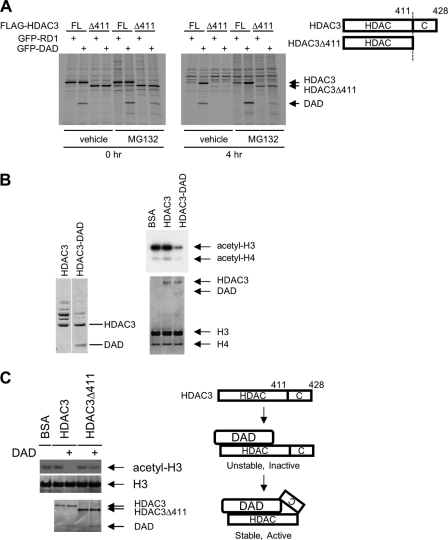
Given previous studies showing that the unique C-terminal region of HDAC3 is important for the binding of corepressors to HDAC3 (18) and for the regulation of HDAC3 function by phosphorylation (31) and caspase-mediated cleavage (32), we asked whether deleting this region would affect HDAC3 degradation or affect its protection by DAD. We first constructed a FLAG-tagged HDAC3 mutant lacking the C-terminal region after amino acid 411 (HDAC3Δ411). Using pulse-chase assays similar to that shown in Fig. 2B, we compared the stability of wild-type HDAC3 (FL) and the HDAC3Δ411 (Δ411) mutant as well as their protection by DAD. Consistent with previous results (18), HDAC3Δ411 was unable to bind DAD in cells grown in the absence of proteasome inhibitor, and was highly unstable like the wild-type HDAC3 (Fig. 3A). Remarkably, MG132 treatment allowed the detection of an interaction between HDAC3Δ411 and DAD (Fig. 3A). However, this binding was insufficient to protect HDAC3Δ411 from degradation, which occurred similarly to that observed in RD1-transfected cells (Fig. 3A). Similar results were also observed with another C-terminal-truncated HDAC3 mutant, HDAC3Δ390, which lacks the region following amino acid 390 (data not shown).
FIGURE 3.
Deletion of the C terminus of HDAC3 uncouples DAD binding from its ability to stabilize and activate HDAC3. A, pulse-chase assays performed as described in Fig. 2 using cells transfected with either full-length FLAG-HDAC3 (FL) or FLAG-HDAC3Δ411, along with GFP-RD1 or GFP-DAD. The right panel shows the schematic representation of HDAC3 and HDAC3Δ411 proteins. B, HDAC assays of purified HDAC3 and HDAC3-DAD complex showing that HDAC3-DAD, but not HDAC3, has histone deacetylase activity. Left and right bottom panels, Coomassie Blue staining of indicated proteins. Right top panel, autoradiographic image. C, in vitro HDAC assays performed using the HDAC3 or HDAC3Δ411 protein alone, or their respective complexes with GFP-DAD, purified from transfected 293T cells following treatment with MG132. The arrows denote the bands corresponding to HDAC3, HDAC3Δ411, DAD, total H3 protein, and the acetylated H3 protein. Top panel, autoradiographic image. Middle and bottom panels, Coomassie Blue staining. Right panel, the proposed stepwise model of HDAC3 stabilization and activation. The C-terminal region of HDAC3 is required for the stabilization and activation of HDAC3 by DAD. In the absence of the C terminus, the binding of DAD to HDAC3 yields an unstable complex that may be quickly degraded or dissociated in the absence of MG132.
These results revealed that the binding of DAD to HDAC3 is not sufficient to stabilize HDAC3 and that additional C-terminal region-dependent step(s) are required. DAD binding has been implicated in the activation of HDAC3 (13, 18), and it has been suggested that deletion of the C terminus of HDAC3 abolishes binding to DAD, precluding activation by DAD (18). Because we found that blocking proteasome function by treatment with MG132 stabilized the interaction between DAD and C-terminal truncated HDAC3, we were able to test whether the C-terminal region of HDAC3 is involved directly in its activation. Using a cell-free HDAC assay, we first confirmed that the purified HDAC3-DAD complex but not uncomplexed HDAC3 was active enzymatically (Fig. 3B). Next, we purified HDAC3-DAD and HDAC3Δ411-DAD complexes by anti-FLAG immunoprecipitation from transfected, MG132-treated, 293T cells. Using the HDAC assay, we found that HDAC3-DAD but not HDAC3Δ411-DAD was active enzymatically (Fig. 3C, left). This result demonstrates that the C-terminal region of HDAC3 is involved directly in the activation of HDAC3 by DAD. Based on these results, we propose a stepwise model for the formation of a stable and active HDAC3-corepressor complex (Fig. 3C, right). The initial binding of HDAC3 to DAD is independent of the HDAC3 C-terminal region. This binding yields an intermediate complex that is unstable, inactive, and sensitive to proteasomal degradation. Subsequent step(s) involving the C-terminal region of HDAC3, which may engage a direct association with DAD, is required to convert this complex into a stable and active form.
