Background: The role of PICK1 in regulating trafficking of its PDZ domain binding partners (e.g. AMPA receptors) remains unclear.
Results: PICK1 clusters and reduces recycling only of PDZ binding partners sorted to Rab11-dependent recycling.
Conclusion: Contrary to other PDZ domain proteins, which regulate postendocytic sorting, PICK1 determines the trafficking rate through an endocytic compartment.
Significance: This function might explain the role of PICK1 in synaptic plasticity.
Keywords: G Protein-coupled Receptors (GPCRs); Glutamate Receptors, Ionotropic (AMPA, NMDA); Intracellular Trafficking; Monoamine Transporters; Protein Sorting; Receptor Recycling; BAR Domains; PDZ Domains; PICK1; Protein-Protein Interactions
Abstract
The scaffolding protein PICK1 (protein interacting with C kinase 1) contains an N-terminal PSD-95/Discs large/ZO-1 (PDZ) domain and a central lipid-binding Bin/amphiphysin/Rvs (BAR) domain. PICK1 is thought to regulate trafficking of its PDZ binding partners but different and even opposing functions have been suggested. Here, we apply ELISA-based assays and confocal microscopy in HEK293 cells with inducible PICK1 expression to assess in an isolated system the ability of PICK1 to regulate trafficking of natural and engineered PDZ binding partners. The dopamine transporter (DAT), which primarily sorts to degradation upon internalization, did not form perinuclear clusters with PICK1, and PICK1 did not affect DAT internalization/recycling. However, transfer of the PICK1-binding DAT C terminus to the β2-adrenergic receptor, which sorts to recycling upon internalization, led to formation of PICK1 co-clusters in Rab11-positive compartments. Furthermore, PICK1 inhibited Rab11-mediated recycling of the receptor in a BAR and PDZ domain-dependent manner. In contrast, transfer of the DAT C terminus to the δ-opioid receptor, which sorts to degradation, did not result in PICK1 co-clusters or any change in internalization/recycling. Further support for a role of PICK1 determined by its PDZ cargo was obtained for the PICK1 interaction partner prolactin-releasing peptide receptor (GPR10). GPR10 co-localized with Rab11 and clustered with PICK1 upon constitutive internalization but co-localized with the late endosomal marker Rab7 and did not cluster with PICK1 upon agonist-induced internalization. Our data suggest a selective role of PICK1 in clustering and reducing the recycling rates of PDZ domain binding partners sorted to the Rab11-dependent recycling pathway.
Introduction
Protein interacting with C kinase 1 (PICK1)2 is a widely distributed dimeric scaffolding protein characterized by the presence of a single N-terminal PSD-95/Discs large/ZO-1 (PDZ) homology domain in each protomer (1, 2). The PDZ domain was originally found to bind the extreme C terminus of protein kinase Cα (PKCα) (3, 4); however, later, the PDZ domain was shown to also bind the C termini of several other proteins (2, 5). These include a number of receptor and transporter proteins expressed predominantly in the CNS (see data in PDZbase (6)), such as the GluA2/3 subunits of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type ionotropic glutamate receptors (AMPARs), the metabotropic glutamate receptor mGluR7, and various neurotransmitter transporters including the glutamate GLT1b transporter (7) and the dopamine transporter (DAT) (2, 5, 8–10). The PICK1 dimer contains a single lipid-binding Bin/amphiphysin/Rvs (BAR) domain in addition to its PDZ domains (11, 12), and PICK1 is so far the only scaffolding protein identified in which PDZ domains are present together with a BAR domain. The BAR domain is an ∼200-residue dimeric α-helical module that forms a crescent-shaped elongated functional unit (13). Together with an N-terminal amphipathic helix, this functional unit is critical for the ability of BAR domains to bind preferentially to curved lipid membranes and/or to promote formation of lipid membrane curvature (14).
PICK1 has been suggested to regulate the function of its PDZ domain binding partners in several different ways. These include promoting clustering of the binding partners in cellular subcompartments (8, 10, 15–17), regulation of their trafficking (10, 18–20), and acting as a scaffold that brings PKCα in close proximity to other binding partners to govern their phosphorylation (9, 21–23).
The role of PICK1 in trafficking of its binding partners seems complex and not yet settled. PICK1 was reported to increase surface expression of DAT, mGluR7, and ASIC1a (10, 19, 20). In contrast, PICK1 has repeatedly been associated with down-regulation of GluA2 surface expression under basal conditions (12, 23–27), although GluA2 surface expression is unaffected in PICK1 knock-out mice (28, 29). Moreover, PICK1 has consistently been shown to be necessary for down-regulation of GluA2 surface expression during long term depression (LTD) in cerebellum and hippocampus (12, 24, 28, 30–32). However, one study found no PICK1 dependence of hippocampal LTD (33), and it was recently demonstrated in studies of knock-out mice that the PICK1 dependence is age- and protocol-specific (29).
PICK1 was previously shown also to be involved in NMDA receptor-mediated long term potentiation (28, 29) and suggested to have a role in several types of plasticity at different synapses involving changes in the ratio of GluA2-containing and GluA2-lacking receptors (34–36). Furthermore, there is evidence indicating that PICK1 is critical for the removal of GluA2-containing receptors in ventral tegmental area neurons in response to a single dose of cocaine (37).
It has been highly difficult to reconcile these observations with a single trafficking function of PICK1. Accordingly, PICK1 has been suggested to be directly involved in cluster-mediated surface stabilization of DAT and ASIC1a (10, 20), stabilization of extrasynaptic GluA2-containing AMPARs at the plasma membrane (25, 35, 38), internalization of GluA2-containing receptors (39–41), and reduction of recycling rates of GluA2-containing AMPARs upon activity-induced internalization (38, 42).
To further clarify the intrinsic role of PICK1 in regulating trafficking of its PDZ domain binding partners, we here used quantitative ELISA-based trafficking assays and confocal microscopy in heterologous cells with inducible PICK1 expression. This offers a valuable isolated system for analyzing the specific effects of PICK1 on both natural and engineered co-expressed interaction partners. We obtained unambiguous evidence for a general ability of PICK1 to cluster and reduce the recycling rates of PDZ domain binding partners after activity-dependent internalization. However, the data also suggest that this role depends on the inherent postendocytic sorting properties of the binding partners; i.e. PICK1 clustered and inhibited recycling of binding partners that independently sorted to the Rab11-mediated “slow” or “long loop” recycling pathway. This further implies that PICK1 by itself neither resides in nor targets its PDZ cargo into a recycling pathway. Of additional interest, our data do not indicate any effects of PICK1 on surface expression or internalization rates of its binding partners, and the inhibition of recycling rates appears to be strictly dependent on PDZ binding and the activity of the BAR domain. Taken together, the results support the notion of PICK1 as a highly versatile scaffolding protein capable of mediating cargo-determined distinct cellular functions depending on the nature and inherent properties of the PDZ domain binding partner.
EXPERIMENTAL PROCEDURES
Molecular Biology
The generation of plasmids encoding FLAG-tagged TacDAT (pcDNA3 TacDAT) and TacDAT C24 (pcDNA3 TacDAT C24) and mycPICK1 (pCMV mycPICK1) was described previously (43, 44). The eGFP-tagged Rab constructs (pEGFP-C1 Rab7 and pEGFP-C1 Rab11) were a kind gift from Dr. Katherine W. Roche, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD (45). N-terminally signal FLAG-tagged GPR10 in pcDNA3 was a kind gift from Dr. Birgitte Holst, Department of Neuroscience and Pharmacology, University of Copenhagen, Copenhagen, Denmark. N-terminally FLAG-tagged δ-opioid receptor (DOR) and β2-adrenergic receptor (β2AR) with C-terminal polyhistidine in pcDNA3 were kind gifts from Dr. Mark von Zastrow, Departments of Psychiatry and Cellular and Molecular Pharmacology, University of California, San Francisco, CA. In GPR10, the C-terminal isoleucine was changed to aspartate by use of the QuikChange® method (Stratagene, La Jolla, CA), yielding GPR10 D. In FLAG-β2ARHis6, the C-terminal histidine tag was removed by PCR-mediated mutagenesis and either substituted by the 8 C-terminal residues of the human DAT (-TLRHWLKV) or substituted by the 8 C-terminal residues of the human DAT with an additional alanine that disrupts the PDZ binding to PICK1 (46) (-TLRHWLKVA). The resulting fragments were cleaved with KpnI and BamHI and ligated into pcDNA3 FLAG-β2ARHis6 using these sites to make the three constructs. The L412A mutation reported to disrupt the N-ethylmaleimide-sensitive fusion protein (NSF) interaction was introduced in β2AR and β2DAT8 with the QuikChange method (Stratagene). Finally, the C-terminal 8 residues of DAT were appended to DOR with the QuikChange method (Stratagene). For generation of the Flp-In T-REx 293 cell line (see below), the sequence encoding PICK1 tagged at the N terminus with enhanced yellow fluorescent protein (eYFP) was amplified by PCR from peYFP-C1-eYFP-PICK1 (44) and inserted into the pcDNA5/FRT/TO vector (Invitrogen) using NotI and AflII. All constructs were sequenced before use.
Cell Cultures and Transfections
To generate a cell line with stable tetracycline-inducible expression of YFP-tagged PICK1, we used the Flp-In T-REx system and the Flp-In T-REx 293 cell line (Invitrogen). The cells were maintained in DMEM 1965 with Glutamax (l-alanyl-l-glutamine) containing 10% fetal calf serum at 37 °C in a humidified 5% CO2 atmosphere. Before transfection, cells were selected using 15 μg/ml blasticidin and 100 μg/ml Zeocin (both from Invitrogen). Cells (90% confluent) were transfected using Lipofectamine 2000 (Invitrogen) with a total of 3 μg of DNA in a 1:9 ratio of the pcDNA5/FRT/TO with the eYFP-PICK1 insert and pOG44 vector (Invitrogen) in Opti-MEM® (Invitrogen) overnight. Cells were then split to 50% confluence and grown for an additional 24 h with no antibiotics before selection was induced using 15 μg/ml blasticidin and 150 μg/ml hygromycin. Cells were maintained until visible foci appeared after which the cells were harvested, pooled, and further maintained as a polyclonal cell line (Flp-In T-REx 293 eYFP-PICK1). Inducible eYFP-PICK1 cell lines with the K251E,K252E,K257E (3KE) mutation and the A87L mutation were generated using the same protocol.
