Background: O-GlcNAc occupies histone H3, but specific sites and functions are unknown.
Results: Threonine 32 is O-GlcNAcylated, and O-GlcNAc inhibits mitosis-specific phosphorylations on H3. Impaired removal of O-GlcNAc inhibits efficient cell cycle transition from G2 to mitosis.
Conclusion: The reciprocal relationship between O-GlcNAc and phosphorylation on H3 contributes to a G2-M transition checkpoint.
Significance: A mechanistic link between O-GlcNAc on H3 and mitosis has been revealed.
Keywords: Glycobiology, Histones, O-GlcNAc, O-GlcNAcylation, Phosphorylation, H3
Abstract
O-Linked β-N-acetylglucosamine, or O-GlcNAc, is a dynamic post-translational modification that cycles on and off serine and threonine residues of nucleocytoplasmic proteins. The O-GlcNAc modification shares a complex relationship with phosphorylation, as both modifications are capable of mutually inhibiting the occupation of each other on the same or nearby amino acid residue. In addition to diabetes, cancer, and neurodegenerative diseases, O-GlcNAc appears to play a significant role in cell growth and cell cycle progression, although the precise mechanisms are still not well understood. A recent study also found that all four core nucleosomal histones (H2A, H2B, H3, and H4) are modified with O-GlcNAc, although no specific sites on H3 were reported. Here, we describe that histone H3, a protein highly phosphorylated during mitosis, is modified with O-GlcNAc. Several biochemical assays were used to validate that H3 is modified with O-GlcNAc. Mass spectrometry analysis identified threonine 32 as a novel O-GlcNAc site. O-GlcNAc was detected at higher levels on H3 during interphase than mitosis, which inversely correlated with phosphorylation. Furthermore, increased O-GlcNAcylation was observed to reduce mitosis-specific phosphorylation at serine 10, serine 28, and threonine 32. Finally, inhibiting OGA, the enzyme responsible for removing O-GlcNAc, hindered the transition from G2 to M phase of the cell cycle, displaying a phenotype similar to preventing mitosis-specific phosphorylation on H3. Taken together, these data indicate that O-GlcNAcylation regulates mitosis-specific phosphorylations on H3, providing a mechanistic switch that orchestrates the G2-M transition of the cell cycle.
Introduction
O-Linked-β-N-acetylglucosamine (O-GlcNAc)2 is a highly dynamic post-translational modification that cycles on and off serine and threonine residues of nucleocytoplasmic proteins (1, 2). Many studies revealed an extensive cross talk between O-GlcNAc and phosphorylation, usually through mutual inhibition of the same or nearby residue (3). The enzyme responsible for covalently linking O-GlcNAc to nucleocytoplasmic proteins is β-N-acetylglucosaminyltransferase (O-GlcNAc transferase, OGT) (4), and its removal is mediated by β-N-acetylglucosaminidase (O-GlcNAcase, OGA) (5). UDP-GlcNAc, the donor substrate, is a nutrient sensor, as its intracellular concentration directly correlates with fluctuating glucose concentrations (6). O-GlcNAc plays significant roles in many functions and diseases, such as diabetes, cancer, and neurodegeneration (1, 6).
O-GlcNAc has also begun to emerge as a major modulator of the cell cycle (7, 8). Disrupting O-GlcNAc cycling leads to defects in cellular growth rate, cell cycle progression, and cytokinesis. Biochemical analysis revealed dynamic cycling of O-GlcNAcylation and phosphorylation of nuclear proteins throughout the cell cycle. Nuclear proteins are highly O-GlcNAcylated during interphase, but become phosphorylated during mitosis (9). In addition, OGT and OGA were found to be physically associated with Aurora B and PP1, a kinase and phosphatase that target histone H3 during mitosis (10).
DNA is coiled around an octamer of nucleosomal histones consisting of H2A, H2B, H3, and H4 (11, 12). Histones are modified with many types of post-translational modifications that regulate DNA condensation, which subsequently modulates a wide range of processes, such as transcriptional activity, DNA recombination and repair, and chromosomal compaction and segregation during mitosis (13). Because of the numerous variety and combination of modifications that occupy histones, the unique, synergistic functions of these modifications have been termed the histone code (12).
Histone H3, the most widely modified of all four nucleosomal histones, has several phosphorylation sites on its N-terminal tail (11). H3 is phosphorylated on threonine 3, serine 10, serine 28, and threonine 32 when cells enter mitosis (11, 14–17). Aurora B and PP1, enzymes that are physically associated with OGT and OGA, phosphorylate and dephosphorylate H3 serine 10 and serine 28 during mitosis, respectively (10, 18–23). Most importantly, a recent study revealed that all four nucleosomal histones are modified with O-GlcNAc (24). However, no O-GlcNAc sites on H3 were identified.
Here, we used a histone extraction method that differs from the previous study to confirm the existence of O-GlcNAc on H3, and identified a novel O-GlcNAc site on the H3 tail with mass spectrometry. The cumulative data generated from a series of additional experiments examining how O-GlcNAc affects mitosis-specific phosphorylation on H3 are consistent with a model in which the interplay between O-GlcNAc and phosphorylation contributes to a significant regulatory mechanism that allows cells to transition from G2 to mitosis.
EXPERIMENTAL PROCEDURES
Cell Culture, Mitotic Synchronization, and OGA Inhibition
HeLa cells were grown in Dulbecco's modified Eagle's media with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin (Invitrogen).