Knockdown of SMRT Does Not Increase Complex Formation between N-CoR and HDAC3
Several independent studies have indicated that the N-CoR complex does not contain SMRT and that the SMRT complex does not contain N-CoR (13–15). This also was confirmed here by the failure to detect SMRT in the FLAG-N-CoR-HDAC3 complex purified from the FLAG-N-CoR-expressing cells (Fig. 4A). Because SMRT and N-CoR are expected to compete for binding to HDAC3, we asked whether knockdown of SMRT would allow increased formation of the N-CoR-HDAC3 complex. We tested this idea using the FLAG-N-CoR-expressing cells. We found that knockdown of SMRT in the FLAG-N-CoR-expressing cells reduced the total level of HDAC3 expression (Fig. 4B), similar to the effect of knocking down N-CoR in normal HeLa cells observed earlier. SMRT knockdown did not reduce and may even slightly increase the mRNA level of HDAC3 in HeLa cells (see Fig. 4E below). These results indicate that both N-CoR and SMRT contribute to the maximal steady-state level of HDAC3 protein expression. Surprisingly, we found that the level of HDAC3 contained in the FLAG-N-CoR complex essentially was unaffected by the knockdown of SMRT (Fig. 4B). This result, showing that the level of the N-CoR-HDAC3 complex is stable and insensitive to changes in SMRT corepressor levels, indicates that levels of corepressor complexes are regulated independently.
Steady-state Level of Free HDAC3 Is Unaffected by Corepressor Knockdown
Because the steady-state level of the corepressor-HDAC3 complex is a function of the steady-state level of the uncomplexed free HDAC3, we hypothesized that a buffering mechanism may exist, which enables cells to maintain a constant level of free HDAC3, thus preventing competitive interference between the two corepressor complexes. To test this hypothesis, we sought to directly measure the cellular level of free, uncomplexed HDAC3. To this end, we examined an HDAC3 antibody (N-19) that recognizes the N terminus of HDAC3. We reasoned that because the N-terminal region of HDAC3 is required for corepressor association (18), its binding to corepressors might interfere with its recognition by the N-19 antibody. To test this possibility, we compared immunoprecipitates obtained using N-19 and anti-FLAG antibodies from 293T cells transfected with FLAG-HDAC3 in the absence or presence of GFP-DAD. The results confirmed that the N-19 antibody was unable to immunoprecipitate the DAD-bound form of HDAC3 (Fig. 4C). In contrast, the FLAG antibody captured both free and GFP-DAD-bound forms of FLAG-HDAC3 (Fig. 4C). These studies also confirmed that HDAC3 is stabilized by binding DAD. This is shown by the increased expression of FLAG-HDAC3 in GFP-DAD-expressing cells (Fig. 4C, input lanes), and the selective enrichment of HDAC3 in its DAD-bound form in these cells (Fig. 4C, IP lanes), compared with GFP-RD1-expressing cells.