For transient transfections of Flp-In T-REx 293 eYFP-PICK1 cells, cells were seeded in 25-cm2 cell flasks (1 × 106 cells) or 75-cm2 cell flasks (3 × 106 cells) and grown in medium without selection for ∼20 h to reach ∼70% confluence. Cells were transfected with Lipofectamine 2000 (Invitrogen) for 16 h in medium using 1 μg of DNA/75-cm2 flask except for the β2AR constructs for which we used 0.1 μg/75-cm2 flask to obtain lower expression. For double and triple transfections, we used 1 μg of DNA of each construct. In general, we transfected >80% of the cells. The transfected cells were trypsinized and seeded on polyornithine-coated coverslips in 6-well plates (300,000 cells/well) or 96-well ELISA plates (35,000 cells/well) for 48–72 h prior to experiments. After ∼20 h, medium was changed to new medium without or with tetracycline (1 μg/ml) to induce expression of eYFP-PICK1.
Immunocytochemistry
In all trafficking experiments, FLAG-tagged surface receptors or transporters were labeled in DMEM 1965 for 30 min at 4 °C with M1 mouse anti-FLAG antibody (1 μg/ml) conjugated with Alexa Fluor® 568 or 647 carboxylic acid succinimidyl esters (Invitrogen) according to the manufacturer's protocol. For GPR10 sorting studies, Flp-In T-REx 293 eYFP-PICK1 cells were cotransfected with the plasmids encoding the receptor and either eGFP-Rab7 or eGFP-Rab11. Surface-labeled GPR10 receptors were allowed to internalize constitutively at 37 °C in DMEM 1965 or in the presence of prolactin-releasing peptide (PrP) (Thr-Pro-Asp-Ile-Asn-Pro-Ala-Trp-Tyr-Ala-Ser-Arg-Gly-Ile-Arg-Pro-Val-Gly-Arg-Phe) (1 nm) for 60 min before fixation in 4% paraformaldehyde for 10 min at 4 °C followed by 10 min at RT and mounting in Prolong® Gold antifade reagent (Invitrogen). For trafficking studies of surface-labeled receptors and transporters together with eYFP-PICK1, we adopted the following generalized scheme. Surface-expressed FLAG-tagged proteins were labeled as described above, and to visualize surface protein (named “Surface” in figures), cells were immediately fixed. To visualize internalization (named “Internalization” in figures), surface-labeled proteins were allowed to internalize constitutively at 37 °C in DMEM 1965 or in the presence of agents that stimulate internalization for 25 min (or as indicated in the legends) before fixation in 4% paraformaldehyde for 10 min at 4 °C followed by 10 min at RT and mounting in Prolong Gold antifade reagent (Invitrogen). For the receptors, we stimulated internalization with their respective agonists (GPR10, PrP (1 nm); β2AR, isoproterenol (Iso; 10 μm); DOR, DADLE peptide (10 μm)), and for TacDAT, we stimulated internalization with 1 μm phorbol 12-myristate 13-acetate (PMA). To enable detection of subsequent receptor/transporter trafficking (i.e. recycling or degradation), the action of the internalizing agent was terminated by incubation for an extended period at 37 °C with antagonists (β2AR, alprenolol (Alp; 10 μm); DOR, naxolone (Naxo; 10 μm); TacDAT, the protein kinase C inhibitor staurosporine (1 μm)). For GPR10, we had no potent antagonist, and cells were instead washed and left in DMEM 1965 at 37 °C for prolonged trafficking before fixation and mounting. To study the effect of blocking lysosomal degradation, Flp-In T-REx 293 eYFP-PICK1 cells were transfected with plasmids encoding TacDAT, GPR10, or DOR DAT8 and incubated with leupeptin (100 μm) together with Alexa Fluor 647-conjugated M1 for 16 h before fixation and mounting. To study colocalization of β2DAT8 with eYFP-Rab11 and mycPICK1, we triple transfected Flp-In T-REx 293. FLAG-tagged surface receptors were labeled with Alexa Fluor 568-conjugated M1 antibody as described above and internalized with 10 μm isoproterenol for 25 min before fixation. Cells were permeabilized with 0.2% saponin in PBS and 5% goat serum before labeling with primary M1 and rabbit anti-Myc (1:1000) (Upstate). Cell were washed three times in PBS + 5% goat serum and labeled with secondary antibodies (Alexa Fluor 568 goat anti-rabbit and Alexa Fluor 647 goat anti-mouse) (1:500).
Confocal Imaging
All imaging was performed with a Zeiss LSM 510 inverted confocal laser-scanning microscope using an oil immersion numerical aperture 1.4 63× objective (Zeiss, Jena, Germany). GFP and eYFP were excited with the 488 nm laser line from an argon-krypton laser, and the emitted light was detected using a 505–550-nm band pass filter. The Alexa Fluor 568 dye was excited at 543 nm with a helium-neon laser, and the emitted light was detected using a 560–615-nm band pass filter. The Alexa Fluor 647 was excited at 633 nm with another helium-neon laser, and the emitted light was detected using a 650-nm long pass filter. Channels were imaged separately. Resulting images were combined using ImageJ software.3 All experiments were done at least three times, imaging at least 15 cells/condition in each session.
Quantification of colocalization of GPR10 with eGFP-tagged Rab7 and Rab11 was done using the RG2B colocalization ImageJ plug-in as described (47, 48). Single cells were defined as regions of interest to avoid noise from untransfected cells and nonspecific staining. A minimum threshold pixel intensity of 100 was set for each channel in the eight-bit pictures to focus on clustering, and the minimum ratio for pixel intensity between the two channels was set to 0.5. Results are displayed as percent colocalization as determined by dividing the area of colocalization pixels by the total area over the threshold in the green channel reporting GFP-Rab localization. About 50 cells were used for quantification in each condition. Statistical significance was determined using Student's t test.
Surface ELISA
For ELISA-based trafficking experiments, FLAG-tagged surface proteins were labeled with 1 μg/ml M1 mouse anti-FLAG antibody for 30 min at 4 °C in parallel in two 96-well plates. In half of the wells on each plate, internalization was stimulated at 37 °C for 25 min as indicated. The wells on the other half of the plate were left at 37 °C and are referred to as non-treated. Subsequently, the action of the internalizing agent was terminated as indicated (β2AR, alprenolol (10 μm); DOR, naxolone (10 μm); TacDAT, staurosporine (1 μm)) using 37 °C medium on one plate and 4 °C medium on the other. One plate was left at 4 °C for 1 h to stop further trafficking, and the other plate was left at 37 °C to allow further trafficking. For GPR10, we had no potent antagonist, and cells were instead washed and left in DMEM 1965 for prolonged trafficking. Subsequently, cells were washed in DMEM 1965, fixed for 10 min at 4 °C, and washed twice in PBS before 30-min blocking in PBS + 5% goat serum and incubation with 0.5 μg/ml horseradish peroxidase-conjugated goat anti-mouse IgG (Thermo Scientific). Finally, cells were washed twice in PBS + 5% goat serum and twice in PBS before addition of SuperSignal® ELISA Femto Maximum Sensitivity Substrate (Thermo Scientific). The luminescence was detected in a Wallac Victor2 plate reader after 2 min. Internalization is expressed as the ratio of the surface signal from agonist- or PMA-internalized protein relative to the non-treated cells on the 4 °C plate. The percentage of recycling was calculated from the proportion of internalized receptor that was recovered at the cell surface. Statistical significance was determined using Student's t test.
RESULTS
PICK1 Co-clusters Only with Subset of Its Interaction Partners
It is well established that PICK1 can form characteristic juxtanuclear or perinuclear co-clusters with PDZ domain interaction partners, such as the GluA2 subunit of the AMPAR (8) and mGluR7 (16) in heterologous expression systems. In a recent analysis, we confirmed that indeed GluA2 can co-cluster with PICK1 in COS7 cells and demonstrated that the single transmembrane-spanning protein Tac (the α-subunit of the interleukin 2 receptor (49)) with the GluA2 tail fused to its C terminus (TacGluR2 C24) effectively co-clustered with PICK1 (44). When the last 24 residues from the C terminus of the DAT were fused to the Tac C terminus (TacDAT C24) (Fig. 1a), we also observed co-clustering with PICK1 in COS7 cells (44). To further explore the conditions determining clustering of PICK1, we established, using the Flp-In T-REx system from Invitrogen, an HEK293-derived cell line with tetracycline-inducible expression of eYFP-PICK1 (PICK1 with eYFP fused to the N terminus). In these cells (Flp-In T-REx 293 eYFP-PICK1), we observed marked clustering of eYFP-PICK1 when transfected with TacDAT C24 as in COS7 cells (Fig. 1b, upper panel). However, when we fused the entire DAT sequence to Tac and co-expressed the resulting construct (TacDAT) (43) (Fig. 1a) in the Flp-In T-REx 293 eYFP-PICK1 cells, we did not see any signs of eYFP-PICK1 clustering (Fig. 1b, lower panel). The absence of juxtanuclear clusters was most likely not because TacDAT was a non-functional, incorrectly folded protein (i.e. previous studies supported that this fusion protein has Vmax and Km values for [3H]dopamine uptake similar to WT DAT and undergoes, like WT DAT, constitutive and PMA-induced internalization (43)). Although non-tagged DAT was suggested to cluster with PICK1 (10), we also did not observe clusters of eYFP-PICK1 when we expressed non-tagged DAT in the Flp-In T-REx 293 eYFP-PICK1 cells (data not shown). Likewise, in one of our earlier studies, we did not see any convincing signs of clustering when DAT tagged at the N terminus with green fluorescent protein (GFP) was co-expressed with PICK1 (46).
