Background: Alix participates in fundamental cellular processes, but how it is regulated remains unknown.
Results: Alix is ubiquitinated by the Ozz-E3 ligase and participates in actin cytoskeleton remodeling, filopodia formation, and myoblast migration.
Conclusion: Ozz influences Alix conformation and in turn the extent of ubiquitination in Alix.
Significance: Ozz-E3 ligase regulates Alix concentration at sites where the actin cytoskeleton undergoes remodeling.
Keywords: Cell Migration, Myogenesis, Skeletal Muscle, Ubiquitin Ligase, Ubiquitination, Alix, F-actin, Ozz-E3 Ubiquitin Ligase, Skeletal Muscle Cells
Abstract
Alix/AIP1 is a multifunctional adaptor protein that participates in basic cellular processes, including membrane trafficking and actin cytoskeleton assembly, by binding selectively to a variety of partner proteins. However, the mechanisms regulating Alix turnover, subcellular distribution, and function in muscle cells are unknown. We now report that Alix is expressed in skeletal muscle throughout myogenic differentiation. In myotubes, a specific pool of Alix colocalizes with Ozz, the substrate-binding component of the muscle-specific ubiquitin ligase complex Ozz-E3. We found that interaction of the two endogenous proteins in the differentiated muscle fibers changes Alix conformation and promotes its ubiquitination. This in turn regulates the levels of the protein in specific subcompartments, in particular the one containing the actin polymerization factor cortactin. In Ozz−/− myotubes, the levels of filamentous (F)-actin is perturbed, and Alix accumulates in large puncta positive for cortactin. In line with this observation, we show that the knockdown of Alix expression in C2C12 muscle cells affects the amount and distribution of F-actin, which consequently leads to changes in cell morphology, impaired formation of sarcolemmal protrusions, and defective cell motility. These findings suggest that the Ozz-E3 ligase regulates Alix at sites where the actin cytoskeleton undergoes remodeling.
Introduction
During the early stages of vertebrate limb development, skeletal muscle progenitor cells migrate from the lateral lip of the dermomyotome into the limb buds, where they differentiate and fuse to form the primary myotubes of limb muscles (1). Similarly, in response to muscle lesions or disease, migration of satellite cells is required to lead the regeneration of the adult skeletal muscle. To migrate, individual cells must correctly sense or respond to migratory cues and translate a number of different inputs into appropriate cellular responses. First, a cell must discern the direction in which to move and must orient itself (i.e. polarize) so to effect directed migration. This process is largely coordinated by membrane receptors, which interpret local migratory cues and transfer them to the underlying cytoskeleton (2). Lamellipodia and filopodia are then extended from the leading edge of the cell in the direction of migration. New adhesions to the extracellular matrix are initiated at the leading edge, and these serve to pull the cell forward (3). Central to this process is, on one hand, the actomyosin contractile machinery and the microtubules (4, 5), and on the other hand the formation of membrane protrusions (6, 7). The latter process requires actin polymerization followed by the stabilization of the actin filaments in the area of membrane extensions (6, 7). Although the mechanisms regulating cell migration remain poorly understood, this cellular process has been linked to various components of the endocytosis and the actin-polymerization machinery (8, 9).
Alix/AIP1 (ALG-2 interacting protein X or 1) is an evolutionary conserved adaptor protein that was first identified as an interactor of ALG-2 (apoptosis-linked gene 2), and in this capacity it was found to cooperate with ALG-2 in promoting apoptosis (10, 11). More recent reports have implicated Alix in other basic cellular processes, which all hinge on membrane trafficking, endosomal sorting, and remodeling of the actin cytoskeleton (12–14). These multitasking properties of Alix derive from the ability of the protein to interact with a plethora of partner proteins, which are themselves components of large oligomeric complexes. By interacting with the endosomal sorting complexes required for transport, ESCRT I and III, Alix synergistically coordinates endocytosis and recycling of membrane receptors, viral budding, and cytokinesis (13–20). Relevant to this study are the findings that Alix also participates in cytoskeleton remodeling by binding with F-actin, α-actinin, cortactin, and focal adhesion kinase (14, 21). These multiple and seemingly diverse interactions of Alix are made possible by the primary structure of the protein that includes at least three distinct protein-protein interaction domains as follows: the N-terminal Bro1 domain; a middle region called the V-domain and containing a coiled-coil motif; and a C-terminal proline-rich region (PRR),5 which is a potential docking site for proteins containing Src homology 3 domains (22–24). Both the Bro1 and the PRR domains of Alix bind F-actin, whereas the N-terminal half of its V-domain interacts with α-actinin and cortactin. Given that Alix directly interacts with α-actinin and promotes its association with F-actin in fibroblast, a direct role of Alix in the F-actin bundling step of stress fiber assembly has been proposed (21).
It is noteworthy that among the proteins known to interact directly or indirectly with Alix are components of the ubiquitin pathway, including the deubiquitinating enzyme Doa4 and the E3 ubiquitin ligases Cbl, POSH (plenty of Src homology 3), and Nedd4-1 (25–28). The latter two ligases have been shown to ubiquitinate and cooperate with Alix during the process of HIV-1 release. However, the functional significance of Alix ubiquitination by these ligases is still poorly understood.
Despite the wealth of information on the proteins that interact with Alix and the physiological processes to which they contribute, the mechanisms regulating the turnover and subcellular distribution of Alix are still largely unknown. We now report that in muscle cells Alix is a substrate of the RING-type ubiquitin ligase, Ozz-E3. Ozz (also known as Neurl2 or Neutralized-like protein 2) is a member of the SOCS (suppressor of cytokine signaling) family of proteins and is the substrate-recognition module of Ozz-E3 (29). This ligase complex includes, beside Ozz, Elongin B/C (Elo B/C), Rbx1, and Cullin 5 (Cul5) (29). We have shown previously that Ozz-E3 plays an active role in myofiber differentiation and maturation, during embryogenesis and muscle regeneration, by targeting and ubiquitinating two proteins within macromolecular complexes as follows: the sarcolemma-associated pool of β-catenin, and the sarcomeric embryonic myosin heavy chain (MyHCemb) (29, 30). Ozz−/− mice develop overt sarcomeric defects, which we attribute in part to impaired turnover of cadherin-associated β-catenin and to incomplete exchange of embryonic MyHC isoforms with their neonatal and adult counterparts within the sarcomere, during myofiber differentiation (29, 30). These findings point to distinct functions of Ozz to maintain the integrity of at least two cell compartments, containing actin filaments, the sarcolemmal-associated cortical cytoskeleton and the sarcomere. Here, we demonstrate that Alix modulates actin and membrane dynamics in muscle cells and propose that Ozz intervention in this process in required at specific subcellular sites of the differentiated myotubes.
EXPERIMENTAL PROCEDURES
Ethics Statement
The animal work was first approved by the Animal Care and Use Committee at St. Jude Children's Research Hospital that governs animal research at this Institution (approval ID 388). The research was then conducted in compliance with appropriate guidelines from the Institutional Animal Care and Use Committee, the Association for Assessment and Accreditation of Laboratory Animal Care International, the United States Public Health Service, and United States Department of Agriculture animal welfare regulations. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Antibodies and Reagents
Rabbit anti-Ozz antibody was prepared as described (29). The antibody was diluted 1:500 for immunoblotting and 1:10 for immunofluorescence. Polyclonal antibodies against Alix were generated by Rockland Immunochemicals Inc. (Gilbertsville, PA). Alexa Fluor 555-conjugated polyclonal anti-Alix was labeled and purified according to the manufacturer's procedure. Commercial antibodies included the following: mouse anti-AIP1/Alix for immunoblotting (BD Transduction Laboratories); anti-Alix (clones 1A12, 3A9, and 2H12, Santa Cruz Biotechnology) for immunofluorescence; anti-GAPDH (Millipore); anti-pan-actin (Cell Signaling); anti-α-enolase (Santa Cruz Biotechnology); anti-β-catenin (BD Transduction Laboratories); anti-polyubiquitin (Thermo Scientific); anti-cortactin (Millipore); anti-FLAG (Sigma); normal rabbit IgG (Santa Cruz Biotechnology); Cy3-conjugated anti-mouse IgG and Cy3-conjugated anti-rabbit (Jackson ImmunoResearch); Alexa Fluor 488-conjugated anti-mouse 1:500 (Invitrogen); and FITC-phalloidin (Sigma). Synthetic siGENOME, ON-TARGETplus SMARTpool siRNAs targeting Alix, standard negative controls (siCONTROL NonTargeting siRNAs and ON-TARGETplus siCONTROL NonTargeting Pool), and the transfection reagent DharmaFECT3 were purchased from Dharmacon. Ozz was transiently expressed in C2C12 using the MSCV-based bicistronic retroviral vector encoding full-length Ozz and the green fluorescent protein (MSCV-Ozz-IRES-GFP) (31).