Having demonstrated that the N-19 antibody specifically immunoprecipitated the free, uncomplexed form of HDAC3, we then asked whether the level of free HDAC3 was refractory to a change in corepressor expression. To better test our hypothesis, we reduced both corepressor levels by simultaneously depleting N-CoR and SMRT in HeLa cells. This caused a dramatic reduction of the total protein expression of HDAC3 (Fig. 4D, input lanes). Its mRNA level, however, was unaffected (Fig. 4E), consistent with the protective function of N-CoR and SMRT against HDAC3 degradation. Using the N-19 antibody, we then immunoprecipitated the free HDAC3 from both control and corepressor knockdown cells. As expected, the N-19 antibody failed to immunoprecipitate any corepressors from these cells, consistent with its specificity for the free form of HDAC3 (Fig. 4D). Despite the dramatic reduction of the total HDAC3 level in corepressor knockdown cells, the level of free HDAC3 detected by the N-19 antibody was similar in both control and knockdown cells (Fig. 4D), thus confirming our hypothesis. In Fig. 4D, the detection of free HDAC3 requires a much longer exposure as compared with the detection of total HDAC3 (left), indicating that free HDAC3 exists at a low level in cells. This is consistent with the pronounced reduction of the total HDAC3 level following corepressor depletion.
To further confirm the ability of cells to maintain a stable level of free HDAC3, we co-transfected 293T cells with the Gal4-DAD fusion protein and a Gal4-responsive luciferase reporter, along with control, HDAC3-, and corepressor-specific shRNAs. Repression of luciferase expression in this system has been shown previously (18) and is thought to be dependent on the recruitment of endogenous, free HDAC3 (not bound to corepressors). Thus, the ability of Gal4-DAD to repress luciferase expression is a measurement of the cellular level of free HDAC3. We found that knocking down HDAC3 reduced Gal4-DAD-mediated repression (Fig. 4F). In contrast and confirming the existence of similar levels of free HDAC3 in control and corepressor knockdown cells, knocking down both N-CoR and SMRT corepressors did not significantly change the level of Gal4-DAD-mediated repression (p = 0.3) (Fig. 4F). Together with the immunoprecipitation result, this result also shows that the ability to maintain a stable free HDAC3 concentration is cell type-independent.
Knockdown of Corepressors Accelerates Free HDAC3 Degradation
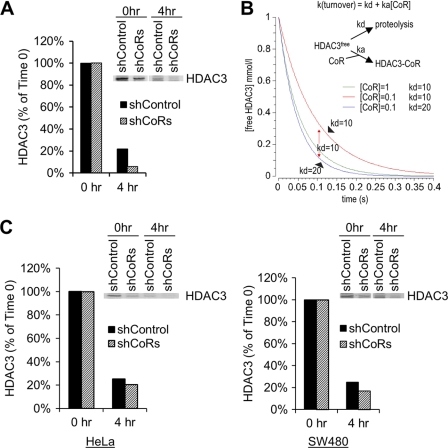
A stable level of free HDAC3 should allow each corepressor to independently maintain its optimal level of complex formation. We next sought to gain further insight into the mechanism underlying the ability of cells to maintain a stable level of free HDAC3. Given that HDAC3 is under dynamic equilibrium between degradation and complex formation, along with the finding that the mRNA level of HDAC3 is unaffected by corepressor knockdown, we hypothesized that reducing corepressor levels may increase the rate of free HDAC3 clearance, thereby allowing cells to maintain a stable steady-state level of free HDAC3. This also would explain why the complex formation between N-CoR and HDAC3 was not increased in SMRT knockdown cells (Fig. 4B). To test this hypothesis, we performed pulse-chase assays using the N-19 antibody in both control and corepressor knockdown cells. We found that the turnover rate of free HDAC3 was significantly increased by knocking down corepressors in 293T cells (Fig. 5A). The increased turnover of free HDAC3 in corepressor-depleted cells should result from increased degradation based on the following considerations. First, because degradation of free HDAC3 and the formation of HDAC3-corepressor complexes are competing reactions (Fig. 5B), the overall rate constant of free HDAC3 turnover, k(turnover) (Fig. 5B), should be the sum of both reactions. Second, because complex formation is reduced in corepressor-depleted cells, the accelerated turnover of free HDAC3 in corepressor knockdown cells should, therefore, result from an increased rate of degradation (i.e. an increased kd, Fig. 5B). This idea was also confirmed by the results from modeling of the pulse-chase reaction using COPASI software (complex pathway simulator) (33) under different corepressor levels using different kd values (Fig. 5B). Increased turnover of free HDAC3 upon corepressor depletion was observed in other cell types (Fig. 5C), although the extent varied, possibly related to the different levels of corepressor knockdown or to cell type-specific effects. Taken together, the above studies indicate that the corepressors are not only capable of forming a complex with HDAC3 to prevent its degradation, but are also capable of exerting a more direct inhibition on the degradation of free HDAC3. A reduction in one corepressor is predicted to increase the degradation rate allowing more HDAC3 to be degraded, thereby avoiding an increase in the formation of the other corepressor complex.