FIGURE 1.
PICK1 co-clusters with Tac DAT C24 but not with TacDAT. a, schematic representation of the fusion proteins Tac DAT C24 and TacDAT. TacDAT C24 is a fusion protein between Tac and the 24 C-terminal residues of DAT with an N-terminal M1 antibody FLAG epitope. TacDAT is a head-to-tail fusion of Tac (α-subunit of the IL2 receptor) (green) with an N-terminal M1 antibody FLAG epitope (DYKDDDDK) and full-length DAT (blue) (43). b, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells induced with tetracycline and transiently expressing TacDAT C24 (top panel) or TacDAT (bottom panel). Cells were surface-labeled with Alexa Fluor 568-conjugated anti-FLAG antibody to visualize surface TacDAT C24 (top middle panel) and TacDAT (middle bottom panel) in transfected cells. The eYFP-PICK1 signal (green) is shown in the left panels and illustrates the lack of clustering and partial plasma membrane recruitment in TacDAT-transfected cells. In contrast, profound eYFP-PICK1 clustering was seen in TacDAT C24-transfected cells. Nuclei were highlighted in the top panel to demonstrate the juxtanuclear localization of the eYFP-PICK1 clusters. c, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells induced with tetracycline and transiently expressing TacDAT. Cells were surface-labeled with Alexa Fluor 568-conjugated anti-FLAG antibody and internalized with PMA for 25 min (top middle panel). Subsequently, the cells were treated with staurosporine (Stau; 1 μm) for 60 min to allow potential recycling (bottom middle panel). The eYFP-PICK1 signal (green) is shown in the left panel and illustrates the lack of marked clustering in the TacDAT-internalized cells. Representative images are shown from ∼15 cells visualized per condition in each experiment and over three separate experiments. The small squares mark areas that are shown enlarged inside large squares. Scale bar, 15 μm.
We showed earlier that when clustered intracellularly with PICK1 TacDAT C24 originates from the plasma membrane and thus that it is not sorted directly to the clusters from the Golgi apparatus (44). Because the constitutive internalization rate of TacDAT C24 is considerably higher than that of full-length DAT,4 we speculated that the internalized pool of full-length transporter (wild type DAT or TacDAT) could be too low to cause clustering of PICK1. To test this, we surface-labeled TacDAT with Alexa Fluor 543-conjugated M1 antibody and induced rapid internalization of the transporter by activating PKC with the phorbol ester PMA (50, 51). After 25 min of stimulation, substantial redistribution of TacDAT to vesicular compartments was seen in most cells; however, the massive clustering of eYFP-PICK1 seen when co-expressed with TacDAT C24 was never observed (Fig. 1c, upper panel). To allow the internalized transporter to travel further into the endosomal pathways, we treated cells with staurosporine to terminate the effects of PMA and waited for an additional 60 min. The transporter remained in vesicular compartments, but no major accumulation of YFP-PICK1 associated with these compartments was detected (Fig. 1c). Taken together, despite a C terminus with high affinity for the PICK1 PDZ domain, full-length DAT appears not to form the co-clusters that are observed with other interaction partners.
Only Interaction Partners Sorted to Rab11-positive Compartments Co-cluster with PICK1
Recently, we demonstrated that upon constitutive internalization both TacDAT and DAT with an HA tag in the second extracellular loop (HA-DAT) are sorted primarily to late endosomes/lysosomes and in part to a Rab4'positive “short loop” recycling pathway (43). Earlier it was also demonstrated that DAT internalized in response to PKC activation sorts to the lysosomal pathway and ends up being degraded (51, 52). However, according to our previous data, constitutively internalized TacDAT C24 is sorted to a Rab11-positive compartment and co-clusters with PICK1 in this compartment (44). Consequently, an explanation for the difference between TacDAT and TacDAT C24 in their ability to co-cluster with PICK1 would be that their postendocytic sorting properties are different and thus that it is the sorting properties of the PDZ binding partner and not PICK1 that determine whether co-clustering will occur or not.
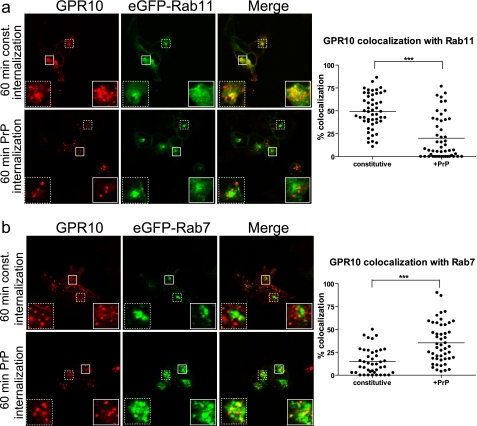
To further investigate this possibility, we turned to another interaction partner of PICK1, the G protein-coupled prolactin-releasing peptide receptor, or GPR10 (53). We introduced in this receptor an N-terminal M1 antibody FLAG epitope and transiently expressed the resulting construct in HEK293 cells. The receptor displayed a high degree of constitutive internalization as determined by exposure to Alexa Fluor 543-conjugated anti-FLAG M1 antibody for 60 min (Fig. 2a). The internalized receptor showed considerable colocalization with co-expressed eGFP-Rab11, a marker of the slow recycling pathway, and rather low colocalization with eGFP-Rab7, a marker of late endosomes (Fig. 2, compare a and b). Interestingly, when internalization was promoted by the agonist prolactin-releasing peptide (Thr-Pro-Asp-Ile-Asn-Pro-Ala-Trp-Tyr-Ala-Ser-Arg-Gly-Ile-Arg-Pro-Val-Gly-Arg-Phe), the receptor colocalized predominantly with eGFP-Rab7 and only occasionally with eGFP-Rab11 (Fig. 2, compare a and b). Quantification of the colocalization after thresholding both channels to intensities >100 (eight-bit pictures) showed a significant shift in sorting upon agonist treatment away from the recycling pathway (identified by eGFP-Rab11) (Fig. 2a) and toward the degradative pathway (identified by eGFP-Rab7) (Fig. 2b).
FIGURE 2.
Agonist-induced internalization of GPR10 by PrP redirects endocytic sorting away from Rab11-positive pathway toward Rab7-positive pathway. a, left, confocal laser scanning micrographs of HEK293 cells transiently expressing GPR10 and GFP-Rab11. Cells were surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C to label GPR10 before 60 min of constitutive (const.) internalization at 37 °C (top panel) or 60 min of agonist-induced internalization (1 nm PrP) (lower panel) at 37 °C. The GPR10 signal is shown in red, and the eGFP-Rab11 signal is in green. Small white squares mark areas that are shown enlarged inside large white squares. Right, quantification of colocalization between internalized GPR10 and eGFP-Rab11 after 60 min of constitutive (n = 52) and agonist-induced internalization (n = 52). Each dot represents a single cell, and horizontal bars indicate means. ***, p < 0.001, unpaired t test. Data were collected on three separate experimental days. b, left, confocal laser scanning micrographs of HEK293 cells expressing eGFP-Rab7 and GPR10. Cells were surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C to label GPR10 before 60 min of constitutive internalization at 37 °C (top panel) or 60 min of agonist-induced internalization (1 nm PrP) (lower panel) at 37 °C. The GPR10 signal is shown in red, and the eGFP-Rab7 signal is in green. Small white squares mark areas that are shown enlarged inside large white squares. Right, quantification of colocalization between internalized GPR10 and eGFP-Rab7 after 60 min of constitutive (n = 42) and agonist-induced internalization (n = 50). Each dot represents a single cell, and horizontal bars indicate means. ***, p < 0.001, unpaired t test. Data were collected on three separate experimental days.
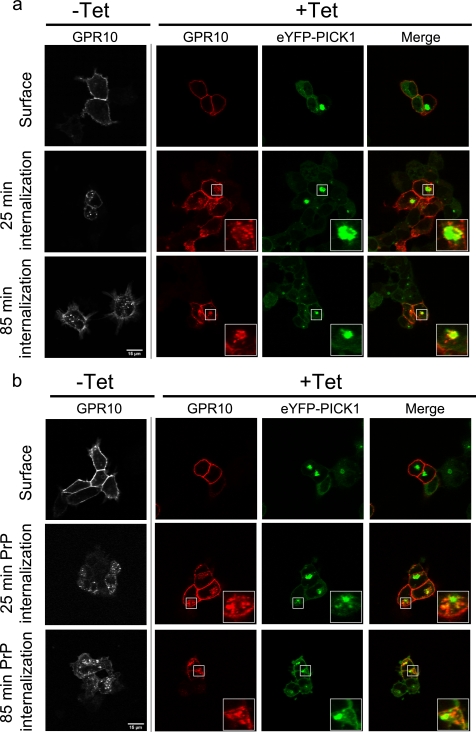
This remarkable property of GPR10 allowed us specifically to ask whether postendocytic sorting of the PDZ binding partner determines whether co-clustering will occur or not. To this end, GPR10 was expressed in the Flp-In T-REx 293 eYFP-PICK1 cells, and surface receptors were labeled by incubating with Alexa Fluor 543-conjugated anti-FLAG M1 antibody at 4 °C (Fig. 3, a and b, upper panels). Both in cells with tetracycline induction (+Tet) and without induction of eYFP-PICK1 expression (−Tet), we observed clear plasma membrane staining for GPR10 (Fig. 3, a and b, upper panels). In induced cells, eYFP-PICK1 (in green) was seen diffusely expressed in the cytoplasm as well as partly localized to the plasma membrane (Fig. 3, a and b, upper panels). In addition, eYFP-PICK1 was seen in large clusters, presumably formed as a result of the constitutive internalization of GPR10 prior to antibody labeling (Fig. 3, a and b, upper panels). Note that in the pictures shown the receptor is not visible in the clusters because the staining was done without membrane permeabilization.
FIGURE 3.