For mitotic synchronization, HeLa cells were incubated for 16 h with colcemid N-methyl-N-deacetyl-colchine (Roche) used at 1:100 from stock concentration. Cells synchronized at the G2-M border were incubated for 20 h with 9 μm RO-3306 (Enzo, ALX-270–463). For G2-M release, RO-3306 media was removed and replaced by RO-3306-free media.
For OGA inhibition, asynchronous cells were incubated with 100 μm PUGNAc (Toronto Research Chemicals A157250) for 16 h. Mitotically synchronized cells were incubated with 100 μm PUGNAc for 6 h prior to the addition of colcemid, then incubated in 100 μm PUGNAc and colcemid for 16 h. HeLa cells synchronized at the G2-M border were incubated with 100 μm PUGNAc for 4–6 h prior to the addition of RO-3306, and throughout the incubation with RO-3306.
Nuclear Extracts
For collection, asynchronized cells were washed in ice-cold PBS, scraped from culture dish, and pelleted by gentle centrifugation. Mitotic cells were collected by shake off and pelleted by gentle centrifugation. After pelleting, cells were lysed with 10 mm Tris-HCl, 150 mm NaCl, 1.5 mm MgCl2, 3% glycerol, and 10 mm EDTA, and spun through 10 mm Tris-HCl, 1.5 mm MgCl2, 25% glycerol solution. Next, the pellet was sonicated in 1% SDS, 10 mm EDTA, 50 mm NaCl, protease inhibitor mixture, phosphatase inhibitor mixture, 50 μm PUGNAc, and 1 μm okadaic acid. After sonication, the extract was diluted 1:10 to a final concentration of 0.1% SDS, with all other reagents remaining at the same concentration.
Western Blot
Samples were mixed with Laemmli Sample Buffer and boiled at 100 °C for 5 min. SDS-PAGE was performed with Bio-Rad 18% Criterion Tris-HCl Precast gel or “Any kDa” Mini-PROTEAN TGX Precast gel. Proteins were transferred onto Bio-Rad Trans-Blot Transfer Medium 0.2 μm nitrocellulose membrane, and blocked with 3% BSA, 0.02% Tween-20 in TBS for 1 h. The following antibodies were used: H3 serine 10 phosphorylation (Upstate Technology 06–570); H3 serine 28 phosphorylation (Abcam Ab5169); H3 threonine 32 phosphorylation (Abcam Ab4076); mouse total H3 (Abcam ab10799); rabbit total H3 (Abcam Ab1791); O-GlcNAc CTD 110.6 (kind gift from Gerald Hart, John Hopkins University School of Medicine); O-GlcNAc RL2 (Abcam ab2739). O-GlcNAc competition was performed with 1 m GlcNAc (Sigma 2423432). After incubation, membranes were washed three times in 0.05% Tween-20 in TBS, and incubated in the appropriate secondary antibody linked to horseradish peroxidase for 1 h. Chemiluminescent signals were detected using Thermo Scientific SuperSignal West Pico solution.
Immunoprecipitation
Immunoprecipitation was performed with magnetic Dynabeads from Invitrogen, as per the manufacturer's protocol. Nuclear extracts were obtained as described above. Antibodies were incubated in PBS (total H3 antibody) or TBS (phospho-H3 antibodies) with Dynabeads at room temperature. Proteins were eluted from the beads by boiling in Bio-Rad Laemmli Sample Buffer at 100 °C for 10 min.
Immunofluorescence
HeLa cells were seeded onto acid-washed coverslips overnight. After treatment, cells were washed in TBS, fixed in 3.7% formaldehyde in TBS for 10 min at room temperature, washed again. Cells were permeabilized in 0.5% Triton X-100, washed, then blocked in 3% BSA, 0.05% Tween-20 in TBS. Nuclei were stained with DAPI “FluoroPure grade” (Invitrogen Molecular Probes D21490) as per manufacturer's recommendations. Coverslips were mounted onto microscope slides with ProLong Gold antifade reagent (Invitrogen Molecular Probes P36934).
OGT Overexpression
EGFP- and HA-tagged human OGT were cloned into pEF-GW51 expression vector. GFP and HA-OGT were overexpressed in HeLa cells by DNA transfection, performed using the Fugene 6 Transfection Reagent (Roche Scientific), per the manufacturer's protocol.
Mass Spectrometry
H3 immunoprecipitated from asynchronized HeLa cells was reduced with 1 m DTT and carboxyamidomethylated with 55 mm iodoacetamide, and digested overnight with Arg-C. Next, the peptides were acidified with 1% trifluoroacetic acid, desalted, dried, and resuspended with 38 μl of mobile phase A (0.1% formic acid in water) and 2 μl of mobile phase B (80% acetonitrile and 0.1% formic acid in water). Samples were loaded onto a nanospray tapered capillary column/emitter self-packed with C18 reverse-phase resin via a nitrogen pressure bomb for 15 min at 1000 psi for each run.
Each run consisted of a 160 min gradient of increasing mobile phase B at a flow rate of ∼200 nl/minute. H3 was identified with a LC-MS/MS analysis, and an instrument method was used to collect and generate MS/MS spectra for the 8 most intense peaks using collision-induced dissociation. To map modifications, LC-MS/MS analysis was performed as described above. An instrument method was used to collect a full MS spectrum and generate MS/MS spectra in FT mode for the 10 most intense peaks using Higher Energy Collision Dissociation utilizing dynamic exclusion and rejecting peaks assigned a charge state of +1. Each MS/MS spectra were searched for signature peaks associated with the loss of HexNAc. Each precursor mass that showed this loss was selected for fragmented in the ion trap using ETD with supplemental activation (25).