FIGURE 5.
Knockdown of corepressors accelerates free HDAC3 degradation. A, pulse-chase analysis using the N-19 antibody to detect free HDAC3 in control and corepressor-depleted 293T cells. B, kinetic modeling of pulse-chase reactions of free HDAC3 under conditions of variable corepressor concentration ([CoR] = 1 or 0.1) and variable degradation rate constant of free HDAC3 (kd = 10 or 20). ka, association rate constant of HDAC3 and corepressors. The results confirmed that reduced expression of corepressors will slow the process of free HDAC3 turnover unless the degradation rate has been increased concurrently. C, pulse-chase analysis as performed in A in control and corepressor-depleted HeLa (left panel) and SW480 (right panel) cells.
N-CoR/SMRT Controls HDAC3 Protein Expression in Cell Type-independent Manner
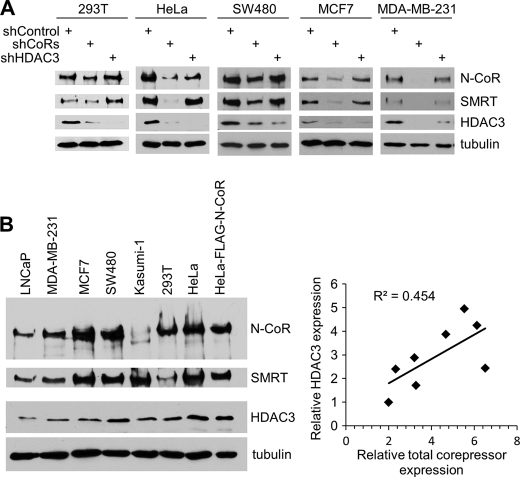
Consistent with the cell type-independent regulation of HDAC3 degradation, we confirmed that knockdown of corepressors generally reduced the total protein levels of HDAC3 in multiple cell types, including SW480 colon cancer cells, which overexpress HDAC3 (Fig. 6A) (34). To directly correlate HDAC3 level with corepressor expression, we performed Western blot analysis and quantified their relative expression levels from multiple cell types by using densitometry and NIH ImageJ software (30). The results confirmed a linear correlation between HDAC3 expression and the overall expression of N-CoR and SMRT corepressors (Fig. 6B and supplemental Table S1), which further underscored a dominant role of corepressors in the control of HDAC3 protein levels. Interestingly, between the two breast cancer cell lines, MDA-MB-231 and MCF7, the more aggressive MDA-MB-231 cells showed a higher expression ratio of HDAC3 to corepressors (Fig. 6B and supplemental Table S1). This suggests an interesting possibility that the reduced HDAC3 degradation rate (kd), which should increase the ratio of HDAC3 versus corepressors, may be linked to tumor progression.
FIGURE 6.
Corepressor expression generally correlates with HDAC3 protein level in multiple cell types. A, Western blot analysis of indicated proteins expressed in cells following control knockdown, corepressor knockdown, or HDAC3 knockdown. B, Western blot analysis of N-CoR, SMRT, and HDAC3 expression in the cell lines indicated. The level of expression was quantified by densitometry and NIH ImageJ software. Relative expression, after normalization to the expression observed in LNCaP cells, is shown in the supplemental Table S1. The bottom right panel shows a scatter plot between relative HDAC3 expression and the overall level of corepressor expression (N-CoR, SMRT). The squared correlation coefficient (R2) is shown.
DISCUSSION
This is the first study to show that the formation of multisubunit complexes can be protected from mutual interference through regulated clearance of the uncomplexed form of the shared subunit. There are other heterogeneous transcriptional complexes that contain shared subunits but possess distinct functions, such as the various HDAC1 complexes mentioned earlier, the BRM- and BRG1-containing SWI/SNF complexes (35), and the various TATA-binding protein-associated factor-containing complexes (36–38). Therefore, our study should have broad implications in the regulation of these complexes as well.