Constitutively internalized GPR10 co-clusters with eYFP-PICK1 in contrast to GPR10 internalized by agonist stimulation. a, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing GPR10. The pictures show constitutive internalization of GPR10 that was surface-labeled with Alexa Fluor 568-conjugated anti-FLAG antibody at 4 °C without tetracycline (−Tet) and with (+Tet) induced expression of eYFP-PICK1 (green) prior to constitutive internalization for 25 or 85 min at 37 °C. Panels starting from the left show the GPR10 signal in non-induced cells, GPR10 signal in tetracycline-induced cells (red), eYFP-PICK1 signal (green), and merge of the red and green signals. b, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing GPR10. The pictures show agonist-induced internalization of GPR10 that was surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C without (−Tet) and with (+Tet) induced expression of eYFP-PICK1 (green). Internalization was stimulated by agonist treatment (1 nm PrP) for 25 or 85 min at 37 °C. Panels starting from the left show the GPR10 signal in non-induced cells, GPR10 signal in tetracycline-induced cells (red), eYFP-PICK1 signal (green), and merge of red and green signals. Representative images are shown from ∼15 cells visualized per condition in each experiment and over three separate experiments. The small squares mark areas that are shown enlarged inside the large squares. Scale bars, 15 μm.
Constitutive internalization was next allowed by incubating the cells at 37 °C for 25 and 85 min (Fig. 3a, middle and lower panels). The antibody-labeled constitutively internalized GRP10 (shown to colocalize with eGFP-Rab11; see Fig. 2) trafficked to the eYFP-PICK1 clusters already after 25 min, and after an additional 60 min, the localization of GPR10 to these clusters was even more pronounced (Fig. 3a, middle and lower panels). In non-induced cells (−Tet), constitutively internalized GPR10 showed no tendency to accumulate in larger clusters (Fig. 3a, middle and lower panels).
Interestingly, when internalization was promoted by the agonist PrP for 25 min, vesicular GPR10 (shown to colocalize with GFP-Rab7; see Fig. 2) largely resided outside the eYFP-PICK1 clusters produced by previous constitutive internalization (Fig. 3b, middle panels). Stimulation for an additional 60 min still left most internalized GPR10 outside eYFP-PICK1 clusters (Fig. 3b). Importantly, the formation of co-clusters between GPR10 and eYFP-PICK1 was strictly dependent on an intact C-terminal PDZ binding sequence in GPR10 because clustering was completely abolished by disrupting the PDZ interaction by changing the C-terminal Val-Val-Leu sequence to Val-Val-Asp (supplemental Fig. S1). In summary, the data provide support for the idea that co-clustering with PICK1 is determined by the postendocytic sorting pattern of the PDZ domain binding partner and that PICK1 does not affect this sorting pattern.
Only Interaction Partners Recycling through Rab11-mediated Slow Recycling Pathway Cause PICK1 to Cluster
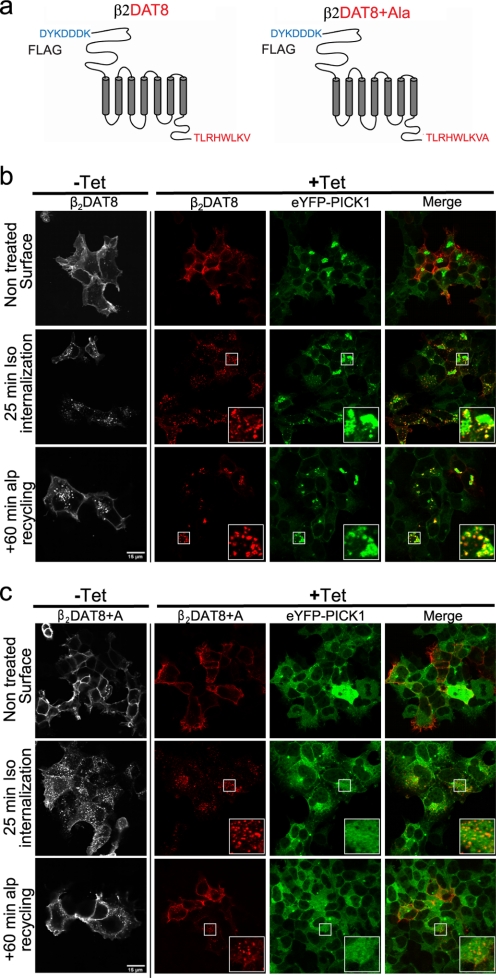
To address further whether the sorting properties of the PDZ binding partners determine the formation of PICK1 co-clusters, we fused the C-terminal 8 residues of DAT, which are sufficient to mediate interaction with PICK1,5 to the C terminus of FLAG-tagged β2AR (β2DAT8) (Fig. 4a) and of FLAG-tagged δ-opioid receptor (DOR DAT8) (Fig. 5a). The β2AR is known to recycle efficiently via Rab4- and Rab11-positive pathways (54, 55), whereas the δ-opioid receptor is sorted almost entirely to lysosomal degradation via the Rab7-positive pathway (56).
FIGURE 4.
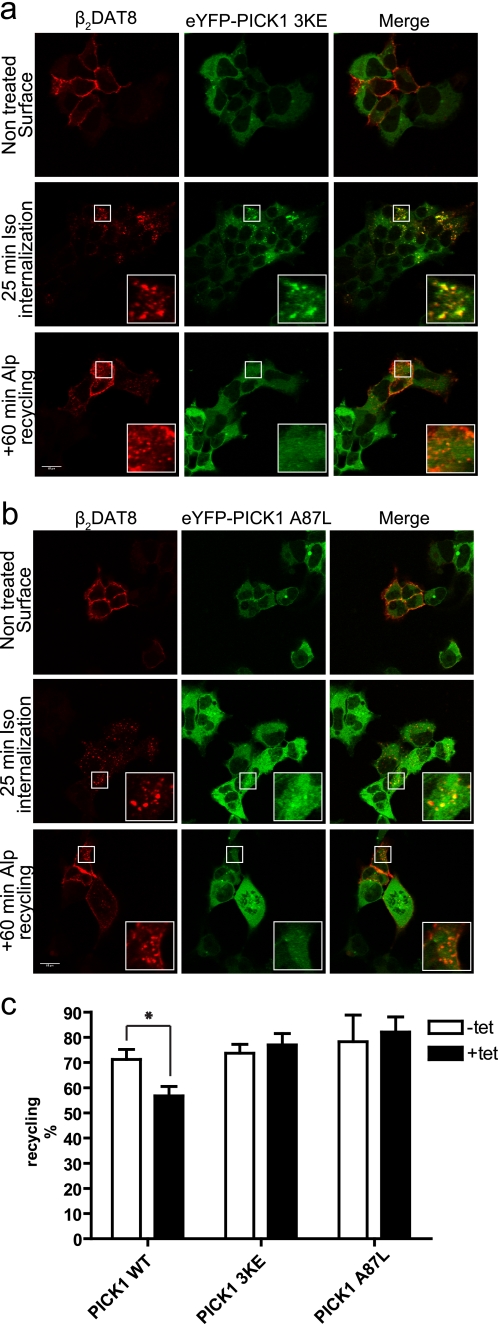
PICK1-binding chimeric receptor β2DAT8 clusters eYFP-PICK1 and traffics to PICK1-positive clusters upon agonist-induced internalization. a, schematic representation of β2DAT8 and β2DAT8 +Ala showing the N-terminal FLAG tag sequence followed by the full-length β2 adrenergic receptor and the last 8 PICK1-binding C-terminal residues of DAT. In β2DAT8 +Ala, PICK1 binding is disrupted due to the addition of an extra C-terminal alanine. b, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing β2DAT8. The pictures show agonist (isoproterenol)-induced internalization of β2DAT8 that was surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C without (−Tet) and with (+Tet) induced expression of eYFP-PICK1 (green). Internalization was stimulated by 10 μm Iso for 25 min at 37 °C or by 10 μm Iso for 25 min followed by a 60-min incubation at 37 °C with the antagonist Alp to allow recycling of receptors. Panels starting from the left show the β2DAT8 signal in non-induced cells, β2DAT8 signal in tetracycline-induced cells (red), eYFP-PICK1 signal (green), and merge of red and green signals. c, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing β2DAT8 +Ala. The pictures show conditions similar to those described for β2DAT8 above. Notably, β2DAT8 +Ala does not promote formation of perinuclear eYFP-PICK1 clusters. Pictures are representative of ∼25 cells visualized per condition in each experiment and over six separate experiments. The small squares mark areas that are shown enlarged inside the large squares. Scale bars, 15 μm.
FIGURE 5.
Chimeric receptor DOR DAT8 does not cluster eYFP-PICK1. a, schematic representation of DOR DAT8 showing the N-terminal FLAG tag sequence followed by the full-length DOR and the last 8 PICK1 binding C-terminal residues of DAT. b, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing DOR DAT8. The pictures show agonist-induced internalization of DOR DAT8 (compared with untreated control) that was surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C without (−Tet) and with (+Tet) induced expression of eYFP-PICK1 (green). Internalization was stimulated by 10 μm δ-opioid receptor agonist peptide DADLE for 25 min at 37 °C followed by a 60-min incubation at 37 °C with 10 μm antagonist Naxo to allow potential recycling of receptors. Panels starting from the left show the DOR DAT8 signal in non-induced cells, DOR DAT8 signal in tetracycline-induced cells (red), eYFP-PICK1 signal (green), and merge of red and green signals. Small white squares mark areas that are shown enlarged inside large white squares. Pictures are representative of ≈15 cells visualized per condition in each experiment and over five separate experiments. Scale bars, 15 μm.
In agreement with our hypothesis, expression of β2DAT8 caused eYFP-PICK1 to cluster very strongly (Fig. 4b, upper panels) (note again that the receptor is not seen in the clusters because the antibody labeling was done without membrane permeabilization). As for GPR10, these clusters are presumably formed as a result of constitutive receptor internalization prior to antibody labeling. Importantly, although the β2AR shows rather low constitutive internalization, exposure to Alexa Fluor 568-conjugated M1 antibody for an extended period (16 h) demonstrated considerable constitutive trafficking of β2DAT8 from the plasma membrane to eYFP-PICK1 clusters (supplemental Fig. S2A).