The data were searched against a human data base and histone only database using a Sequest Algorithm, with parameters allowing for oxidation of methionine, alkylated cysteine, acetylated lysine and arginine, methylated arginine, dimethylated lysine, and arginine, glycosylation of serine and threonine, and phosphorylated serine, threonine and tyrosine. Precursor mass tolerance was set at 20 ppm and fragment ion tolerance was set at 1 Dalton. Results were filtered at a false discovery rate (FDR) of 1%.
RESULTS
Histone H3 Is Modified with O-GlcNAc
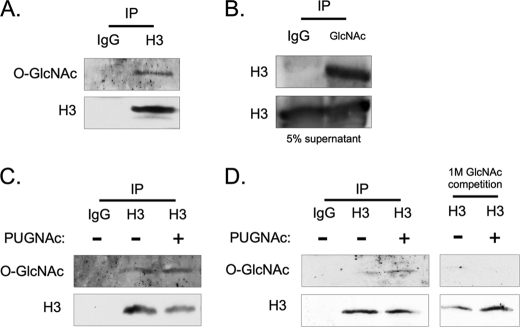
An antibody against total H3 was used to immunoprecipitate the corresponding endogenous protein from HeLa cell nuclear extracts. Antibody CTD 110.6 was then used in Western blotting to successfully detect attached O-GlcNAc (Fig. 1A). A reciprocal immunoprecipitation with an anti-O-GlcNAc antibody confirmed the attachment to histone H3 (Fig. 1B). The O-GlcNAc modification on H3 was not exclusive to the HeLa cell line, as it was also detected in H3 immunoprecipitates from UCD melanoma cell line (Fig. 1C). Here, we also observed an increased signal for O-GlcNAc on H3 when the cells were incubated with PUGNAc, an OGA inhibitor (26). HeLa cells incubated with PUGNAc also displayed an increased signal for O-GlcNAc on H3, and co-incubation of the antibody with 1 m GlcNAc prevented detection (Fig. 1D), validating the specificity of the detection method.
FIGURE 1.
Histone H3 is modified with O-GlcNAc. Biochemical validation of O-GlcNAc on histone H3. A, H3 was immunoprecipitated, and subsequently blotted for O-GlcNAc using the CTD 110.6 antibody. B, reciprocal IP was performed using the O-GlcNAc antibody RL2, and blotted for H3. C, O-GlcNAc was also detected on histones immunoprecipitated from UCD melanoma cells, in addition to an increased signal for O-GlcNAc after incubation with PUGNAc. D, 1 m GlcNAc competition during primary antibody incubation was used to validate the specificity of the CTD 110.6 antibody in HeLa cells.
O-GlcNAc Cycles on Histone H3 in a Mitosis-dependent Manner
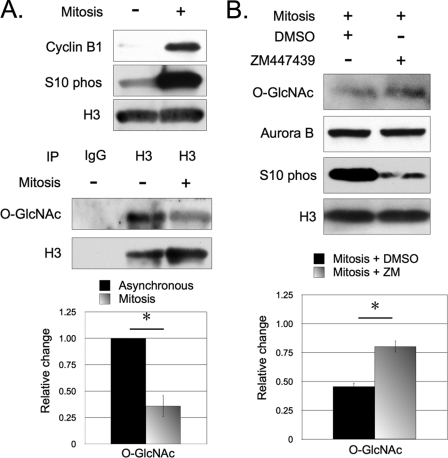
O-GlcNAc has been found to inversely cycle with phosphorylation in a mitosis dependent manner (9). Because H3 phosphorylation occurs during mitosis (27), we examined whether O-GlcNAc on H3 also cycles in a mitosis-dependent manner. For this purpose, we compared the signal for O-GlcNAc between H3 immunoprecipitated from asynchronous HeLa cells and mitotically synchronized cells. O-GlcNAc on H3 consistently followed the same cycling pattern as global fluctuations of O-GlcNAcylation, displaying a stronger signal on H3 from asynchronous cells, and a weaker signal from mitotic cells (Fig. 2A).
FIGURE 2.
O-GlcNAc cycling on H3 is associated with mitosis. Relative O-GlcNAcylation of H3 was compared between asynchronous and mitotic cells. A, H3 immunoprecipitated from mitotic cells displayed a weaker signal for O-GlcNAc when compared with H3 immunoprecipitated from asynchronous cells. The increase of cyclin B1 and H3S10ph were used to validate mitotic synchronization. B, histone H3 extracted from mitotic cells incubated with Aurora B inhibitor ZM447439 displayed a stronger signal for O-GlcNAc when compared with DMSO control. As expected, ZM447439 did not alter Aurora B expression but suppressed H3S10ph. Relative signals for O-GlcNAc were normalized to asynchronous HeLa cells (not shown). *, p < 0.05, as calculated by paired two-sample t test.
To further characterize the relationship between phosphorylation and O-GlcNAc on H3, we asked whether preventing mitotic phosphorylation alters O-GlcNAcylation. Aurora B, the kinase that phosphorylates H3S10 and H3S28, was specifically targeted in this experiment due to its physical association with OGT and OGA (9, 18, 28). HeLa cells were incubated with ZM447439, an Aurora B kinase inhibitor (29), for 6 h before mitotic arrest (30). Here, mitotically synchronized HeLa cells incubated with ZM447439 displayed higher levels of O-GlcNAc on total H3 from nuclear extracts when compared with the mitotically synchronized control cells (Fig. 2B).