Yeast Two-hybrid Screening
Screening of an E14.5 mouse cDNA library (gift of Dr. P. McKinnon, Dept. of Genetics, St. Jude Children's Research Hospital) for putative Ozz-binding partners was performed as described previously (29, 30). Three cDNA clones encoding Alix were isolated, the smallest of which encoded a fragment from 424 to 868 amino acids. To map the minimal region mediating the interaction with Ozz, three deletion mutants of this shorter Alix fragment were generated by PCR using Pfu DNA polymerase (Stratagene) and appropriate primers (see below). These PCR fragments were digested with SalI and NotI and subcloned into the prey vector pEXP-AD502 in-frame with the GAL4 activation domain. The full-length (bp 1–845) Ozz was subcloned in-frame with the GAL4 DNA binding domain of the bait vector pDBLeu. All resulting constructs were confirmed by sequence analysis. The bait and prey constructs were co-transformed into yeast strain Mav203. Two reporter genes (lacZ and HIS3) were employed to examine the interaction (Invitrogen). For the construction of the deletion mutants of Alix to be tested in the yeast two-hybrid assay, the following primers were used: M-533–868, forward AAAAAGTCGACCGCCATCCCCTCTGCTAACCC and reverse TAATTGCGGCCGCCTACTGCTGTGGATAGTAAGACTGCTGC; M-667–868, forward AAATTGTCGACCGTAGCTAACTTGAAGGAGGGC and reverse TAATTGCGGCCGCCTACTGCTGTGGATAGTAAGACTGCTGC; M-771–868, forward AAATTGTCGACCGCTTCTGCTGCTGCTGCCCCTGC and reverse TAATTGCGGCCGCCTACTGCTGTGGATAGTAAGACTGCTGC.
Cell Culture Methods
The mouse myoblast cell line C2C12 was maintained in high glucose DMEM supplemented with 20% fetal calf serum (FCS), sodium pyruvate, l-glutamine, penicillin, and streptomycin (proliferation medium) at 37 °C in a humidified atmosphere with 5% CO2. Myotube differentiation was induced when the medium was supplemented with 2% horse serum in place of FCS (differentiation medium). Plasmid and siRNAs were transfected using DharmaFECT3 (Dharmacon) according to the manufacturer's instructions. Briefly, C2C12 cells were plated 1 day before transfection in 10-cm dishes (∼2 × 105 cells/dish). On the day of the transfection, the cells were ∼50% confluent. 250 μl of siRNA duplex (2 μm) was diluted in 250 μl of DMEM (without FCS, sodium pyruvate, l-glutamine, penicillin, and streptomycin) and mixed with 20 μl of DharmaFECT3 (Dharmacon). The mixture was added to cells grown in DMEM (without FCS, penicillin, and streptomycin). After 2–3 h, FCS was added to a final concentration of 20%. The cells were incubated for 48 h in the presence of the siRNA oligonucleotides. Then cells from each dish were re-plated into two new dishes and incubated for another 24 h in proliferation medium (for D0) or 72 h in differentiation medium (for D3), in the absence of siRNA. For the three-dimensional cultures, C2C12 cells were seeded inside a 50-mm thick collagen I gel (diluted 1:10; BD Biosciences), in inserts for 24-well plates (50,000 cells/insert; BD Biosciences). The three-dimensional culture was then submerged in proliferating or differentiating medium and incubated for 24 or 72 h. Cells were fixed with 2.5% glutaraldehyde in sodium cacodylate buffer and processed for electron microscopy (32). Primary myoblast cultures were established as described previously (29).
F-actin FACS Assay
For FACS assay, mock-treated and Alix-silenced C2C12 cells were trypsinized, and 106 cells were fixed in 4% paraformaldehyde/PBS before permeabilization with 0.03% Triton X-100 and stained with 33 nm FITC-phalloidin. Mean cellular F-actin content, as determined by phalloidin staining, was quantified using the FACScan (BD Biosciences).
In Vitro Wound Healing and Cell Attachment Assays
For wound healing assay, siRNA- and mock-treated C2C12 cells were replated, and after 24 h of incubation, cells were scratched with a micropipette tip. The culture medium was replaced with completed DMEM, and after culture for 24 h, cell migration was observed with a Leica inverted microscope. To analyze the process of adhesion, the rate of attachment to plastic of siRNA- versus mock-treated C2C12 cells was tested using the in vitro cell attachment assays. Trypsinized cells were washed once and resuspended in prewarmed DMEM containing 20% FBS at a density of 104 cells/cm2. Single cell suspensions were added to 6-well dishes. At different times (6, 10, 20, 40, and 60 min), the wells were gently washed three times under constant agitation with phosphate-buffered saline (PBS) to remove nonadherent cells; the adherent cells were then trypsinized and counted. Results were expressed as percent of attachment as compared with initial seeded cells that represented 100% of attachment.
Transmission Electron Microscopy
Cells were cultured on 24-well plate inserts (BD Biosciences) and were treated as described previously (33).
Subcellular Fractionation and Analysis of in Vivo Alix Ubiquitination
Cytoplasmic, membrane/organelle, nuclear, and cytoskeletal fractions were prepared using a ProteoExtract® subcellular proteome extraction kit (Calbiochem, Merck) according to the manufacturer's instructions. Protein concentrations were determined at A595, using BSA as standard. 20–30 μg of cytoplasmic protein or cell equivalent amounts of the other fractions were electrophoresed (100 V, 60 min) on SDS-polyacrylamide gels, and wet-blotted overnight at 90 mA. Membranes were probed with specific antibodies, followed by HRP-conjugated goat anti-rabbit or anti-mouse IgG (Amersham Biosciences). The specific secondary antibody binding was detected with a SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Each of the immunoblots included in the figures was representative of results obtained in at least three independent experiments. To test the in vivo ubiquitination of Alix, 0.5–1 mg of cytosolic or membrane proteins were adjusted to 2 mm CaCl2 (final concentrations) in a final volume of 1 ml, pre-cleared by 1 h of incubation at room temperature with 25 μl of protein A/G Plus-agarose beads (Santa Cruz Biotechnology), and then spun at 1000 × g. 2 μg of the indicated antibodies or equal amounts of control IgG were added to the supernatant, incubated overnight at 4 °C, and followed by immunoprecipitation for 2 h of incubation at room temperature with protein A/G Plus-agarose beads (Santa Cruz Biotechnology). The beads were washed five times with Hypotonic buffer supplemented with 0.1% Nonidet P-40, and the bound proteins detached and run on SDS-polyacrylamide gels under denaturing conditions. Membranes were blocked and probed with specific antibodies and then washed before incubation with species-appropriate secondary antibodies for 1 h at room temperature. Where indicated, blots were analyzed using an Odyssey Infrared Imaging System (LI-COR), and relevant signal intensity was determined using LI-COR imaging software. Each of the immunoblots included in the figures was representative of results obtained in at least three independent experiments.