This clustering of eYFP-PICK1 in β2DAT8 cells was dependent on PDZ domain interactions because a non-PDZ binding mutant with an alanine added to the C terminus of the DAT C8 tail (β2DAT8 +Ala) (46) did not cause clustering of eYFP-PICK1 (Fig. 4c, upper panels). Upon treatment with the agonist Iso (25 min), surface-labeled β2DAT8 was internalized to the periphery of the clusters (Fig. 4b, middle panel), and after prolonged incubation (Iso for 25 min + 60 min in the presence of the antagonist Alp), β2DAT8 was found almost completely colocalized in clusters with eYFP-PICK1 (Fig. 4b, bottom panel) (44). Importantly, this was similar to the pattern observed for the constitutive trafficking of the endogenous PICK1 interaction partner GPR10 (see Fig. 3) and for TacDAT C24 (44). It is also important to note that β2DAT8 displayed the same postendocytic sorting pattern as β2AR itself; i.e. internalized β2DAT8 was mainly found in Rab11-positive endosomes (supplemental Fig. S2B).
In contrast to β2DAT8, DOR DAT8 (Fig. 5a) did not cause eYFP-PICK1 to cluster any more than it did on its own (Fig. 5b, upper panel). In the plasma membrane, we did see co-localization of DOR DAT8 and eYFP-PICK1; however, after internalization in response to the peptide DOR agonist DADLE (25 min; 10 μm), we did not see any signs of recruitment of eYFP-PICK1 to DOR DAT8-positive vesicles or any indication of eYFP-PICK1 clustering (Fig. 5b, middle panels), and the same was true after an additional 60 min of antagonist treatment (25 min DADLE + 60 min Naxo) (Fig. 5b, bottom panels). Interestingly, this pattern was similar to the trafficking pattern observed for GPR10 in the presence of agonist; in fact, it was clearer because both the constitutive internalization and agonist-induced internalization of DOR DAT8 were directed to the degradative pathway. Moreover, it was similar to what was seen for Tac-DAT, which also was unable to cluster with eYFP-PICK1.
One possible reason for the absence of co-clustering between PICK1 and interaction partners sorting to the degradative pathway could be that these complexes only form transiently before they are degraded. To test this, we incubated cells co-expressing TacDAT or DOR DAT8 and eYFP-PICK1 with leupeptin (100 μm) for 16 h to block lysosomal degradation. As expected, permeabilization and immunostaining revealed marked intracellular accumulation of vesicular TacDAT and DOR DAT8 inside the cells, however, still without showing recruitment of eYFP-PICK1 to these structures. In contrast, eYFP-PICK1 was still effectively clustered by GPR10 in the presence of leupeptin (supplemental Fig. S3).
PICK1 Promotes Intracellular Accumulation of Recycling Interaction Partners
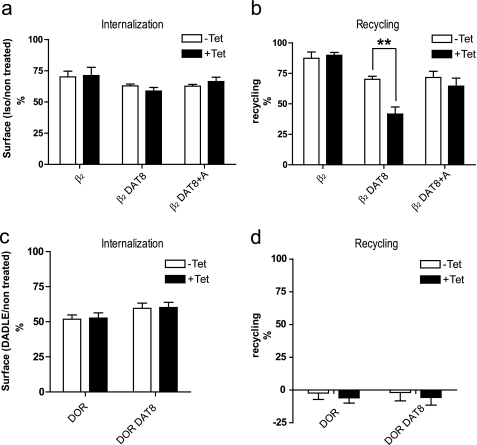
PICK1 was recently suggested to impair recycling of AMPARs internalized by NMDA in hippocampal neurons (38, 42). Based on the clustering of internalized β2DAT8 but not of internalized DOR DAT8 with eYFP-PICK1, we speculated that PICK1 might be able to selectively impair recycling of binding partners sorted to Rab11-positive recycling endosomes. To address this, we performed quantitative surface ELISA recycling experiments according to previously published protocols (57, 58) using the extracellular N-terminal FLAG tag in β2DAT8 and asked whether inducible expression of eYFP-PICK1 affected this process. β2AR, β2DAT8, and β2DAT8 +Ala showed similar surface expression, and induction of eYFP-PICK1 expression (+Tet) did not affect their basal surface expression (data not shown). Because binding of NSF (59, 60) and the PDZ domain protein NHERF1 (Na+/H+ exchanger regulatory factor 1) (61, 62) to the extreme β2AR C terminus was suggested previously to regulate recycling of the β2AR, we first compared the trafficking of the fusion proteins β2DAT8 and β2DAT8 +Ala with β2AR. Just like β2AR itself, the fusion proteins were internalized robustly in response to 10 μm Iso (25 min 10 μm), and notably, this was not affected by induction of eYFP-PICK1 expression (+Tet); i.e. both with and without tetracycline induction, well over 25% of the surface-expressed receptors were internalized during stimulation over 25 min in response to 10 μm Iso (Fig. 6a).
FIGURE 6.
PICK1 impairs recycling of agonist-internalized β2DAT8 but not DOR DAT8, which sorts to degradation. a and b, flp-In T-REx 293 eYFP-PICK1 cells transiently expressing β2, β2DAT8, or β2DAT8 +Ala with (black bars; +Tet) and without (white bars; −Tet) tetracycline-induced expression of eYFP-PICK1 were surface-labeled with anti-FLAG M1 antibody prior to stimulation of internalization with agonist (10 μm Iso for 25 min). Subsequently, cells were treated with the antagonist Alp (60 min at 10 μm) to allow recycling. Surface receptor immunoreactivity was determined by surface ELISA as described under “Experimental Procedures.” Internalization (a) refers to the fractional reduction of surface receptor in response to 25 min of agonist exposure compared with non-treated cells. Recycling (b) refers to the fractional recovery of surface receptor following antagonist incubation for 1 h. Data represent means ± S.E. from four independent experiments. c and d, Flp-In T-REx 293 eYFP-PICK1 cells transiently expressing DOR or DOR DAT8 with (black bars; +Tet) and without (white bars; −Tet) tetracycline-induced expression of eYFP-PICK1 were surface-labeled with anti-FLAG M1 antibody prior to stimulation of internalization with 10 μm δ-opioid receptor agonist peptide DADLE for 25 min followed by a 60-min incubation with 10 μm antagonist Naxo to allow potential recycling of receptors. Surface receptor immunoreactivity was determined by surface ELISA as described under “Experimental Procedures.” Internalization (c) refers to the fractional reduction of surface receptor in response to 25 min of agonist exposure compared with non-treated cells. Recycling (d) refers to the fractional recovery of surface receptor following antagonist incubation for 1 h. Data represent means ± S.E. from four independent experiments. **, p < 0.01, unpaired t test.
Upon incubation with the antagonist alprenolol for an additional 60 min (25 min with 10 μm Iso + 60 min with 10 μm Alp) all three constructs demonstrated potent recycling (expressed as the percentage of internalized receptors recovered at the surface) (Fig. 6b). However, both DAT8 and DAT8 +Ala showed reduced recycling as compared with wild type β2AR (β2DAT8, 70 ± 2% and β2DAT8, 71 ± 5% versus β2AR, 87 ± 5%; means; n = 4) (Fig. 6b). This impairment of recycling is in accordance with the previous results and likely reflects the compromised interaction with NHERF and/or NSF (59–62). Nonetheless, β2DAT8 and β2DAT8 +Ala were still recycled rather efficiently, meaning that it would be possible also to assess the ability of PICK1 to regulate this process. As suggested by the immunofluorescence, induction of eYFP-PICK1 expression indeed reduced the recycling of β2DAT8 without affecting β2DAT8 +Ala and β2AR (25 min with 10 μm Iso + 60 min with 10 μm Alp); i.e. recycling of β2DAT8 was reduced from ∼70% down to ∼40% with no effect on β2DAT8 +Ala and β2AR (Fig. 6b). Interestingly, this ability of PICK1 to impair recycling after active internalization of the receptor is fully analogous to the recently reported function of PICK1 in relation to AMPAR trafficking (38, 42).
We then performed analogous experiments with DOR and DOR DAT8 and confirmed that both constructs were internalized in response to incubation with agonist (10 μm DADLE for 25 min) (Fig. 6c); however, no recovery of surface receptors could be detected upon incubation for an additional 60 min with the antagonist Naxo (25 min with 10 μm DADLE + 60 min with 10 μm Naxo) for either DOR or DOR DAT8 (Fig. 6d). This is in agreement with little or no recycling and thus in agreement with previously published results for this receptor (56). Similar to what we observed for β2DAT8, induction of eYFP-PICK1 expression did not affect surface expression or internalization (10 μm DADLE for 25 min) of DOR and DOR DAT8, but unlike β2DAT8, eYFP-PICK1 expression also did not affect surface levels after an additional 60 min of incubation with antagonist (25 min with 10 μm DADLE + 60 min with 10 μm Naxo).
We performed similar experiments with the endogenous binding partners DAT and GPR10. For both DAT (TacDAT) and GPR10, induction of PICK1 expression had no detectable effect on their basal surface expression (supplemental Fig. S4). For internalization of TacDAT, we treated the cells for 25 min with PMA, which lead to an ∼35% reduction in surface levels (supplemental Fig. S5). No surface expression was recovered after treatment with the PKC inhibitor staurosporine for 60 min (supplemental Fig. S5) in agreement with targeting of PMA-internalized transporter to degradation (43, 51, 52). For internalization of GPR10, we incubated the cells with PrP for 20 min, and as suggested by the predominant localization of internalized receptors to Rab7-positive compartments in the presence of the agonist PrP (see above), we also saw no recovery of surface expression (supplemental Fig. S5). For both TacDAT and GPR10, induction of eYFP-PICK1 expression was unable to affect trafficking of the two different constructs similar to what we observed for the chimeric PICK1 binding partner DOR DAT8 above (supplemental Fig. S5). Taken together, the data suggest that PICK1 specifically regulates trafficking of interaction partner trafficking through the long loop Rab11-positive recycling pathway.