O-GlcNAc Occupies Threonine 32 on H3
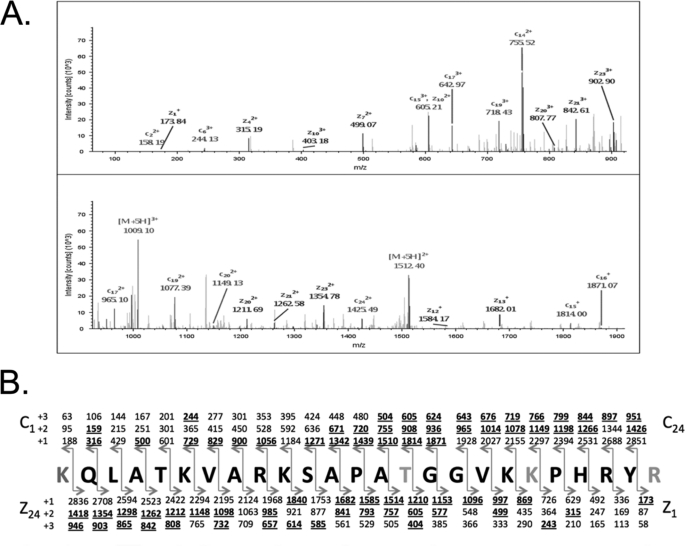
To identify specific sites on H3 modified with O-GlcNAc, we immunoprecipitated H3 from asynchronous HeLa cells and performed LC-MS/MS analysis on the H3 immunoprecipitates. From the analysis, 25 variations of the same polypeptide backbone of H3 were identified with glycosylation of threonine 32 (T32Glc) (Fig. 3A). The peptide backbone fragmentation produced a nearly complete series of c and z ions for each variation allowing confident assignment of O-GlcNAc modification (Fig. 3B). We also noted the lack of phosphorylation at serine 28 (S28) in all peptides identified with T32Glc.
FIGURE 3.
Threonine 32 is an O-GlcNAc site on H3. A, ETD MS2 spectra of the [M+5H]5+ peptide range lysine 18 to arginine 42 (KQLATKVARKSAPATGGVKKPHRYR mass range 0–925 and 925–1900). ETD product ion peaks are labeled as well as charge-reduced species and species resulting from water loss. B, sequence of the most frequently observed variation of the O-GlcNAcylated peptide KQLATKVARKSAPATGGVKKPHRYR with all of the predicted c and z product ions shown in singly, doubly, and triply charged forms. Observed product ions are displayed in bold and underlined.
O-GlcNAc Displays a Reciprocal Relationship with Mitosis-specific Phosphorylations on Histone H3
Next, we examined whether O-GlcNAc inhibits phosphorylation of Ser-28 and Thr-32 on H3 due to our previous finding (17, 23). We also examined Ser-10 because both Ser-10 and Ser-28 are phosphorylated by Aurora B. Moreover, H3 is predicted to exhibit multiple O-GlcNAc sites, including serine 10, threonine 6, and threonine 11. These predictions are based on the Homo sapiens sequence for histone H3 calculated by the algorithms OGlcNAcScan and YinOYang.3,4
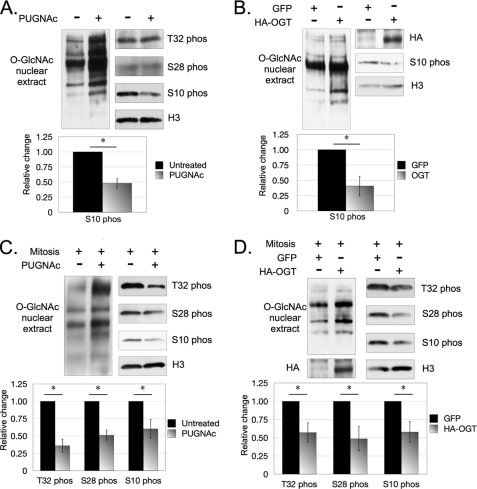
Asynchronous cells incubated with PUGNAc did not display differences in S28ph and T32ph when compared with untreated cells (Fig. 4A). However, PUGNAc-incubated cells displayed a weaker signal for S10ph when compared with untreated cells. To further investigate this observation, we overexpressed OGT in asynchronous cells and examined whether there was an effect on S10ph. We found that S10ph is also reduced in asynchronous cells overexpressing OGT when compared with the transfection control cells (Fig. 4B). Here, we did not examine S28ph or T32ph because no changes were observed in the previous experiment.
FIGURE 4.
O-GlcNAc inhibits mitosis-specific phosphorylations on H3. O-GlcNAc inhibits several histone H3 phosphorylation sites. Asynchronous cells incubated with PUGNAc (A) or overexpressing HA-tagged OGT (B) displayed a weaker signal for H3S10ph when compared with the respective controls. Mitotic cells incubated with PUGNAc (C) or overexpressing HA-tagged OGT (D) displayed weaker signals for S10ph, S28ph, and T32ph as compared with their relative controls. *, p < 0.05, as calculated by paired two-sample t test.