Alix/Ozz Coimmunoprecipitation
The Ozz-Alix complex was detected in post-nuclear fractions of muscle cells by immunoprecipitation with either the anti-Ozz or the anti-Alix antibodies. Cultured C2C12 or primary myoblasts (D0) and myotubes (D3) were lysed with Hypotonic buffer, incubated at 4 °C for 30 min, and then disrupted by Dounce homogenization (40 strokes). Cellular debris and nuclei were removed by 10 min of centrifugation at 800 × g, and 0.5–1 mg of post-nuclear proteins were adjusted to 2 mm CaCl2 (final concentrations) in a final volume of 1–1.3 ml and pre-cleared by 1 h of incubation at room temperature with 25 μl of protein A/G Plus-agarose beads (Santa Cruz Biotechnology) and then spun at 1000 × g. Anti-Alix (Santa Cruz Biotechnology) or polyclonal anti-Ozz antibodies or the appropriate antibodies used as negative controls were added to the supernatant and incubated overnight at 4 °C, followed by immunoprecipitation with protein A/G Plus-agarose beads for 2 h of incubation at room temperature. The beads were washed five times with Hypotonic buffer supplemented with 0.3% Nonidet P-40, and the bound proteins were detached by boiling the beads in the presence of sample buffer. 50 μg of the post-nuclear fraction and the whole immunoprecipitates were run on SDS-polyacrylamide gels under denaturing conditions.
In Vitro Ubiquitination of Alix
In vitro ubiquitination was performed as described previously (29, 30). Briefly, the entire Ozz-E3 complex was reconstituted in vitro by coinfecting Tni-Pro insect cells with baculoviruses encoding His-tagged Ozz/Elongin B/Elongin C or His-tagged Rbx1/Cullin5. The two subcomplexes were then purified by consecutive nickel-agarose chromatography, ion exchange chromatography, and size-exclusion chromatography. 1.0 μg of the purified Ozz-E3 ubiquitin ligase, 150 ng of purified recombinant E1 (Calbiochem, Merck Biosciences), 200 ng of UbcH5b (a gift of Dr. B. Schulman, Dept. of Structural Biology, St. Jude Children's Research Hospital), and 7.5 μg of ubiquitin (Calbiochem, Merck) were incubated with 1 μg of a bacterially expressed GST-Alix in a final volume of 30 μl of ubiquitination buffer. Incubation was performed at 30 °C for 1 h in the presence of 1.5 mm ATP (GE Healthcare). To analyze the ubiquitinated products, the ubiqitination reaction mixtures were diluted in 500 μl of RIPA buffer, immunoprecipitated with anti-Alix (Santa Cruz Biotechnology), resolved on a 7.5% SDS gels, and immunoblotted with anti-ubiquitin.
Immunofluorescence and Imaging
Muscle cells were fixed in 3% paraformaldehyde, permeabilized with 0.1% saponin in 1× PBS, and immunostained with the indicated primary antibodies. Cy3 anti-rabbit IgG (The Jackson Laboratory) and Alexa Fluor 488 anti-mouse IgG (Invitrogen) were used as secondary antibodies. Actin was detected by staining with FITC-phalloidin (1 mm) for 40 min.
Images were acquired on a Nikon C1si confocal microscope, with a Plan Apo 40×, NA 1.3 and/or Plan Apo 60×, NA 1.45 objective (Melville, NY). Computational analyses of confocal images were performed with the NES-Elements AR 3.1 (Melville, NY), as described previously (30).
Statistical Analysis
Data were expressed as mean ± S.D. and were evaluated using Student's t test. Mean differences were considered statistically significant when p values were less than 0.005 (***) or 0.05 (*).
RESULTS
Ozz Interacts with Alix in Skeletal Muscle Cells
By yeast two-hybrid screening of an E14.5 mouse cDNA library, we identified Alix as a putative Ozz-interacting partner. Either full-length Ozz or the N-terminal half of the protein (residues 1–229), including the neutralized homologous region 1 (NHR1) domain (residues 14–104) and most of the NHR2 domain (residues 208–242), was used as bait. Both screens yielded three different clones with 93–96% homology to Alix that shared a region starting at amino acid 425 until the C terminus (Fig. 1A). The shortest clone encoded a fragment that encompassed part of the V domain and the PRR (from amino acid 424 until the end of the Alix protein). To map the minimal region mediating the interaction with Ozz, deletion mutants of Alix were generated and tested in a yeast two-hybrid assay, using two different growth selections. Besides full-length Alix, three of the four deletion mutants, M(424–868), M(533–868), and M(677–868), retained sustained interaction with Ozz. The lack of interaction between Ozz and the Alix fragment M(771–868) allowed us to conclude that a minimal interacting region between amino acids 677 and 771 of Alix is sufficient to mediate interaction with Ozz. This Alix region contains part of the V-domain arm2(677–702), the full V-PRR hinge region (703–716), and the N terminus of the PRR (717–771).
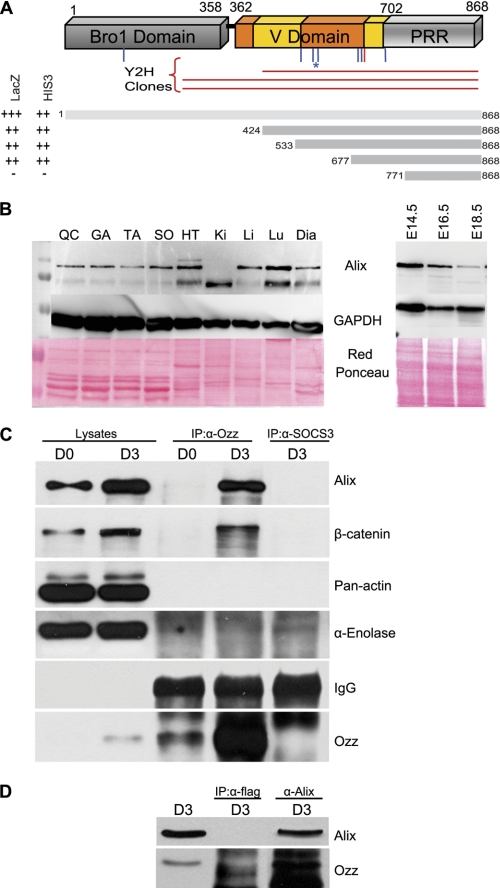
FIGURE 1.
Endogenous Ozz and Alix interact in muscle cells. A, yeast two-hybrid screening of an E14.5 mouse cDNA library identified three clones encoding Alix (Y2H clones) that shared a region starting at amino acid 425 until the C terminus. Highlighted are the eight putative ubiquitination sites predicted with medium (blue) and high (red) confidence; asterisk indicates the most conserved Lys-574 residue. The indicated Alix fragment was used in a yeast two-hybrid assay, using two different reporter genes (HIS3 and lacZ). The minimal Ozz-interacting region of Alix includes amino acids 677–771. B, expression of Alix in adult tissues (QC, quadriceps; GA, gastrocnemius; TA, tibialis anterior; SO, soleus; HT, heart; Ki, kidney; Li, liver; Lu, lung; Dia, diaphragm) and embryos at different embryonic stages (E14.5, E16.5, and E18.5). C, post-nuclear fractions of proliferating (D0) and differentiated (D3) C2C12 were immunoprecipitated (IP) with anti-Ozz and with anti-SOCS3, used as negative control. Immunoblotting with anti-Alix showed that endogenous Alix and Ozz interact in differentiated C2C12. D, post-nuclear fractions of differentiated primary myotubes (D3) were immunoprecipitated with anti-Alix, or with an isotype matching control antibody (anti-FLAG), used as a negative control. Immunoblotting with anti-Ozz showed a pattern of Ozz and Alix interaction identical to C2C12.