To further test whether PICK1 affected internalization or recycling rates of β2DAT8, we repeated the trafficking ELISA in the presence of the recycling inhibitor monensin (63, 64). Monensin had little effect on internalization (compare supplemental Fig. S6A with Fig. 6a). However, monensin strongly reduced the recycling of β2DAT8, β2DAT8 +Ala, and β2AR (compare supplemental Fig. S6B with Fig. 6b). Moreover, the effect of eYFP-PICK1 expression on β2DAT8 trafficking was blunted by monensin, strongly supporting that PICK1 specifically interferes with recycling (compare supplemental Fig. S6B with Fig. 6b).
As described above, the fusion of DAT8 to the C terminus of β2AR was expected to compromise the binding of NSF and NHERF, which are both involved in regulating the recycling of β2AR (59–62). The decrease in recycling of the fusion proteins (see Fig. 6b) is consistent with a compromised interaction with these proteins. However, to substantiate that PICK1 actively decreases recycling and does not just compete with these endogenous β2AR binding partners, we performed the recycling experiment with a 10-fold higher expression of the receptors (as determined from surface expression in the ELISA) by increasing the DNA concentration in the transfections. At this expression level, recycling was no longer significantly different for β2DAT8, β2DAT8 +Ala, and β2AR, suggesting that endogenous factors affecting recycling were stoichiometrically outnumbered by receptors. Nonetheless, induction of eYFP-PICK1 overexpression could still selectively decrease recycling of β2DAT8 (supplemental Fig. S7). The fusion of the DAT C-terminal sequence to β2AR should block the interaction of the extreme β2AR C terminus with the PDZ domains of NHERF because NHERF has a strong preference for type I PDZ ligands and ligands terminating with leucine (5, 65). We also specifically disrupted the interaction of β2ARwith NSF using a known single mutation, L412A (59). In our hands, this mutation showed a tendency toward decreased recycling, suggesting that β2DAT8 retained some NSF binding. Nonetheless, eYFP-PICK1 expression still reduced recycling of β2DAT8 L412A as efficiently as that of β2DAT8 (supplemental Fig. S8). In summary, these data show that PICK1 does not reduce recycling of the β2DAT8 by interfering with binding of other interaction partners, such as NHERF or NSF.
Inhibition of Recycling by PICK1 Is Dependent on PDZ and BAR Domains
PICK1 contains two important functional domains, the protein-binding PDZ domain and the lipid-binding BAR domain. We next asked whether these domains were critical for the ability of PICK1 to inhibit recycling of its interaction partners. To this end, we made two new cells lines, one with inducible expression of eYFP-PICK1 containing the 3KE mutation in the BAR domain (11, 44) (Flp-In T-REx 293 eYFP-PICK1 3KE) and one with inducible expression of eYFP-PICK1 containing a mutation in the ligand binding pocket of the PDZ domain (A87L) (5) (Flp-In T-REx 293 eYFP-PICK1 A87L).
The eYFP-PICK1 3KE mutation did not cluster in cells expressing β2DAT8 (Fig. 7a). It was, however, initially recruited quite efficiently to internalized β2DAT8-containing vesicles (Iso for 25 min), but after prolonged trafficking of β2DAT8 (Iso for 25 min + Alp for 60 min), no clustering was seen (Fig. 7a). In agreement with these imaging data, eYFP-PICK1 3KE was unable to slow down β2DAT8 recycling according to the ELISA (Fig. 7c). These observations demonstrate the unequivocal importance of the BAR domain for the ability of PICK1 to regulate recycling of an interaction partner. Furthermore, they suggest that the BAR domain serves its function after the interaction partner reaches the recycling endosomes.
FIGURE 7.
Impairment of β2DAT8 recycling by PICK1 is dependent on both intact PDZ binding crevice and BAR domain. a, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 3KE cells induced with tetracycline and transiently expressing β2DAT8. The pictures show untreated cells (top) and agonist (isoproterenol at 10 μm)-induced internalization of β2DAT8 that was surface-labeled with Alexa Fluor 568-conjugated anti-FLAG M1 antibody at 4 °C prior to stimulation of internalization at 37 °C by 10 μm Iso for 25 min (middle) or with 10 μm Iso for 25 min followed by a 60-min incubation with the antagonist Alp (bottom) to allow recycling of receptors. Left panels, β2DAT8 signal (red); middle panels, eYFP-PICK1 3KE signal (green); right panels, merge of red and green signals. b, confocal laser scanning micrographs of Flp-In T-REx 293 eYFP-PICK1 A87L cells induced with tetracycline and transiently expressing β2DAT8. The pictures show conditions similar to those described for eYFP-PICK1 3KE above. Pictures are representative of ≈20 cells visualized per condition in each experiment and over four separate experiments. Scale bars, 15 μm. Small white squares mark areas that are shown enlarged inside large white squares. c, Flp-In T-REx 293 eYFP-PICK1 cells, Flp-In T-REx 293 eYFP-PICK1 3KE cells, or Flp-In T-REx HEK293 eYFP-PICK1 A87L cells transiently expressing β2DAT8 with (black bars; +Tet) and without (white bars; −Tet) tetracycline induction were surface-labeled with anti-FLAG M1 antibody prior to stimulation of internalization with agonist (10 μm Iso for 25 min). Subsequently, cells were treated with the antagonist Alp (60 min at 10 μm) to allow recycling. Surface receptor immunoreactivity was determined by surface ELISA as described under “Experimental Procedures.” Data are shown as recycling expressed as the fractional recovery of surface receptor following antagonist incubation for 1 h. Neither PICK1 3KE nor PICK1 A87L retained the ability to impede recycling of β2DAT8. Data represent means ± S.E. from four independent experiments. *, p < 0.05, unpaired t test.
The eYFP-PICK1 A87L mutation, which previously was shown to disrupt binding of the DAT C-terminal peptide (5), also completely blocked PICK1 function (Fig. 7b). The immunofluorescence pictures did not show any sign of colocalization of β2DAT8 with eYFP-PICK1 A87L (Fig. 7b). In addition, induction of eYFP-PICK1 A87L expression had no effect on β2DAT8 recycling, altogether confirming that indeed binding to the PDZ domain is essential for the observed effects of PICK1 on binding partner trafficking.
DISCUSSION
PICK1 has been proposed to regulate the function of its PDZ domain binding partners in several different ways. In particular, there has been focus on the role of PICK1 in regulating trafficking of its binding partners including modulating their surface stability (10, 12, 19, 20, 23–27), promoting their internalization (39–41), or inhibiting their recycling rates (38, 42). It is possible that the picture is blurred because PICK1 has been studied in many different and often complex integrated systems, such as brain slices and primary neuronal cultures. It is desirable to study the function of proteins in their natural environment, but multiple simultaneous protein-protein interactions and methodological limitations can make it difficult to dissect specific roles of the individual protein.
Here, we decided to study the functional role of PICK1 in a heterologous cell line with inducible expression of PICK1, which should permit delineation of PICK1 functions in a more isolated context. Taken together, our data provide evidence that at least in part the functional role of PICK1 is determined by the cargo of the PDZ domain. First, we found that only binding partners sorted to the slow Rab11-positive recycling pathway formed characteristic perinuclear co-clusters with PICK1. This suggested a function of PICK1 specifically in the Rab11 pathway, and in agreement, we observed that PICK1 failed to affect trafficking of the natural PDZ binding partners DAT and GPR10 as well as the artificial binding partner DOR DAT8, which all sorted to degradation. In contrast, PICK1 expression markedly reduced recycling rates of β2DAT8, which recycled via a Rab11-mediated pathway. Such a compartmentalized function of PICK1 might represent one possible explanation for the observed discrepancies in PICK1 function; i.e. only binding partners trafficking through a Rab11-positive recycling compartment will be affected by this function of PICK1.
It remains a prominent idea that PICK1 actively internalizes AMPARs (18, 39–41, 66). However, PICK1 has not been shown to internalize any other of its several interaction partners, and consonant with this, PICK1 did not increase internalization rates of any of the natural or artificial interaction partners in the present study. This also concurs with other recent studies. Using a fusion construct between the pH-sensitive GFP variant pHluorin and the AMPAR GluA2 subunit (pHluorinGluR2), it was found that internalization in response to NMDA receptor stimulation was identical in dissociated hippocampal neurons from wild type and PICK1 knock-out mice (38). In contrast, recycling of pHluorinGluR2 was accelerated in neurons from knock-out mice in agreement with a role of PICK1 in diminishing recycling (38). More recently, this was further supported by our observation that a small molecule inhibitor of the PICK1 PDZ domain accelerated recycling of pHluorinGluR2 in the same setup (67). In this study, we generalize these observations by showing that the effect on recycling also can be observed for an unrelated integral membrane protein (β2AR) in a heterologous cell line if an appropriate PDZ binding sequence is transferred to the C terminus of the protein and if the protein is sorted to a Rab11-positive recycling pathway. Indeed, previous observations support that AMPARs sort to a Rab11-positive compartment (68), and although the trafficking of AMPAR and β2AR has slightly different intrinsic time courses, the effect of PICK1 appears similar for the two proteins. Thus, in essence, we have generated a setup where PICK1 gives rise to an agonist-dependent decrease in surface expression of a membrane protein that lasts for more than 85 min. Although it is difficult to compare heterologous cells with differentiated neurons, it seems reasonable to consider that inhibition of membrane reinsertion should fully be able to explain the role of PICK1 in AMPAR LTD (12, 24, 28–32).
Our data also suggest a correlation between regulation of recycling and formation of Rab11-positive intracellular clusters. Conceivably, the clusters represent fused elements of the recycling endosomes; however, whether cluster formation is required for the effect of PICK1 on recycling or it represents an associated event is unclear. In relation to the AMPAR, it is tempting to speculate that the clusters not only contain the AMPAR accumulated intracellularly during LTD but also could be considered an equivalent of the functional reservoirs of AMPARs that are recruited to the plasma membrane during long term potentiation. Thus, in the absence of PICK1, an intracellular recruitable pool of AMPAR will never develop in concurrence with the recent finding that long term potentiation is compromised in PICK1 knock-out mice and can be reduced by a specific blocker of the PICK1 PDZ domain (28, 29, 67).