Because O-GlcNAc cycles on H3 in a mitosis-dependent manner, we asked whether perturbing O-GlcNAc cycling in mitotic cells alters T32ph, S28ph, and S10ph. We found that mitotic cells incubated with PUGNAc displayed a weaker signal for phosphorylation at all three sites when compared with untreated cells (Fig. 4C), suggesting an inhibitory role for O-GlcNAc. This experiment was also repeated using OGA inhibitor thiamet-G, and the same reductions in T32ph, S28ph, and S10ph were also observed (supplemental Fig. S2). In addition, mitotic cells overexpressing OGT displayed lower levels of T32ph, S28ph, and S10ph when compared with mitotic transfection control cells (Fig. 4D).
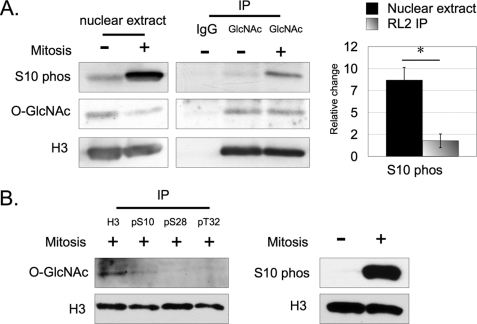
Because O-GlcNAc was not identified on or near Ser-10, we continued to test whether S10ph is regulated by O-GlcNAc. To further investigate this relationship, we first quantified the fold increase in S10ph on total H3 from nuclear lysates between asynchronous and mitotic cells. Next, we quantified the fold increase in S10ph on O-GlcNAc-immunoprecipitated H3 (H3Glc-IP). Lastly, we compared the change of S10ph from the first set (total H3 from nuclear lysate) to the second set (H3Glc-IP). All samples (total H3 versus H3Glc-IP) were derived from the same cell population and tested in parallel.
We consistently detected a stronger signal for S10ph from total H3 over H3Glc-IP in both asynchronous and mitotic cells (Fig. 5A). The normalized signal for S10ph from the H3Glc-IP was much lower when compared with H3 probed directly from nuclear extracts. We also detected the expected reduction in O-GlcNAc on total H3 from mitotically-synchronized cells compared with asynchronous cells.
FIGURE 5.
The reciprocal relationship between O-GlcNAc and phosphorylation on H3. Complementary immunoprecipitations support the reciprocal relationship hypothesis between O-GlcNAc and phosphorylation on H3. A, relative increase of normalized H3S10ph between asynchronous HeLa cells to mitotically synchronized cells was compared between total H3 from nuclear lysates (black bar) and O-GlcNAc-immunoprecipitated H3 (gradient bar). O-GlcNAc-immunoprecipitated histone H3 displayed much less S10ph when compared with total H3. B, H3 immunoprecipitated with an antibody against total H3 from mitotically synchronized HeLa cells displays a stronger signal for O-GlcNAc when compared with H3 immunoprecipitated with antibodies against phosphorylated H3. *, p < 0.05, as calculated by paired two-sample t test.
Furthermore, the O-GlcNAc bands in the H3Glc-IP group represent equal amounts of O-GlcNAcylated protein from the immunoprecipitation. This result was expected since the experiment was designed to obtain relatively similar concentrations of O-GlcNAcylated protein between asynchronous and mitotically synchronized cells rather than maximizing efficient immunoprecipitation. These results indicate that the O-GlcNAcylated H3 exhibits significantly diminished amounts of S10ph.
Next, we immunoprecipitated H3 using antibodies against total H3, S10ph, S28ph, and T32ph, and compared the signal for O-GlcNAc between each of the immunoprecipitates. In this experiment, the amount of antibody used in each respective immunoprecipitation was optimized for the purpose of obtaining relatively equal concentrations of histone H3 across all samples, and not for the purpose of maximizing efficient histone H3 immunoprecipitation. Relative to the immunoprecipitate from the antibody against total H3, we found that the normalized signal for O-GlcNAc was lower or undetectable in immunoprecipitates from antibodies against each of the phosphorylated H3 sites (Fig. 5B).
The O-GlcNAcylation to Phosphorylation Switch Is a Checkpoint for Entering Mitosis
Because H3 phosphorylation is necessary for efficient entry into mitosis, we asked whether the removal of O-GlcNAc is required during the G2-M transition. Because of the difficulty in targeting specific O-GlcNAc sites on a single protein, this experiment addresses the question whether O-GlcNAc cycling affects the G2-M transition on a global level. To answer this question, we synchronized HeLa cells at the G2-M transition phase by incubating cells in media that contains compound RO-3306 (33). RO-3306 is a reversible small molecule enzymatic inhibitor of Cdk1, a kinase that phosphorylates many targets during the transition into mitosis. Blocking phosphorylation by Cdk1 inhibits entry into mitosis and arrests cells at the G2-M transition. Removing RO-3306 from culture media allowed the cells to immediately enter mitosis.