To gain insights into the significance of Ozz-Alix interaction in vivo, we first assessed the expression pattern of Alix in different muscle lysates, compared with other mouse tissues, and in embryos (Fig. 1B). We found that a predominant Alix band of ∼100 kDa was present in the adult skeletal muscles as well as in the diaphragm and heart muscle. In the latter tissues and in the systemic organs an additional band of lower molecular weight was also detected. The ratio between these two bands varied in different tissue samples with kidney containing only the smallest protein (Fig. 1B, left panels). In embryos, the 100-kDa Alix form was expressed at high levels during the early stages of development (E14.5), and its expression decreased slightly later in embryogenesis (Fig. 1B).
To verify the interaction between endogenous Alix and Ozz, post-nuclear fractions of proliferating (D0) and differentiated (D3) C2C12 muscle cells were immunoprecipitated with a polyclonal anti-Ozz antibody (Fig. 1C). Immunoblotting of the immunoprecipitates with anti-Alix antibody confirmed the interaction between the two proteins in differentiated C2C12 (Fig. 1C); no such interaction was seen with anti-SOCS3, used as a negative control. Similar results were obtained in post-nuclear fractions from primary myotubes immunoprecipitated with anti-Alix and probed on immunoblots with anti-Ozz (Fig. 1D). In this case the isotype-matching antibody anti-FLAG (IgG1) was used as negative control. Coimmunoprecipitation of the two proteins was more efficiently driven by anti-Alix than anti-Ozz antibodies. This might depend on differences in the affinity of the two antibodies to their native proteins or on the stoichiometry of the Alix-Ozz complex in the post-nuclear fraction. We then tested the cellular localization of the two endogenous proteins by indirect immunofluorescent labeling of primary myoblasts (D0) and myotubes (D3), followed by confocal microscopy and computational analyses of the two fluorescent signals (Fig. 2). In line with its pattern of expression during myogenesis (29), Ozz was undetectable by immunofluorescence in the proliferating (D0) primary myoblasts, although Alix was expressed in good amounts in the same cells (Fig. 2A, upper panels). In contrast, Alix and Ozz colocalized in discrete puncta throughout the cytosol of myotubes at D3 of differentiation (Fig. 2A, lower panel, merged image). Both the Pearson's correlation coefficient and the cytofluorograms confirmed a strong colocalization of the Alix and Ozz in D3 fibers (Fig. 2B, left and right panels, respectively); a similar analysis performed on the Ozz null D3 myotubes served as negative control. Together these results indicate that Alix is a bona fide interacting partner of Ozz and that interaction between the two native proteins occurs in the differentiated muscle fibers.
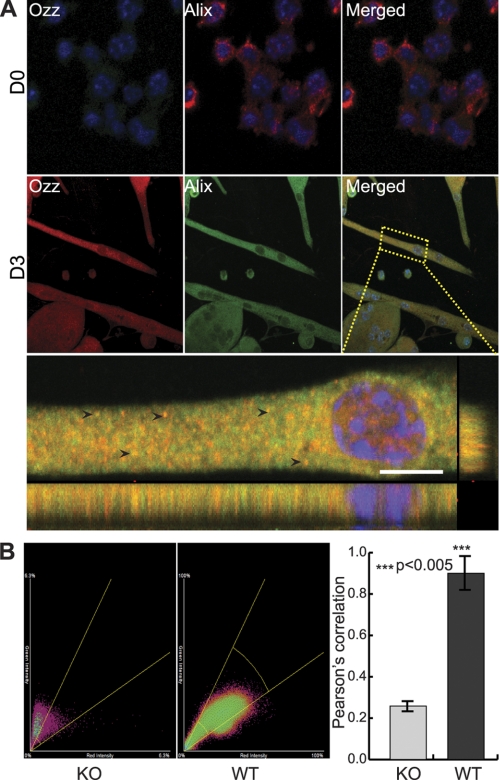
FIGURE 2.
Localization of Alix and Ozz in primary myotubes. A, representative confocal microscopy images showing Alix and Ozz localization in proliferating (D0) and differentiated (D3) primary myoblasts. In myotubes, Ozz and Alix displayed a punctated distribution pattern and were colocalized in discrete puncta (arrowheads) and in the proximity of the nucleus. Scale bar, 10 μm. B, computational analyses of several confocal images from Ozz wild-type or knock-out D3 myotubes (n = 5–6) confirmed the colocalization of the two fluorescent signals. The right panel shows the Pearson's correlations. The colocalization between Alix (green fluorescence) and Ozz (red fluorescence) is also shown as scatter plots (cytofluorograms) in the center and left panels. Ozz−/− cells were included in this analysis as negative control. Standard deviations (error bars) and p values (***, p < 0.005) confirmed that the analysis was statistically significant.
Ozz Mediates Ubiquitination of Alix
Alix and Ozz interaction implied that Alix can be a substrate of the Ozz-E3 ligase. In this regard, it is noteworthy that the minimal region of Alix needed for its interaction with Ozz encompasses and is in close proximity with seven lysines that are predicted to be sites of ubiquitination with medium (six) and high confidence (one) (72% accuracy claimed), by the random forest predictor of ubiquitination sites, UbPred (Fig. 1A) (34). Two of these lysines (Lys-574 and Lys-699) are completely conserved among evolutionary distant species, which make these residues a likely target of ubiquitination.
Alix ubiquitination analysis was first performed in C2C12 transfected with an MSCV-based bicistronic retroviral vector encoding full-length Ozz and the green fluorescent protein (GFP) (MSCV-Ozz) or a similar vector carrying only GFP (MSCV-GFP). Subcellular fractions of transfected cells at D3 of differentiation were immunoprecipitated, in the presence of calcium, with two commercial monoclonal anti-Alix antibodies (clones 3A9 and 2H12) or an isotype-matching antibody (anti-FLAG), and probed on immunoblots with anti-ubiquitin, anti-Ozz, or anti-Alix antibodies. We found that Alix was ubiquitinated in the cytosolic fraction of MSCV-GFP mock-transfected cells (Fig. 3A, 2nd and 4th lanes). However, overexpression of Ozz induced a significant increase of Alix ubiquitination levels (Fig. 3A, 3rd and 5th lanes), suggesting that Ozz mediates ubiquitination of endogenous Alix in myotubes. Both the 3A9 and 2H12 monoclonal antibodies did not efficiently precipitate Alix from the membrane fractions, likely because of the detergent that was used in these preparations (data not shown). However, we chose to test the 2H12 monoclonal antibody because it was shown to recognize an epitope within the hydrophobic pocket of Alix that is exposed only in its “open” conformation (35). The use of this antibody would allow us to discriminate between different forms of Alix and to assess whether interaction with Ozz could modify the conformation of Alix. In the presence of calcium, the 2H12 clone immunoprecipitated Alix from the cytosol of mock-transfected cells, although with lower efficiency than clone 3A9 (Fig. 3A, 4th versus 2nd lane). In contrast, overexpression of Ozz resulted in a significant increase in the amount of native Alix immunoprecipitated by the 2H12 clone, suggesting that Ozz promotes the open conformation of Alix (Fig. 3A, 5th versus 4th lane).
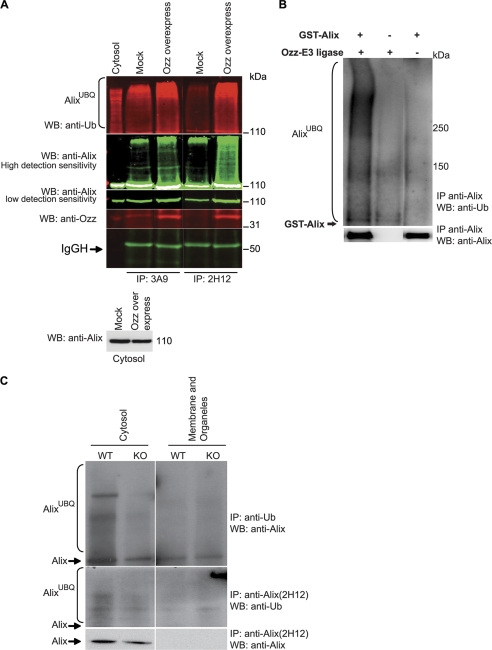
FIGURE 3.