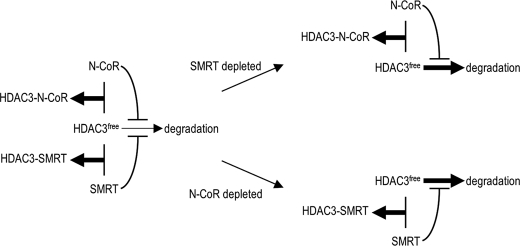
Based on our findings, we propose a model to explain how each corepressor self-maintains its complex level independent of changes in levels of the other corepressor. Although the uncomplexed free HDAC3 is intrinsically unstable, the extent of degradation is determined by the balance between complex formation and the degradation potential of HDAC3. The ability of corepressors both to protect HDAC3 from degradation and to inhibit the rate of degradation (Fig. 7) drives the equilibrium toward complex formation and allows both corepressors to establish their independent and optimal levels of complex assembly (Fig. 7). When the level of a particular corepressor is reduced, its inhibition of HDAC3 degradation is relieved, causing a readjustment of the balance between complex formation and degradation. The fraction of HDAC3 that would otherwise form a complex with the (now) reduced corepressor is directed into the degradation pathway, preventing formation of additional complex with the other corepressor (Fig. 7). It should also be noted that, although our study described here has focused on SMRT depletion to confirm the independent formation of corepressor complexes, given that the level of free HDAC3 is unaffected by the change of expression of both corepressors, depletion of N-CoR similarly should have no effect on the complex formation between SMRT and HDAC3.
FIGURE 7.
Proposed model showing that regulated HDAC3 degradation serves as a buffering mechanism to protect independent formation of N-CoR and SMRT corepressor complexes. The thickness of arrows toward complex formation or degradation indicates the level of complex formation or the speed of HDAC3 degradation, respectively. See text for details.
Previous studies have shown that the expression of corepressors can be regulated depending on cell cycle status (39), hormone stimulation (40), or signaling-dependent phosphorylation (41–43). Under these circumstances, when the expression of one corepressor is affected, the proposed mechanism of insulating the two corepressor complexes would protect the independent function of each corepressor. It would minimize possible compensating effects from increased or decreased formation of the other corepressor complex, while also ensuring that the independent function of the other corepressor (whose expression is unaffected) is not compromised.
Our results show that corepressor binding protects HDAC3 from both proteasomal and non-proteasomal degradation pathways. Consistent with this, we found that free HDAC3 is polyubiquitinated in transfected 293T cells and that the formation of HDAC3-DAD complex prevents this polyubiquitination (data not shown). Although ubiquitination can mediate both proteasome-dependent and -independent degradation (44), it remains to be determined whether ubiquitination is the sole driver of HDAC3 degradation. It also will be important to determine if HSP70 and/or TRiC chaperones are involved in the regulation of HDAC3 degradation, as these proteins have been shown to bind HDAC3 only prior to the formation of a stable corepressor complex (45). We speculate that factors that mediate HDAC3 degradation might also mediate cross-talk between degradation and corepressor expression. Conceivably, these factors may be sequestered by binding to corepressors and become released upon reduced corepressor expression. Candidates include TBL1 and TBLR1, which can function as ubiquitin ligases and have been shown to be involved in the degradation of HDAC3 (46, 47).
Finally, our studies have implications for the therapeutic targeting of HDAC3. It is known that HDAC3 is commonly up-regulated in cancers (34). Although histone deacetylase inhibitors are useful in the treatment of these cancers, they are associated with nonspecific effects. It has been reported that treatment with histone deacetylase inhibitors can induce concomitant degradation of HDAC3 and corepressors in cancer cells (48), confirming the inverse correlation between HDAC3 clearance and corepressor expression as observed here. Along with the presently observed HDAC3 reduction upon corepressor depletion in all cancer cell lines tested here, these studies suggest that the pathways driving HDAC3 degradation in tumors are intact. Therefore, further analysis of the mechanisms underlying, and the factors involved in, the degradation of HDAC3 may reveal new targets allowing specific inactivation of HDAC3 in cancers and leukemias.
Supplementary Material
Acknowledgments
We thank Dr. Jerry Lingrel and Dr. Peter Stambrook for critical reading and comments.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL093195 (to J. Z.) and Training Grant T32CA059268 (to A. G.). This work was also supported by University of Cincinnati Cancer Center and University Research Council grants (to J. Z.).