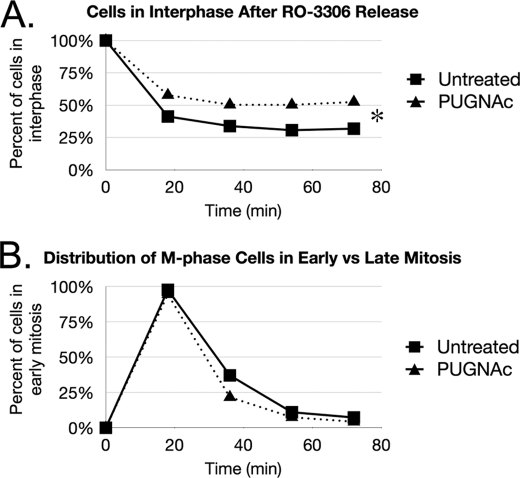
After releasing the cells from RO-3306 and fixing them across various time points, we stained the nuclei with DAPI and counted the number of cells in each phase of mitosis on the basis of DNA condensation. For this study, cells in prophase and prometaphase were categorized as early mitosis, and cells in metaphase, anaphase, and telophase were categorized as mid-late mitosis. Cells that did not enter mitosis, including ones in G1, S, G2, and the G2-M transition, were allocated into the interphase category. With this method, we compared the percentage of cells in each phase of mitosis between a PUGNAc-incubated group to an untreated group.
After release from RO-3306, there was a consistently larger percentage of PUGNAc-incubated cells retained in interphase at all time points when compared with the untreated group (Fig. 6A). The statistical significance between the two curves was validated through binomial regression analysis. This cell cycling defect parallels the original studies that revealed the significance of H3 serine 10 phosphorylation during mitosis. Interestingly, when we analyzed the population of cells that entered M-phase, PUGNAc did not appear to have a significant effect on the rate of progression from early to late mitosis (Fig. 6B). Thus, we concluded that removal of O-GlcNAc contributes to the required checkpoint for entering mitosis during the G2-M transition phase.
FIGURE 6.
O-GlcNAc regulates the G2-M transition during the cell cycle. Perturbing O-GlcNAc cycling alters the G2-M transition phase of the cell cycle. HeLa cells were incubated with or without PUGNAc before and during arrest at the G2-M border with RO-3306. A, after release from G2-M arrest, PUGNAc treated cells had a higher proportion remaining in interphase at all time points when compared with the untreated control. B, PUGNAc did not appear to alter the rate of mitotic progression for cells that were able to enter M-phase. *, p < 0.05, as calculated by binomial regression analysis.
DISCUSSION
The presence of O-GlcNAc on nucleosomal histones was first revealed by Sabake et al. (24). Although O-GlcNAc sites on H2A, H2B, and H4 were discovered, O-GlcNAc sites on H3 had not been identified. Importantly, we are the first to report the novel O-GlcNAc site threonine 32 on histone H3 as identified by mass spectrometry. With this discovery, we found that increased O-GlcNAcylation reduces mitosis-specific phosphorylation of Thr-32, Ser-28, and Ser-10 (17). We attribute the decrease in T32ph and S28ph as a direct result of O-GlcNAc occupying Thr-32. Thus, we propose that T32Glc regulates mitosis-specific Ser-28 and Thr-32 phosphorylation on H3.
The results from this study also indicate that S10ph is regulated by O-GlcNAc. Because S10ph and S28ph appear to be regulated in a functionally distinct manner (16, 23), it is unlikely that the decrease in S10ph is a consequence of diminished S28ph. Because there is still a detectable signal for O-GlcNAc on H3 during mitosis, we postulate that O-GlcNAc on H3 is the regulator that accounts for the temporal and spatial differences in S10ph and S28ph.
We propose a few explanations to account for the relationship between S10ph and O-GlcNAc. The first possibility is that O-GlcNAc also modifies Ser-10 or a nearby amino acid residue. Although we did not detect O-GlcNAc on or near Ser-10, several residues in this region are predicted to have this modification.
An alternative explanation is that O-GlcNAc structurally alters the H3 N-terminal tail in a manner that prevents Aurora B from accessing Ser-10. NMR studies found that phosphorylation can linearize a polypeptide backbone, whereas O-GlcNAcylation can lead to the generation of a coil in the peptide backbone (34–36). These studies support the possibility that T32Glc coils the H3 tail to inhibit Aurora B attachment. Similarly, T32ph may even be a prerequisite modification for S10ph and S28ph, as T32ph may linearize the histone tail to allow Aurora B attachment.
Phosphorylation of H3S10 is a molecular checkpoint for entering mitosis (37). We found that inhibiting OGA prevents the efficient transition from G2 to mitosis. Because of the apparent reciprocal relationship between O-GlcNAcylation and mitosis-specific phosphorylations on H3, we propose that the removal of the glycosylation from H3 contributes to the molecular checkpoint required for entering mitosis. However, because OGA has many target substrates, future studies specifically targeting O-GlcNAc on H3 are required.
During the preparation of this report, a study by Zhang et al. (38) was published, describing their observations of O-GlcNAc on H3. They suggested Ser-10 as a putative O-GlcNAc site, consistent with our hypothesis. However, they also found an increase in S28ph as a result of increased O-GlcNAcylation, as well as a stronger signal for O-GlcNAc on H3 during mitosis when compared with interphase, contradictory to our data. The differences in experimental design may account for the discrepancy observed with S28ph. Zhang et al. used glucosamine to increase O-GlcNAcylation in asynchronous cells. Glucosamine also activates the p38 MAP kinase pathway, which can subsequently phosphorylate H3S28 during interphase (39–42). The increase in S28ph may be the result of activating an alternate pathway independent of O-GlcNAc on H3. In addition, these experiments were performed on an asynchronous cell population, but the S28ph reduction we observed took place in mitotic cells.