The inhibition of β2DAT8 recycling by PICK1 was dependent on an intact PDZ binding sequence because the effect was absent when an extra alanine was added to the C terminus (β2DAT8 +Ala). This was further supported by the PICK1 A87L mutation that discretely disrupts binding of interaction partners due to the larger size of the leucine side chain that fills out the P-2 pocket of the PDZ domain (5). Surprisingly, it was suggested in a recent study that a direct interaction between the C terminus of GluA2 and the PICK1 PDZ domain was not required for the function of PICK1 in NMDA-induced AMPAR trafficking and LTD (42). We have no immediate explanation for this discrepancy. It is possible that the situation is more complex in a neuronal environment; however, our use of an “isolated” cell system demonstrates that indeed PICK1 alone has the capacity to regulate trafficking of its binding partners in a PDZ domain-dependent fashion. Moreover, several previous studies have argued that also in a neuronal environment a direct PDZ-mediated interaction is important (24, 30, 31). The discrepancy might somehow relate to the use of different mutations. Whereas our observations are based on the discrete mutation of Ala-87 to Leu, Citri et al. (42) base their conclusions on mutation of either Lys-27/Glu-28 to alanines or Lys-27 to glutamate.
The inhibition by PICK1 of β2DAT8 recycling was furthermore dependent on the functional integrity of the BAR domain. This finding directly links BAR domain function to the role of PICK1 in regulating trafficking of its interaction partners. It is tempting to conclude that it is the lipid binding capacity of the BAR domain that is critical for the observed effect (11, 13, 69); however, according to a recent study, the BAR domain is also able to bind actin, and the 3KE mutation affects both functionalities (41). BAR domains are furthermore involved in lipid tubule-assisted polymerization (70) and in the facilitation of interaction with small GTPases (71), and it is unknown how the 3KE mutation affects these putative functions.
Our experiments also evaluated in a simple and quantitative way the presumed effects of PICK1 on surface expression and internalization rates of the interacting integral membrane proteins. Interestingly, PICK1 did not significantly affect steady state surface expression of any of the interaction partners tested. In the case of DAT, this is in conflict with previous results claiming that transient co-expression of PICK1 with DAT increased the uptake capacity of DAT presumably due to increased DAT surface levels (10). Previously, we have been unable to reproduce this finding (46), and the present data suggest that PICK1 does not affect DAT surface levels at least in heterologous cells. Conflicting results have also been reported for the ability of PICK1 to regulate the basal level of AMPARs in neurons (12, 23–29). We have no definite explanation for these discrepancies; however, we would suggest that PICK1 has the ability to regulate the basal surface level of interaction partners that undergo constitutive recycling to the plasma membrane but only if reinsertion via the recycling pathway constitutes a considerable part of the total number of receptors inserted into the membrane.
Other PDZ domain proteins have also been implicated in regulating trafficking of their membrane protein binding partners. However, whereas PICK1 appears to be critical for clustering and inhibiting recycling of its PDZ binding partners, these proteins have usually been associated with sorting their membrane protein binding partners either in the biosynthetic pathway (e.g. the SAP97-CASK (calcium/calmodulin-dependent serine protein kinase) complex) (72) or in the postendocytic pathway (e.g. NHERF and SNX27) (61, 73). NHERF is probably one of the best studied of these proteins and is believed to sort the β2AR to recycling through an interaction with actin (62, 74). Because PICK1 does not cluster with DOR DAT8 and does not promote its recycling, it seems that PICK1 is unable to sort interaction partners to a recycling compartment like NHERF despite having a PDZ together with a putative actin binding domain (in this case the BAR domain). Nonetheless, BAR domain proteins of the sorting nexin (SNX) family have been implicated in endosomal sorting (75). Of these, SNX4 functionally resembles NHERF by being important for sorting of e.g. the transferrin receptor from early endosomes to recycling endosomes. An SNX4-like function of PICK1 is unlikely for the same reasons as discussed for NHERF; however, we cannot completely exclude that PICK1 slows down recycling rates of interaction partners by sorting them between the recycling pathways, e.g. by directing recycling from the fast Rab4 to the slow Rab11 recycling pathway as suggested for the GRIP1-associated PDZ-like protein GRASP-1 (76).
The role of PICK1 in regulating recycling might be explained simply by a stabilization of PICK1 in complex with its binding partner in a Rab11-positive recycling compartment in a BAR domain-dependent fashion (Fig. 8). From our data, it follows that PICK1 does not by itself reside in this compartment but is brought there by its PDZ cargo. In light of the previously proposed “autoinhibition” hypothesis for the PICK1 BAR domain (11, 12, 44), this recruitment to a membrane compartment by its binding partner would be predicted to unmask the membrane binding capacity of the BAR domain (44). In this way, PICK1 will function as a compartment-specific anchor only activated by interaction partners sorted to Rab11-dependent recycling (Fig. 8). Finally, it should be considered that via the lipid deforming capacity of the BAR domain, PICK1 might also be capable of affecting the shape of the membranes making up the recycling endosomes and thereby affect fusion or fission events in general. In such a scenario, the trafficking of other integral membrane proteins through the recycling compartments would also be affected by PICK1. In the context of the AMPAR, this would suggest a mechanism by which internalized AMPAR could trigger a general modulation of trafficking in the dendrites via PICK1.
FIGURE 8.
Putative model illustrating PDZ cargo-determined function of PICK1 in regulating trafficking of its binding partners. a, PICK1 PDZ binding partners (“PDZ cargo”), such as the DAT and agonist-internalized GPR10 as well as the DOR engineered to bind PICK1 (DOR DAT8), are sorted upon internalization primarily to late endosomes and subsequently to lysosomal degradation. In contrast, PICK1 PDZ binding partners, such as constitutively internalized GPR10 and AMPAR as well as β2AR engineered to bind PICK1 (β2DAT8), are sorted to the Rab11-dependent long loop recycling pathway. b, PICK1 will be recruited to the plasma membrane by its different PDZ cargos independently of their postendocytic sorting pattern. At the plasma membrane, PICK1 might serve a variety of different functions in relation to these cargos including e.g. bringing PKCα in close proximity to regulate their phosphorylation or bind other PDZ cargos. Our data provide no evidence that PICK1 affects internalization of its PDZ cargo, and our data do not provide any evidence that PICK1 acts as a sorting module and thus affects their postendocytic sorting pattern. Also, our data suggest that PICK1 does not reside by itself in any recycling pathway but is brought there by its cargo. Thus, the role of PICK1 in regulating recycling might be explained simply by a stabilization of PICK1 in complex with its PDZ cargo in a Rab11-positive recycling compartment in a BAR domain-dependent fashion. According to the previously proposed autoinhibition hypothesis for the PICK1 BAR domain, this recruitment to a membrane compartment by its cargo would unmask the membrane binding capacity of the BAR domain, leading to clustering of PICK1 with its cargo. In this way, PICK1 might function as a compartment-specific anchor only activated by interaction partners sorted to Rab11-dependent recycling. It is possible that this function also involves other proteins, such as the small GTPases ARF1/3 or neuronal calcium sensor 1 (NCS1), and/or direct interaction with actin filaments, but further studies are required to clarify this issue. Finally, it should be considered that via the lipid deforming capacity of the BAR domain, PICK1 once brought to the recycling endosomes and activated might also be capable of affecting the shape of the membranes, thereby affecting fusion or fission events in this compartment in general.
Supplementary Material
Acknowledgments
We thank Dr. Mark von Zastrow for the pcDNA3.1 FLAG-β2-adrenergic and opioid receptor plasmids and Dr. Katherine W. Roche for pEGFP-Rab7 and pEGFP-Rab11 plasmids. We thank Nabeela Khadim and Donna Czerny for excellent technical assistance and Mette Rathje for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 DA 12408 (to U. G.). This work was also supported by the Lundbeck Foundation (to U. G.), the Novo Nordisk Foundation, the Danish Medical Research Councils (to U. G. and K. M.), and the University of Copenhagen Program of Excellence (BioScaRT) (to U. G.).

This article contains supplemental Figs. S1–S8.
W. S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD.
J. Eriksen, K. L. Madsen, and U. Gether, unpublished data.
K. L. Madsen and U. Gether, unpublished observation.
- PICK1
- protein interacting with C kinase 1
- DAT
- dopamine transporter
- eGFP
- enhanced GFP
- PDZ
- PSD-95/Discs large/ZO-1
- BAR
- Bin/amphiphysin/Rvs
- β2AR
- β2-adrenergic receptor
- GPR10
- prolactin-releasing peptide receptor
- AMPAR
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type ionotropic glutamate receptor
- LTD
- long term depression
- DOR
- δ-opioid receptor
- NSF
- N-ethylmaleimide-sensitive fusion protein
- eYFP
- enhanced yellow fluorescent protein
- 3KE
- K251E,K252E,K257E
- PrP
- prolactin-releasing peptide
- Iso
- isoproterenol
- DADLE
- [d-Ala2,d-Leu5]enkephalin
- PMA
- phorbol 12-myristate 13-acetate
- Alp
- alprenolol
- Naxo
- naxolone
- Tet
- tetracycline
- NHERF
- Na+/H+ exchanger regulatory factor
- SNX
- sorting nexin.