Ozz-E3 complex ubiquitinates Alix in vitro and in vivo. A, the ubiquitination of Alix was assessed in the cytosolic fractions of C2C12 transfected with an empty vector (Mock) or a vector containing Ozz cDNA (Ozz overexpressing), at D3 of differentiation. Proteins were immunoprecipitated (IP) with two anti-Alix antibodies (clone 3A9 and 2H12) and immunoblotted with the indicated antibodies. The level of Alix in the cytosolic fraction is also reported. Band intensities were acquired using an Odyssey Infrared Imaging System (LI-COR), at low or high detection sensitivity. WB, Western blot. B, in vitro ubiquitination of GST-tagged Alix was performed in presence or absence of the reconstituted Ozz-E3 complex and GST-Alix. Ubiquitinated Alix was immunoprecipitated with anti-Alix and detected on immunoblot probed with anti-ubiquitin. C, Alix level and the extent of Alix ubiquitination was compared in subcellular fractions (cytosol and membrane/organelle) of Ozz+/+ (WT) and Ozz−/− (KO) primary myotubes. Anti-ubiquitin immunoprecipitated more ubiquitinated Alix in the cytosolic fraction of the wild-type myotubes at D3 of differentiation than in the Ozz null myotubes. Similarly, the amount of native Alix immunoprecipitated with the anti-Alix clone 2H12 was significantly more in the cytosolic fraction of the wild-type than in the Ozz null myotubes.
To further ascertain whether Alix was a target of ubiquitination by the Ozz-E3, we performed an in vitro ubiquitination assay, using a reconstituted Ozz-E3 complex and a GST-tagged recombinant Alix (119 kDa) (29, 30). The ubiquitinated products were immunoprecipitated with anti-Alix, and the immunoblots were probed with anti-ubiquitin antibody (Fig. 3B). Recombinant Alix was ubiquitinated only in the presence of the Ozz-E3 (Fig. 3B, compare 1st and 3rd lanes). Three other GST fusion proteins, used as internal controls, were not ubiquitinated in this assay (data not shown), which validated the specificity of the reaction. In an attempt to identify the lysine residue(s) ubiquitinated by the Ozz-E3 complex, we generated two Alix point mutants (K574R and K699R), targeting the two most conserved ubiquitination sites among Alix homologs. These mutant proteins were tested in the ubiquitination assay. However, their rate of ubiquitination by the Ozz-E3 complex differed only marginally to that of wild-type Alix, indicating that additional lysines might be targets of the ligase (data not shown).
We next tested if Alix was ubiquitinated in vivo by Ozz and if this modification occurred in a specific subcellular pool of the protein in the differentiated myotubes. The extent of Alix ubiquitination was assessed in subcellular fractions of Ozz+/+ myotubes, and we compared it with that in Ozz−/− cells (D3). Using 2H12 antibody, we immunoprecipitated a slightly higher amount of ubiquitinated Alix from the cytosolic fraction of wild-type myotubes than from the same fraction of Ozz null myotubes (Fig. 3C). This difference was more evident when the ubiquitinated Alix was immunoprecipitated with anti-ubiquitin antibody. Furthermore, in Ozz null myotubes there was less Alix immunoprecipitable with the 2H12 clone (Fig. 3C). In line with the Ozz overexpression experiments, these results imply that lack of Ozz leads to a decreased amount of Alix in its open conformation, which can be immunoprecipitated by the 2H12 antibody. Ectopic expression of Ozz, or its loss of function, could therefore influence the balance between the open versus the closed Alix conformation. These findings suggest that Ozz may exert a dual function toward Alix, modulating its conformation and ubiquitination status.
Knockdown of Alix Alters the F-actin Levels and Distribution in Muscle Cells
To elucidate the biological significance of the Ozz-Alix interaction in skeletal muscle, we first analyzed the expression pattern of Alix and the effects of its loss of function in C2C12 cells. Subcellular fractions prepared from C2C12 myoblast (D0) and myotubes (D3) showed that Alix was expressed at high levels in both proliferating (D0) and differentiated (D3) cells (Fig. 4A). Alix was mainly localized in the cytoplasmic fraction and in the fraction comprising the sarcolemma and the intracellular membranes (Fig. 4A). Silencing of Alix in C2C12 using two independent siRNA pools (see “Experimental Procedures”) was specific and effective, as we routinely obtained 70–90% knockdown of Alix expression, compared with mock-transfected cells (Fig. 4A). The expression patterns of the early markers of myoblast differentiation (i.e. myogenin and MyHC) did not change in the Alix-silenced C2C12 compared with mock-treated cells nor did the viability, morphology, and migration of cells treated with two different nontargeting siRNA pools (data not shown). Immunofluorescence microscopy showed that mock-transfected C2C12 myoblasts displayed a fibroblastoid migratory shape with numerous stress fibers and protrusions extending from one cell to the adjacent cell (Fig. 4B, left panel). In contrast, the knockdown of Alix resulted in a substantial number of cells acquiring a less motile phenotype and a different morphology, with a clearly reduced number of protrusions and less organized stress fibers (Fig. 4B, right panel). These Alix-silenced myoblasts exhibited a redistribution of F-actin at the cell periphery, in areas juxtaposed to the submembranous cytoplasm.
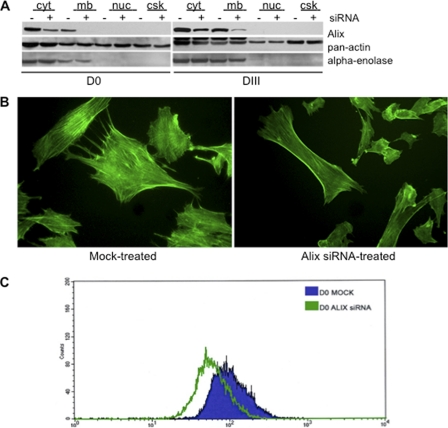
FIGURE 4.
Effect of Alix loss of function on F-actin organization in proliferating and differentiated C2C12. A, treatment of C2C12 myoblasts with Alix-specific double-stranded siRNA pools led to a significant reduction (≈75%) in Alix expression compared with mock-transfected cells, in different subcellular fractions of both proliferating (D0) and differentiated C2C12 (D3). Cellular compartments are as follows: cytosol (cyt); membrane/organelle (3); nuclei (nuc); cytoskeleton (csk). B, C2C12 cells treated with the Alix siRNA pools exhibited an overall reduction of F-actin, in addition to an aberrant localization of F-actin at the cell periphery, most prominent in the submembranous cytoplasmic areas (FITC-phalloidin stained cells). C, to quantify the effects of Alix-depletion on F-actin, FITC-phalloidin stained cells were analyzed by FACS.
To quantify the effects of Alix on F-actin assembly, we compared the mean cellular F-actin content of C2C12 myoblast silenced for Alix with that of mock-transfected cells, using FITC-phalloidin staining followed by FACS analyses. The mean value of phalloidin intensity measured in individual cells treated with Alix siRNA was quantified and compared with that of the mock-treated population. Knockdown of Alix expression induced a significant decrease in the mean cellular content of F-actin, a finding that supports a direct role of Alix in regulating actin dynamics also in muscle cells (Fig. 4C).