The differences found between the groups may also be a testimony to the complexity of understanding O-GlcNAc as part of the histone code. Post-translational modifications are not always distinct switches, but may serve as a sensitive rheostat. Histone modifications can also function in synchrony with each other, as suggested by the histone code hypothesis. Different extraction methods, cell types, plus other unexpected factors may have caused each group to enrich for H3 with a distinctly separate modification code. When considering these factors, we believe the collective reports on O-GlcNAc as part of the histone code are complementary rather than contradictory.
The data presented in this study extend beyond the field of O-GlcNAc and cell cycling. Many chemotherapeutic agents currently in development focus on inhibiting Aurora kinase and Polo kinase to deregulate mitosis and cytokinesis (43). Interestingly, S10ph appears to be a required histone mark for tumor transformations (44, 45). OGT, along with Aurora kinases and PP1 phosphatases (10, 46), localizes to the spindles, suggesting an O-GlcNAcylation phosphorylation interplay on spindle proteins. Disrupting O-GlcNAc cycling during mitosis also leads to severe cytokinesis defects (7, 8). With the reciprocal relationship between O-GlcNAc and phosphorylation on H3 found in this study, perhaps the synergistic disruption of both modifications may be a more effective treatment against cancer. However, this strategy must be pursued carefully, as many powerful oncogenes and tumor suppressors are also regulated by the O-GlcNAc-phosphorylation relationship (47).
In addition to cancer, this study also impacts the field of O-GlcNAc in insulin signaling, metabolic diseases, and aging (48, 49). Because UDP-GlcNAc is a nutrient sensor (6), the identification of an O-GlcNAc sites on histones may lead to new insights regarding how nutritional intake directly affects transcriptional activity of metabolic genes (50, 51). Indeed, studies using ChIP sequencing have revealed that O-GlcNAcylated chromatin-associated proteins regulate transcription of metabolic and aging-related genes (52). The identification of H3T32Glc provides a specific epigenetic marker for future studies in insulin signaling and metabolism.
Furthermore, the results presented in our study will provide a new direction to explore regarding pancreatic beta-cell biology. Unlike many other terminally differentiated cell types, pancreatic beta-cells self-replicate rather than differentiate from stem cell progenitors (31). Although the rate of self-replication is slow, inhibiting the replicative capacity of pancreatic beta-cells drives them toward senescence, decrease insulin production, and leads to type 2 diabetes mellitus. The process of aging and cell senescence is very tightly linked to histone modifications (32). Because of the newly revealed role of O-GlcNAc on H3 and cell cycle progression, we postulate that O-GlcNAc is the pendulum that balances between the decision of whether pancreatic beta-cells produce insulin to meet metabolic needs or self-replicate. Although the appreciation for O-GlcNAc on the histone code is only beginning to emerge, the functional impact of this modification is clearly significant in multiple biological functions and disease applications.
Supplementary Material
Acknowledgments
We thank Drs. Danielle Martinez and Denae Nash for technical assistance and Dr. Scott Pletcher for statistical analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA84282 (to E. E. M).

This article contains supplemental Figs. S1 and S2.
YinOYang Server, YinOYang version 1.2.
dbOGAP: Database of O-GlcNAcylated Proteins and Sites.
- O-GlcNAc
- O-linked-β-N-acetylglucosamine
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase;
REFERENCES
- 1. Hart G. W., Housley M. P., Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 2. Slawson C., Hart G. W. (2003) Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 13, 631–636 [DOI] [PubMed] [Google Scholar]
- 3. Hu P., Shimoji S., Hart G. W. (2010) Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 584, 2526–2538 [DOI] [PubMed] [Google Scholar]
- 4. Kreppel L. K., Blomberg M. A., Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 5. Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 6. Slawson C., Copeland R. J., Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. (2005) Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 8. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakabe K., Hart G. W. (2010) O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slawson C., Lakshmanan T., Knapp S., Hart G. W. (2008) A mitotic GlcNAcylation/phosphorylation signaling complex alters the post-translational state of the cytoskeletal protein vimentin. Mol. Biol. Cell 19, 4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pérez-Cadahia B., Drobic B., Davie J. R. (2009) H3 phosphorylation: dual role in mitosis and interphase. Biochem. Cell Biol. 87, 695–709 [DOI] [PubMed] [Google Scholar]
- 12. Gardner K. E., Allis C. D., Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chi P., Allis C. D., Wang G. G. (2010) Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shoemaker C. B., Chalkley R. (1980) H3-specific nucleohistone kinase of bovine thymus chromatin. Purification, characterization, and specificity for threonine residue 3. J. Biol. Chem. 255, 11048–11055 [PubMed] [Google Scholar]
- 15. Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. (1978) Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. / FEBS 84, 1–15 [DOI] [PubMed] [Google Scholar]
- 16. Goto H., Tomono Y., Ajiro K., Kosako H., Fujita M., Sakurai M., Okawa K., Iwamatsu A., Okigaki T., Takahashi T., Inagaki M. (1999) Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 274, 25543–25549 [DOI] [PubMed] [Google Scholar]
- 17. Caperta A. D., Rosa M., Delgado M., Karimi R., Demidov D., Viegas W., Houben A. (2008) Distribution patterns of phosphorylated Thr 3 and Thr 32 of histone H3 in plant mitosis and meiosis. Cytogenetic Gen. Res. 122, 73–79 [DOI] [PubMed] [Google Scholar]