This article contains supplemental Table S1.
- HDAC
- histone deacetylase
- N-CoR
- nuclear receptor corepressor
- SMRT
- silencing mediator of retinoic acid and thyroid hormone receptor
- CoREST
- corepressor for RE1 silencing transcription factor.
REFERENCES
- 1. Hassig C. A., Schreiber S. L. (1997) Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1, 300–308 [DOI] [PubMed] [Google Scholar]
- 2. Xu L., Glass C. K., Rosenfeld M. G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9, 140–147 [DOI] [PubMed] [Google Scholar]
- 3. Karagianni P., Wong J. (2007) HDAC3: Taking the SMRT-N-CoRrect road to repression. Oncogene 26, 5439–5449 [DOI] [PubMed] [Google Scholar]
- 4. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., Wang W. (1998) NURD, a novel complex with both ATP-dependent chromatin remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 [DOI] [PubMed] [Google Scholar]
- 6. Alland L., Muhle R., Hou H., Jr., Potes J., Chin L., Schreiber-Agus N., DePinho R. A. (1997) Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55 [DOI] [PubMed] [Google Scholar]
- 7. Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 8. Laherty C. D., Yang W. M., Sun J. M., Davie J. R., Seto E., Eisenman R. N. (1997) Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89, 349–356 [DOI] [PubMed] [Google Scholar]
- 9. Ballas N., Battaglioli E., Atouf F., Andres M. E., Chenoweth J., Anderson M. E., Burger C., Moniwa M., Davie J. R., Bowers W. J., Federoff H. J., Rose D. W., Rosenfeld M. G., Brehm P., Mandel G. (2001) Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365 [DOI] [PubMed] [Google Scholar]
- 10. Humphrey G. W., Wang Y., Russanova V. R., Hirai T., Qin J., Nakatani Y., Howard B. H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276, 6817–6824 [DOI] [PubMed] [Google Scholar]
- 11. Kojima T., Murai K., Naruse Y., Takahashi N., Mori N. (2001) Cell-type non-selective transcription of mouse and human genes encoding neural restrictive silencer factor. Brain Res. Mol. Brain Res. 90, 174–186 [DOI] [PubMed] [Google Scholar]
- 12. You A., Tong J. K., Grozinger C. M., Schreiber S. L. (2001) CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. U.S.A. 98, 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Kalkum M., Chait B. T., Roeder R. G. (2002) The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 [DOI] [PubMed] [Google Scholar]
- 14. Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40 repeat protein linked to deafness. Genes Dev. 14, 1048–1057 [PMC free article] [PubMed] [Google Scholar]
- 15. Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen J. D., Evans R. M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457 [DOI] [PubMed] [Google Scholar]
- 17. Hörlein A. J., Näär A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Söderström M., Glass C. K. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 [DOI] [PubMed] [Google Scholar]
- 18. Guenther M. G., Barak O., Lazar M. A. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jepsen K., Hermanson O., Onami T. M., Gleiberman A. S., Lunyak V., McEvilly R. J., Kurokawa R., Kumar V., Liu F., Seto E., Hedrick S. M., Mandel G., Glass C. K., Rose D. W., Rosenfeld M. G. (2000) Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102, 753–763 [DOI] [PubMed] [Google Scholar]
- 20. Jepsen K., Gleiberman A. S., Shi C., Simon D. I., Rosenfeld M. G. (2008) Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 22, 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jepsen K., Solum D., Zhou T., McEvilly R. J., Kim H. J., Glass C. K., Hermanson O., Rosenfeld M. G. (2007) SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450, 415–419 [DOI] [PubMed] [Google Scholar]
- 22. Ghisletti S., Huang W., Jepsen K., Benner C., Hardiman G., Rosenfeld M. G., Glass C. K. (2009) Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 23, 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu X., Li Y., Lazar M. A. (2001) Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell Biol. 21, 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. You S. H., Liao X., Weiss R. E., Lazar M. A. (2010) The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol. Endocrinol. 24, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alenghat T., Meyers K., Mullican S. E., Leitner K., Adeniji-Adele A., Avila J., Buan M., Ahima R. S., Kaestner K. H., Lazar M. A. (2008) Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohen R. N. (2006) Nuclear receptor corepressors and PPARγ. Nucl. Recept. Signal 4, e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peterson T. J., Karmakar S., Pace M. C., Gao T., Smith C. L. (2007) The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor α transcriptional activity. Mol. Cell Biol. 27, 5933–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo C., Hu Q., Yan C., Zhang J. (2009) Multivalent binding of the ETO corepressor to E proteins facilitates dual repression controls targeting chromatin and the basal transcription machinery. Mol. Cell Biol. 29, 2644–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Q., Guo C., Li Y., Aronow B. J., Zhang J. (2011) LMO7 mediates cell-specific activation of the Rho-myocardin-related transcription factor-serum response factor pathway and plays an important role in breast cancer cell migration. Mol. Cell Biol. 31, 3223–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics International 11, 36–42 [Google Scholar]
- 31. Zhang X., Ozawa Y., Lee H., Wen Y. D., Tan T. H., Wadzinski B. E., Seto E. (2005) Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 19, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia Y., Wang J., Liu T. J., Yung W. K., Hunter T., Lu Z. (2007) c-Jun down-regulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Mol. Cell 25, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhal M., Xu L., Mendes P., Kummer U. (2006) COPASI, a complex pathway simulator. Bioinformatics 22, 3067–3074 [DOI] [PubMed] [Google Scholar]
- 34. Spurling C. C., Godman C. A., Noonan E. J., Rasmussen T. P., Rosenberg D. W., Giardina C. (2008) HDAC3 overexpression and colon cancer cell proliferation and differentiation. Mol. Carcinog. 47, 137–147 [DOI] [PubMed] [Google Scholar]
- 35. Miao J., Fang S., Lee J., Comstock C., Knudsen K. E., Kemper J. K. (2009) Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell Biol. 29, 6170–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grant P. A., Schieltz D., Pray-Grant M. G., Steger D. J., Reese J. C., Yates J. R., 3rd, Workman J. L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53 [DOI] [PubMed] [Google Scholar]
- 37. Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H. G., Krasnov A., Georgieva S. G., Schüle R., Takeyama K., Kato S., Tora L., Devys D. (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29, 92–101 [DOI] [PubMed] [Google Scholar]
- 38. Martinez E., Palhan V. B., Tjernberg A., Lymar E. S., Gamper A. M., Kundu T. K., Chait B. T., Roeder R. G. (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell Biol. 21, 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park E. J., Schroen D. J., Yang M., Li H., Li L., Chen J. D. (1999) SMRTe, a silencing mediator for retinoid and thyroid hormone receptors extended isoform that is more related to the nuclear receptor corepressor. Proc. Natl. Acad. Sci. U.S.A. 96, 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frasor J., Danes J. M., Funk C. C., Katzenellenbogen B. S. (2005) Estrogen down-regulation of the corepressor N-CoR: Mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 102, 13153–13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hong S. H., Privalsky M. L. (2000) The SMRT corepressor is regulated by a MEK-1 kinase pathway: Inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell Biol. 20, 6612–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jonas B. A., Privalsky M. L. (2004) SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J. Biol. Chem. 279, 54676–54686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoberg J. E., Yeung F., Mayo M. W. (2004) SMRT derepression by the IκB kinase α: A prerequisite to NF-κB transcription and survival. Mol. Cell 16, 245–255 [DOI] [PubMed] [Google Scholar]
- 44. Clague M. J., Urbé S. (2010) Ubiquitin: Same molecule, different degradation pathways. Cell 143, 682–685 [DOI] [PubMed] [Google Scholar]
- 45. Guenther M. G., Yu J., Kao G. D., Yen T. J., Lazar M. A. (2002) Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 RING complex. Genes Dev. 16, 3130–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perissi V., Aggarwal A., Glass C. K., Rose D. W., Rosenfeld M. G. (2004) A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116, 511–526 [DOI] [PubMed] [Google Scholar]
- 47. Zhao H. L., Ueki N., Hayman M. J. (2010) The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochem. Biophys. Res. Commun. 399, 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rajendran P., Delage B., Dashwood W. M., Yu T. W., Wuth B., Williams D. E., Ho E., Dashwood R. H. (2011) Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: Competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol. Cancer 10, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.