Effect of Alix Loss of Function on Three-dimensional Cultures of Muscle Cells
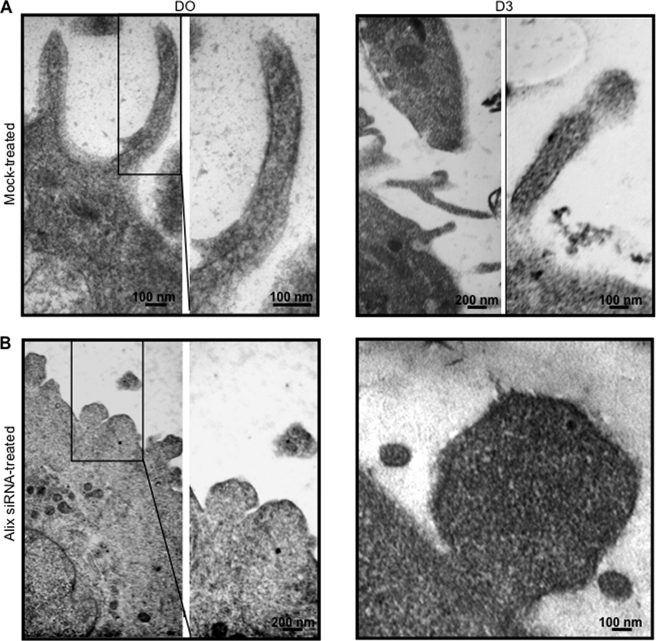
Gross morphological and intracellular changes in muscle cells during differentiation can be readily identified by ultrastructural analysis of three-dimensional cultures, a method that could give insights into skeletal muscle morphogenesis. One particular morphological change that occurs during the transition from monolayer culture to the three-dimensional environment is the appearance of cytoplasmic projections (36). Thus, to investigate the role of Alix in maintaining cell morphology and in the formation of cell projections, Alix-depleted and mock-treated cells were cultured three-dimensionally in a collagen I gel matrix under both proliferating and differentiating conditions. Mock-transfected C2C12 migrated inside the collagen gel matrix within 24–48 h after plating and created a small round mass. In contrast, Alix-depleted cells moved in flat motion and migrated less inside the gel matrix (data not shown). Transmission electron microscopy analysis of both siRNA-treated and mock-treated three-dimensional cultures demonstrated that the latter cells extended their sarcolemma into long filopodium-like projections, whereas Alix-depleted cells had an altered phenotype with aborted protrusions, in both proliferating and differentiating culture conditions (Fig. 5). These results suggest that Alix plays a role in the formation of cellular projections in both proliferating and differentiating muscle cells, which in turn might affect myoblast migration.
FIGURE 5.
Alix depletion affects the formation of filopodia-like structures in three-dimensional culture of C2C12 muscle cells. Transmission electron microscopy of three-dimensional cultures of C2C12 proliferating (D0) and differentiating (D3) cells silenced for Alix showed an altered phenotype, compared with mock-treated cells.
Cell Migration and Rate of Attachment to Substrate Are Altered in Alix-silenced C2C12
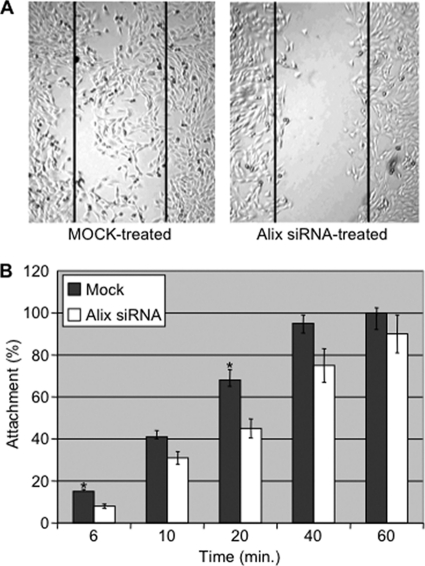
To test this further, the migration of C2C12 silenced for Alix was monitored using an in vitro wound healing assay. A cell monolayer was scraped longitudinally, and the rate of migration of the cells into the scraped surface (wound healing) was evaluated. After 24 h, only 20% of the Alix-silenced C2C12 migrated into the “wound,” whereas mock-transfected cells had virtually closed the gap, with a migratory activity of about 90% (Fig. 6A). Because cell migration also requires the formation of new focal adhesions to the substrates, we tested whether Alix is also required during the adhesion step. For this purpose, the rate of attachment to plastic of siRNA- versus mock-treated C2C12 cells was tested using an in vitro cell attachment assay (Fig. 6B). At 6 min after plating, only 50% of Alix-depleted cells adhered to the plastic surface compared with control cells (p < 0.05). Attachment of Alix-silenced cells was statistically less also at 20 min (p < 0.05) compared with control cells, when the cell-substrate contact stage is concluded and the attachment step is in the logarithmic phase (37). Their reduced ability to adhere was still detected at 60 min when 100% of mock-transfected cells were attached. Thus, in muscle cells one of the functions of Alix could be to regulate cell migration by promoting the formation of protrusions and adhesions to substrate.
FIGURE 6.
Alix negatively modulates muscle cell migration and adhesion to substrate. A, wound healing assays showed that Alix-silenced C2C12 cells have an altered pattern of cell migration, compared with mock-treated cells. B, silencing of Alix expression reduced the capacity of C2C12 cells to adhere to plastic. The percentage of C2C12 cells adhering to plastic was evaluated at each time point. Values are expressed as mean of three independent experiments, and error bars indicate standard deviations. Mean differences were considered statistically significant when p values were <0.05 (*).
Ozz Depletion Affects the Amount and the Subcellular Distribution of Alix in Differentiated Muscle Cells
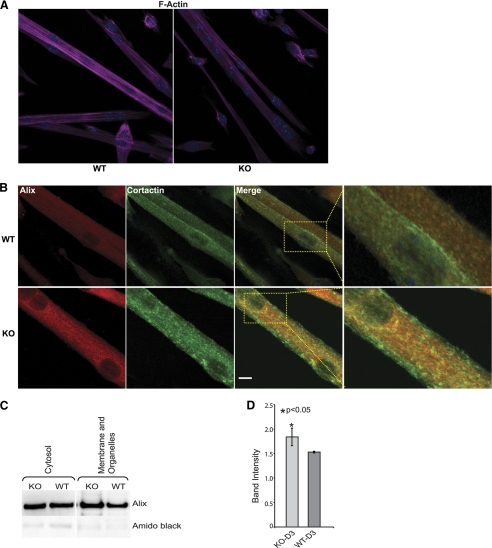
Because Alix silencing affected the levels and distribution of F-actin in C2C12, we asked whether a similar phenotype could be reproduced by the lack of Ozz in differentiated primary myotubes. Indeed, this was confirmed in Ozz null D3 myotubes that showed a decreased amount of phalloidin staining compared with the wild-type myotubes (Fig. 7A). We then determined the effect of the loss of function of Ozz on the expression of Alix and its subcellular distribution in Ozz−/− and Ozz+/+ myotubes. We focused on the pattern of Alix expression in the actin-containing structures. Various actin-associated proteins specify different actin-based compartments and determine their properties and regulation. Given that Alix interacts with cortactin, an actin-binding protein that couples membrane dynamics to the cortical actin cytoskeleton (38–40), we checked the distribution of Alix and cortactin both in wild-type and Ozz null muscle cells. Z-stack analyses of confocal microscopy images showed that Alix and cortactin colocalized in discrete regions of D3 wild-type primary myotubes (Fig. 7B, merged images of WT myotubes). In contrast, Ozz−/− myotubes showed increased amount of Alix throughout their cytosol, with a fraction of the protein accumulating in cortactin positive puncta (Fig. 7B, merged images of KO myotubes). This immunofluorescence staining of Alix was paralleled by an increased amount of the protein in both the cytosolic and membrane/organelle subcellular fractions of Ozz−/− myotubes, as demonstrated by immunoblot analysis of Alix in these fractions (Fig. 7C). These results were confirmed by quantitative analysis of post-nuclear lysates of Ozz+/+ and Ozz−/− myotubes from three independent measurements (Fig. 7D). Together, these data put forward a role of Ozz in modulating the proper local concentration and distribution of Alix.
FIGURE 7.