- 18. Crosio C., Fimia G. M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C. D., Sassone-Corsi P. (2002) Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22, 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murnion M. E., Adams R. R., Callister D. M., Allis C. D., Earnshaw W. C., Swedlow J. R. (2001) Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276, 26656–26665 [DOI] [PubMed] [Google Scholar]
- 20. Sugiyama K., Sugiura K., Hara T., Sugimoto K., Shima H., Honda K., Furukawa K., Yamashita S., Urano T. (2002) Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21, 3103–3111 [DOI] [PubMed] [Google Scholar]
- 21. Giet R., Glover D. M. (2001) Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., Lin R., Smith M. M., Allis C. D. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 [DOI] [PubMed] [Google Scholar]
- 23. Goto H., Yasui Y., Nigg E. A., Inagaki M. (2002) Aurora B phosphorylates histone H3 at serine 28 with regard to the mitotic chromosome condensation. Genes to Cells 7, 11–17 [DOI] [PubMed] [Google Scholar]
- 24. Sakabe K., Wang Z., Hart G. W. (2010) β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao P., Viner R., Teo C. F., Boons G. J., Horn D., Wells L. (2011) Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Prot. Res. 10, 4088–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haltiwanger R. S., Grove K., Philipsberg G. A. (1998) Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 273, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 27. Sauvé D. M., Anderson H. J., Ray J. M., James W. M., Roberge M. (1999) Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J. Cell Biol. 145, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goto H., Yasui Y., Kawajiri A., Nigg E. A., Terada Y., Tatsuka M., Nagata K., Inagaki M. (2003) Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 278, 8526–8530 [DOI] [PubMed] [Google Scholar]
- 29. Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H., Ganguly A., Cabral F. (2010) Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J. Biol. Chem. 285, 32242–32250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tavana O., Zhu C. (2011) Too many breaks (brakes): pancreatic β-cell senescence leads to diabetes. Cell Cycle 10, 2471–2484 [DOI] [PubMed] [Google Scholar]
- 32. Feser J., Tyler J. (2011) Chromatin structure as a mediator of aging. FEBS Lett. 585, 2041–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C., Chen L. (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A. 103, 10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simaneck E. E., Huang D., Pasternack L., Machajewski T., Seitz O., Millar D. S., Dyson H. J., Wong C. (1998) JACS 120, 11567–11575 [Google Scholar]
- 35. Daly N. L., Hoffmann R., Otvos L., Jr., Craik D. J. (2000) Role of phosphorylation in the conformation of tau peptides implicated in Alzheimer's disease. Biochemistry 39, 9039–9046 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y. X., Du J. T., Zhou L. X., Liu X. H., Zhao Y. F., Nakanishi H., Li Y. M. (2006) Alternative O-GlcNAcylation/O-phosphorylation of Ser-16 induce different conformational disturbances to the N terminus of murine estrogen receptor β. Chem. Biol. 13, 937–944 [DOI] [PubMed] [Google Scholar]
- 37. Van Hooser A., Goodrich D. W., Allis C. D., Brinkley B. R., Mancini M. A. (1998) Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 111, 3497–3506 [DOI] [PubMed] [Google Scholar]
- 38. Zhang S., Roche K., Nasheuer H. P., Lowndes N. F. (2011) Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 286, 37483–37495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fülöp N., Zhang Z., Marchase R. B., Chatham J. C. (2007) Glucosamine cardioprotection in perfused rat hearts associated with increased O-linked N-acetylglucosamine protein modification and altered p38 activation. Am. J. Physiol. Heart Circulatory Physiology 292, H2227–H2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldberg H., Whiteside C., Fantus I. G. (2011) O-linked-beta-N-acetylglucosamine supports p38 MAPK activation by high glucose in glomerular mesangial cells. Am. J. Physiol. Endocrinol. Metab. 4, E713–E726 [DOI] [PubMed] [Google Scholar]
- 41. Zhong S., Zhang Y., Jansen C., Goto H., Inagaki M., Dong Z. (2001) MAP kinases mediate UVB-induced phosphorylation of histone H3 at serine 28. J. Biol. Chem. 276, 12932–12937 [DOI] [PubMed] [Google Scholar]
- 42. Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lens S. M., Voest E. E., Medema R. H. (2010) Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer 10, 825–841 [DOI] [PubMed] [Google Scholar]
- 44. Choi H. S., Choi B. Y., Cho Y. Y., Mizuno H., Kang B. S., Bode A. M., Dong Z. (2005) Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 65, 5818–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 46. Wells L., Kreppel L. K., Comer F. I., Wadzinski B. E., Hart G. W. (2004) O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J. Biol. Chem. 279, 38466–38470 [DOI] [PubMed] [Google Scholar]
- 47. Slawson C., Hart G. W. (2011) O-GlcNAc signaling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mondoux M. A., Love D. C., Ghosh S. K., Fukushige T., Bond M., Weerasinghe G. R., Hanover J. A., Krause M. W. (2011) O-linked-N-acetylglucosamine cycling and insulin signaling are required for the glucose stress response in Caenorhabditis elegans. Genetics 188, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rahman M. M., Stuchlick O., El-Karim E. G., Stuart R., Kipreos E. T., Wells L. (2010) Intracellular protein glycosylation modulates insulin mediated lifespan in C. elegans. Aging 2, 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanover J. A., Krause M. W., Love D. C. (2010) The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800, 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Love D. C., Krause M. W., Hanover J. A. (2010) O-GlcNac cycling: emerging roles in development and epigenetics. Sem. Cell Dev. Biol. 21, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., Hanover J. A. (2010) Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.