Subcellular distribution of actin, Alix, and cortactin in wild-type and Ozz null primary myoblasts. A, Ozz null myotubes exhibited an overall reduction of F-actin. B, representative confocal microscopy images showing that Alix and cortactin colocalized in discrete regions in WT myotubes. Loss of Ozz resulted in accumulation of Alix in large puncta positive for cortactin (see inset). Scale bar, 10 μm. C, Alix expression levels were compared on immunoblots containing the cytosolic and membrane/organelle fractions from the Ozz null and wild-type D3 myotubes. D, quantitative analysis of post-nuclear fractions from wild-type and Ozz-null myotubes. Alix expression was significantly higher in differentiated Ozz−/− myotubes compared with Ozz+/+ myotubes. Data are expressed as mean ± S.D. of three independent experiments. Mean differences were considered statistically significant when p values were less than 0.05 (*).
DISCUSSION
Fine-tuning the levels of specific subcellular pools of both structural and regulatory proteins in muscle cells is fundamental to their growth, differentiation, and regeneration. During these processes, ubiquitin modification or ubiquitin-mediated degradation of target substrates plays a central role. We have previously identified a RING-type ubiquitin ligase complex, Ozz-E3, whose substrate recognition component, Ozz, is expressed exclusively in striated muscle and is up-regulated during muscle growth and regeneration (29, 30). We found that Ozz-E3 participates in the processes of myofiber differentiation and sarcomere stability by controlling the levels of cadherin-associated β-catenin (29), and sarcomeric MyHCemb (30). Thus, Ozz-E3 recognizes and ubiquitinates only selected pools of these two substrates, both of which directly or indirectly interact with polymerized actin, either in the sub-cortical region (β-catenin) or in the sarcomere (MyHCemb). We now have identified a new substrate of Ozz in muscle cells, Alix, whose function in muscle has not been investigated before. We found that Alix is expressed throughout the myogenic differentiation and that a specific pool of Alix colocalizes in discrete subcellular regions positive for Ozz.
Alix is an evolutionary conserved, ubiquitously expressed adaptor protein that has been implicated in many cellular processes, including membrane and cytoskeleton remodeling (12). In fact, Alix has been shown to associate with F-actin in epithelial cells and fibroblasts, where it participates in the organization of actin fibers (14). In addition, Alix can bind to acidic phospholipids via its Bro1 domain, which structurally resembles a “boomerang” with the convex face containing a positively charged region (41). This convex surface might function as membrane bending domain to generate or scaffold a negative curvature within the membrane (42). A potential interplay between F-actin and membrane bending domains is thought to occur during the processes of filopodia formation (negative membrane curvature) and endocytosis (positive membrane curvature) (42). By silencing the expression of Alix in C2C12 cells, we were able to demonstrate that loss of Alix does not interfere with the early myogenic program but rather with structural events, such as F-actin levels and distribution, and proper formation of membrane protrusions. The latter process was particularly evident in three-dimensional cultures, which showed aberrant formation of podia upon loss of Alix both in myoblast and myotubes. Thus, Alix could control both cytoskeleton and membrane dynamics in muscle cells by interacting with the F-actin cytoskeleton and at the same time by acting as a membrane bending component. Proper podia formation is also important for cell migration, a fundamental process during skeletal muscle fiber differentiation and regeneration. Because Alix silencing also perturbs muscle cell migration, it is possible that normally Alix takes part in this process by regulating the formation of filopodia and the actin cytoskeletal dynamics.
The function of ubiquitin tagging and degradation, as it pertains to the regulation of assembled and little accessible structural components (e.g. membrane and/or cytoskeletal proteins) in muscle, is still poorly understood. This is further complicated by the existence of multiple pools of protein substrates that interact with a plethora of partners at different subcellular sites. In this respect, Ozz and the ligase complex it specifies represent the means for the controlled regulation of these substrate pools, of which Alix is one constituent.
It has been recently reported that in overexpressing human cell lines, Alix interacts with and is ubiquitinated by the one-chain ligases POSH and Nedd4-1 during the process of virus budding (25, 26). It is noteworthy that the interaction of Alix with these two ligases was detected only in the presence of detergents. In contrast, Ozz is able to recognize Alix in its closed conformation without the aid of detergents, probably functioning as a crowbar to convert it into its open conformation. Ozz uses an analogous mode of action toward its other two substrates (β-catenin and MyHCemb) that are bound by Ozz in their assembled state (29, 30). In the case of MyHCemb, Ozz targets the rod portion of this protein, which forms the core of sarcomeric thick filaments and is not easily accessed by other proteins. Similarly, Ozz recognizes a difficult to access pool of β-catenin located at the sarcolemma, where this protein is assembled in a multiprotein complex, including cadherin and cortical actin.
We have postulated that Ozz binding to MyHCemb does not immediately initiate the ubiquitination and proteolytic degradation of this substrate, but rather renders it accessible for further ubiquitination by the Ozz-E3. Based on our current data, we now propose that Ozz may exert a similar function on Alix by binding to a hydrophobic, low accessible pocket of the protein, thereby changing its conformation and making it accessible for further ubiquitination. In addition, the Ozz-mediated conformational change of Alix appears to affect also the subcellular localization of the protein. In fact, we found that Alix accumulates in a cortactin-positive subcompartment in the absence of Ozz. Our findings are in agreement with the concept suggested by Sundquist and co-workers (22) that opening of the “hinge region” of Alix is driven by post-translational modifications and/or ligand binding. Opening of the V domain would expose different binding sites and consequently modify the function and subcellular localization of Alix (22). This proposed model was recently corroborated by the same authors, who demonstrated that Alix activation/localization requires dissociation of the auto-inhibitory PRR and opening of the V domain arms (43). Therefore, it appears that all the substrates of Ozz identified to date, including Alix, are targeted by this protein in their assembled state and again might require Ozz binding to change conformation and/or to dissociate from their protein partners before their ubiquitination. In this capacity, Ozz may represent a novel regulator of Alix in muscle cells.
We have proposed earlier that the defects in myofibrillogenesis and sarcomere stability observed in the Ozz KO muscle are due to relatively small changes in the concentration of membrane-β-catenin and MyHCemb (29, 30). Considering the involvement of Alix in modulating actin dynamics in muscle cells, we have focused on the effects of Ozz depletion in the organization of the cortical actin cytoskeleton. We found that the abnormal F-actin levels and organization observed in Alix-silenced C2C12 were recapitulated in the primary myotubes lacking Ozz expression. Given the similarity in the way Ozz recognizes and regulates its substrates at specific subcellular sites, we can infer that variations in the local concentration of Alix may also contribute to the overall muscle defects characteristic of the Ozz KO mice.
In conclusion, Alix plays a primary role in modulating actin dynamics in muscle cells, and the role of Ozz in this process is required mainly in the differentiating myotubes when the control of the concentration of Alix at specific sites of the elongating myofiber might be crucial.
Acknowledgments
We thank Gerard Grosveld and Agata Giallongo for helpful suggestions and discussions; A. John Harris for insights in skeletal muscle development; Vito Marcenò and Mario Melis for performing the electron microscopy and FACS analyses; Giovanni Perconti for skilled help with tissue culture; Tommaso Nastasi and James Toy for contributions during the initial phase of this project; and Elida Gomero for invaluable help in maintaining the mouse colonies.
This work was supported, in whole or in part, by National Institutes of Health Grants AR049867 and CA021765 from USPHS and NCI (to A. d'A.). This work was also supported by the Assisi Foundation of Memphis and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital (to A. d'A.).
- PRR
- proline-rich region
- MSCV
- murine stem cell virus.
REFERENCES
- 1. Ordahl C. P., Le Douarin N. M. (1992) Two myogenic lineages within the developing somite. Development 114, 339–353 [DOI] [PubMed] [Google Scholar]
- 2. Orth J. D., Krueger E. W., Weller S. G., McNiven M. A. (2006) A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 66, 3603–3610 [DOI] [PubMed] [Google Scholar]
- 3. Beningo K. A., Dembo M., Kaverina I., Small J. V., Wang Y. L. (2001) Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153, 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunn G. A. (1980) in Cell Adhesion and Motility, BSCB Symposium 3 (Curtis A. S. G., Pitts J. D., eds) pp. 409–423, Cambridge University Press, Cambridge [Google Scholar]
- 5. Nobes C. D., Hall A. (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Condeelis J. (1993) Life at the leading edge. The formation of cell protrusions. Annu. Rev. Cell Biol. 9, 411–444 [DOI] [PubMed] [Google Scholar]
- 7. Welch M. D., Mallavarapu A., Rosenblatt J., Mitchison T. J. (1997) Actin dynamics in vivo. Curr. Opin. Cell Biol. 9, 54–61 [DOI] [PubMed] [Google Scholar]
- 8. Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P. P. (2008) Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 134, 135–147 [DOI] [PubMed] [Google Scholar]
- 9. Somogyi K., Rørth P. (2004) Cortactin modulates cell migration and ring canal morphogenesis during Drosophila oogenesis. Mech. Dev. 121, 57–64 [DOI] [PubMed] [Google Scholar]
- 10. Vito P., Pellegrini L., Guiet C., D'Adamio L. (1999) Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 11. Missotten M., Nichols A., Rieger K., Sadoul R. (1999) Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 6, 124–129 [DOI] [PubMed] [Google Scholar]
- 12. Odorizzi G. (2006) The multiple personalities of Alix. J. Cell Sci. 119, 3025–3032 [DOI] [PubMed] [Google Scholar]
- 13. Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534 [DOI] [PubMed] [Google Scholar]
- 14. Cabezas A., Bache K. G., Brech A., Stenmark H. (2005) Alix regulates cortical actin and the spatial distribution of endosomes. J. Cell Sci. 118, 2625–2635 [DOI] [PubMed] [Google Scholar]
- 15. Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P. D. (2003) Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strack B., Calistri A., Craig S., Popova E., Göttlinger H. G. (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 [DOI] [PubMed] [Google Scholar]
- 17. Carlton J. G., Martin-Serrano J. (2007) Parallels between cytokinesis and retroviral budding. A role for the ESCRT machinery. Science 316, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 18. Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. (2007) Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katzmann D. J., Odorizzi G., Emr S. D. (2002) Receptor down-regulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3, 893–905 [DOI] [PubMed] [Google Scholar]
- 20. Raiborg C., Rusten T. E., Stenmark H. (2003) Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446–455 [DOI] [PubMed] [Google Scholar]
- 21. Pan S., Wang R., Zhou X., He G., Koomen J., Kobayashi R., Sun L., Corvera J., Gallick G. E., Kuang J. (2006) Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J. Biol. Chem. 281, 34640–34650 [DOI] [PubMed] [Google Scholar]
- 22. Fisher R. D., Chung H. Y., Zhai Q., Robinson H., Sundquist W. I., Hill C. P. (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128, 841–852 [DOI] [PubMed] [Google Scholar]
- 23. Pires R., Hartlieb B., Signor L., Schoehn G., Lata S., Roessle M., Moriscot C., Popov S., Hinz A., Jamin M., Boyer V., Sadoul R., Forest E., Svergun D. I., Göttlinger H. G., Weissenhorn W. (2009) A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17, 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmidt M. H., Hoeller D., Yu J., Furnari F. B., Cavenee W. K., Dikic I., Bögler O. (2004) Alix/AIP1 antagonizes epidermal growth factor receptor down-regulation by the Cbl-SETA·CIN85 complex. Mol. Cell. Biol. 24, 8981–8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Votteler J., Iavnilovitch E., Fingrut O., Shemesh V., Taglicht D., Erez O., Sörgel S., Walther T., Bannert N., Schubert U., Reiss Y. (2009) Exploring the functional interaction between POSH and ALIX and the relevance to HIV-1 release. BMC Biochem. 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sette P., Jadwin J. A., Dussupt V., Bello N. F., Bouamr F. (2010) The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 84, 8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikko E., André B. (2007) Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome. Application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell 6, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richter C., West M., Odorizzi G. (2007) Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26, 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nastasi T., Bongiovanni A., Campos Y., Mann L., Toy J. N., Bostrom J., Rottier R., Hahn C., Conaway J. W., Harris A. J., D'Azzo A. (2004) Ozz-E3, a muscle-specific ubiquitin ligase, regulates β-catenin degradation during myogenesis. Dev. Cell 6, 269–282 [DOI] [PubMed] [Google Scholar]
- 30. Campos Y., Qiu X., Zanoteli E., Moshiach S., Vergani N., Bongiovanni A., Harris A. J., d'Azzo A. (2010) Ozz-E3 ubiquitin ligase targets sarcomeric embryonic myosin heavy chain during muscle development. PLoS One 5, e9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Persons D. A., Allay J. A., Allay E. R., Ashmun R. A., Orlic D., Jane S. M., Cunningham J. M., Nienhuis A. W. (1999) Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 93, 488–499 [PubMed] [Google Scholar]
- 32. Di Felice V., Ardizzone N. M., De Luca A., Marcianò V., Gammazza A. M., Macaluso F., Manente L., Cappello F., De Luca A., Zummo G. (2009) OPLA scaffold, collagen I, and horse serum induce an higher degree of myogenic differentiation of adult rat cardiac stem cells. J. Cell Physiol. 221, 729–739 [DOI] [PubMed] [Google Scholar]
- 33. Di Felice V., Cappello F., Montalbano A., Ardizzone N. M., De Luca A., Macaluso F., Amelio D., Cerra M. C., Zummo G. (2007) HSP90 and eNOS partially co-localize and change cellular localization in relation to different ECM components in 2D and 3D cultures of adult rat cardiomyocytes. Biol. Cell 99, 689–699 [DOI] [PubMed] [Google Scholar]
- 34. Radivojac P., Vacic V., Haynes C., Cocklin R. R., Mohan A., Heyen J. W., Goebl M. G., Iakoucheva L. M. (2010) Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou X., Pan S., Sun L., Corvera J., Lin S. H., Kuang J. (2008) The HIV-1 p6/EIAV p9 docking site in Alix is autoinhibited as revealed by a conformation-sensitive anti-Alix monoclonal antibody. Biochem. J. 414, 215–220 [DOI] [PubMed] [Google Scholar]
- 36. Marquette M. L., Byerly D., Sognier M. (2008) The effects of three-dimensional cell culture on single myoblasts. In Vitro Cell. Dev. Biol. Anim. 44, 105–114 [DOI] [PubMed] [Google Scholar]
- 37. Grinnell F. (1978) Cellular adhesiveness and extracellular substrata. Int. Rev. Cytol. 53, 65–144 [DOI] [PubMed] [Google Scholar]
- 38. Weed S. A., Parsons J. T. (2001) Cortactin. Coupling membrane dynamics to cortical actin assembly. Oncogene 20, 6418–6434 [DOI] [PubMed] [Google Scholar]
- 39. Lynch D. K., Winata S. C., Lyons R. J., Hughes W. E., Lehrbach G. M., Wasinger V., Corthals G., Cordwell S., Daly R. J. (2003) Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278, 21805–21813 [DOI] [PubMed] [Google Scholar]
- 40. Kaksonen M., Toret C. P., Drubin D. G. (2006) Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7, 404–414 [DOI] [PubMed] [Google Scholar]
- 41. Kim J., Sitaraman S., Hierro A., Beach B. M., Odorizzi G., Hurley J. H. (2005) Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cory G. O., Cullen P. J. (2007) Membrane curvature. The power of bananas, zeppelins, and boomerangs. Curr. Biol. 17, R455–R457 [DOI] [PubMed] [Google Scholar]
- 43. Zhai Q., Landesman M. B., Chung H. Y., Dierkers A., Jeffries C. M., Trewhella J., Hill C. P., Sundquist W. I. (2011) Activation of the retroviral budding factor ALIX. J. Virol. 85, 9222–9226 [DOI] [PMC free article] [PubMed] [Google Scholar]