REFERENCES
- 1. Dev K. K. (2007) PDZ domain protein-protein interactions: a case study with PICK1. Curr. Top. Med. Chem. 7, 3–20 [DOI] [PubMed] [Google Scholar]
- 2. Xu J., Xia J. (2006) Structure and function of PICK1. Neurosignals 15, 190–201 [DOI] [PubMed] [Google Scholar]
- 3. Staudinger J., Lu J., Olson E. N. (1997) Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-α. J. Biol. Chem. 272, 32019–32024 [DOI] [PubMed] [Google Scholar]
- 4. Staudinger J., Zhou J., Burgess R., Elledge S. J., Olson E. N. (1995) PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 128, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Madsen K. L., Beuming T., Niv M. Y., Chang C. W., Dev K. K., Weinstein H., Gether U. (2005) Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem. 280, 20539–20548 [DOI] [PubMed] [Google Scholar]
- 6. Beuming T., Skrabanek L., Niv M. Y., Mukherjee P., Weinstein H. (2005) PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 21, 827–828 [DOI] [PubMed] [Google Scholar]
- 7. Bassan M., Liu H., Madsen K. L., Armsen W., Zhou J., Desilva T., Chen W., Paradise A., Brasch M. A., Staudinger J., Gether U., Irwin N., Rosenberg P. A. (2008) Interaction between the glutamate transporter GLT1b and the synaptic PDZ domain protein PICK1. Eur. J. Neurosci. 27, 66–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xia J., Zhang X., Staudinger J., Huganir R. L. (1999) Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron 22, 179–187 [DOI] [PubMed] [Google Scholar]
- 9. Dev K. K., Nakajima Y., Kitano J., Braithwaite S. P., Henley J. M., Nakanishi S. (2000) PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J. Neurosci. 20, 7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torres G. E., Yao W. D., Mohn A. R., Quan H., Kim K. M., Levey A. I., Staudinger J., Caron M. G. (2001) Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron 30, 121–134 [DOI] [PubMed] [Google Scholar]
- 11. Lu W., Ziff E. B. (2005) PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron 47, 407–421 [DOI] [PubMed] [Google Scholar]
- 12. Jin W., Ge W. P., Xu J., Cao M., Peng L., Yung W., Liao D., Duan S., Zhang M., Xia J. (2006) Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci. 26, 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 [DOI] [PubMed] [Google Scholar]
- 14. Madsen K. L., Bhatia V. K., Gether U., Stamou D. (2010) BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 584, 1848–1855 [DOI] [PubMed] [Google Scholar]
- 15. Baer K., Bürli T., Huh K. H., Wiesner A., Erb-Vögtli S., Göckeritz-Dujmovic D., Moransard M., Nishimune A., Rees M. I., Henley J. M., Fritschy J. M., Fuhrer C. (2007) PICK1 interacts with α7 neuronal nicotinic acetylcholine receptors and controls their clustering. Mol. Cell. Neurosci. 35, 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boudin H., Doan A., Xia J., Shigemoto R., Huganir R. L., Worley P., Craig A. M. (2000) Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron 28, 485–497 [DOI] [PubMed] [Google Scholar]
- 17. Torres R., Firestein B. L., Dong H., Staudinger J., Olson E. N., Huganir R. L., Bredt D. S., Gale N. W., Yancopoulos G. D. (1998) PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron 21, 1453–1463 [DOI] [PubMed] [Google Scholar]
- 18. Hanley J. G. (2006) Molecular mechanisms for regulation of AMPAR trafficking by PICK1. Biochem. Soc. Trans. 34, 931–935 [DOI] [PubMed] [Google Scholar]
- 19. Suh Y. H., Pelkey K. A., Lavezzari G., Roche P. A., Huganir R. L., McBain C. J., Roche K. W. (2008) Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 58, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin W., Shen C., Jing L., Zha X. M., Xia J. (2010) PICK1 regulates the trafficking of ASIC1a and acidotoxicity in a BAR domain lipid binding-dependent manner. Mol. Brain 3, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leonard A. S., Yermolaieva O., Hruska-Hageman A., Askwith C. C., Price M. P., Wemmie J. A., Welsh M. J. (2003) cAMP-dependent protein kinase phosphorylation of the acid-sensing ion channel-1 regulates its binding to the protein interacting with C-kinase-1. Proc. Natl. Acad. Sci. U.S.A. 100, 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirbec H., Francis J. C., Lauri S. E., Braithwaite S. P., Coussen F., Mulle C., Dev K. K., Coutinho V., Meyer G., Isaac J. T., Collingridge G. L., Henley J. M. (2003) Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 37, 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perez J. L., Khatri L., Chang C., Srivastava S., Osten P., Ziff E. B. (2001) PICK1 targets activated protein kinase Cα to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 21, 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim C. H., Chung H. J., Lee H. K., Huganir R. L. (2001) Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc. Natl. Acad. Sci. U.S.A. 98, 11725–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Terashima A., Cotton L., Dev K. K., Meyer G., Zaman S., Duprat F., Henley J. M., Collingridge G. L., Isaac J. T. (2004) Regulation of synaptic strength and AMPA receptor subunit composition by PICK1. J. Neurosci. 24, 5381–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yao Y., Kelly M. T., Sajikumar S., Serrano P., Tian D., Bergold P. J., Frey J. U., Sacktor T. C. (2008) PKMζ maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J. Neurosci. 28, 7820–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sossa K. G., Court B. L., Carroll R. C. (2006) NMDA receptors mediate calcium-dependent, bidirectional changes in dendritic PICK1 clustering. Mol. Cell. Neurosci. 31, 574–585 [DOI] [PubMed] [Google Scholar]
- 28. Terashima A., Pelkey K. A., Rah J. C., Suh Y. H., Roche K. W., Collingridge G. L., McBain C. J., Isaac J. T. (2008) An essential role for PICK1 in NMDA receptor-dependent bidirectional synaptic plasticity. Neuron 57, 872–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Volk L., Kim C. H., Takamiya K., Yu Y., Huganir R. L. (2010) Developmental regulation of protein interacting with C kinase 1 (PICK1) function in hippocampal synaptic plasticity and learning. Proc. Natl. Acad. Sci. U.S.A. 107, 21784–21789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia J., Chung H. J., Wihler C., Huganir R. L., Linden D. J. (2000) Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron 28, 499–510 [DOI] [PubMed] [Google Scholar]
- 31. Steinberg J. P., Takamiya K., Shen Y., Xia J., Rubio M. E., Yu S., Jin W., Thomas G. M., Linden D. J., Huganir R. L. (2006) Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron 49, 845–860 [DOI] [PubMed] [Google Scholar]
- 32. Hu X. D., Huang Q., Yang X., Xia H. (2007) Differential regulation of AMPA receptor trafficking by neurabin-targeted synaptic protein phosphatase-1 in synaptic transmission and long-term depression in hippocampus. J. Neurosci. 27, 4674–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Daw M. I., Chittajallu R., Bortolotto Z. A., Dev K. K., Duprat F., Henley J. M., Collingridge G. L., Isaac J. T. (2000) PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron 28, 873–886 [DOI] [PubMed] [Google Scholar]
- 34. Liu S. J., Cull-Candy S. G. (2005) Subunit interaction with PICK and GRIP controls Ca2+ permeability of AMPARs at cerebellar synapses. Nat. Neurosci. 8, 768–775 [DOI] [PubMed] [Google Scholar]
- 35. Gardner S. M., Takamiya K., Xia J., Suh J. G., Johnson R., Yu S., Huganir R. L. (2005) Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron 45, 903–915 [DOI] [PubMed] [Google Scholar]
- 36. Mameli M., Balland B., Luján R., Lüscher C. (2007) Rapid synthesis and synaptic insertion of GluR2 for mGluR-LTD in the ventral tegmental area. Science 317, 530–533 [DOI] [PubMed] [Google Scholar]
- 37. Bellone C., Lüscher C. (2006) Cocaine triggered AMPA receptor redistribution is reversed in vivo by mGluR-dependent long-term depression. Nat. Neurosci. 9, 636–641 [DOI] [PubMed] [Google Scholar]
- 38. Lin D. T., Huganir R. L. (2007) PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J. Neurosci. 27, 13903–13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iwakura Y., Nagano T., Kawamura M., Horikawa H., Ibaraki K., Takei N., Nawa H. (2001) N-Methyl-d-aspartate-induced α-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid (AMPA) receptor down-regulation involves interaction of the carboxyl terminus of GluR2/3 with Pick1. Ligand-binding studies using Sindbis vectors carrying AMPA receptor decoys. J. Biol. Chem. 276, 40025–40032 [DOI] [PubMed] [Google Scholar]
- 40. Hanley J. G., Henley J. M. (2005) PICK1 is a calcium-sensor for NMDA-induced AMPA receptor trafficking. EMBO J. 24, 3266–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rocca D. L., Martin S., Jenkins E. L., Hanley J. G. (2008) Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat. Cell Biol. 10, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Citri A., Bhattacharyya S., Ma C., Morishita W., Fang S., Rizo J., Malenka R. C. (2010) Calcium binding to PICK1 is essential for the intracellular retention of AMPA receptors underlying long-term depression. J. Neurosci. 30, 16437–16452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eriksen J., Bjørn-Yoshimoto W. E., Jørgensen T. N., Newman A. H., Gether U. (2010) Postendocytic sorting of constitutively internalized dopamine transporter in cell lines and dopaminergic neurons. J. Biol. Chem. 285, 27289–27301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madsen K. L., Eriksen J., Milan-Lobo L., Han D. S., Niv M. Y., Ammendrup-Johnsen I., Henriksen U., Bhatia V. K., Stamou D., Sitte H. H., McMahon H. T., Weinstein H., Gether U. (2008) Membrane localization is critical for activation of the PICK1 BAR domain. Traffic 9, 1327–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lavezzari G., McCallum J., Dewey C. M., Roche K. W. (2004) Subunit-specific regulation of NMDA receptor endocytosis. J. Neurosci. 24, 6383–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bjerggaard C., Fog J. U., Hastrup H., Madsen K., Loland C. J., Javitch J. A., Gether U. (2004) Surface targeting of the dopamine transporter involves discrete epitopes in the distal C terminus but does not require canonical PDZ domain interactions. J. Neurosci. 24, 7024–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chmelar R. S., Nathanson N. M. (2006) Identification of a novel apical sorting motif and mechanism of targeting of the M2 muscarinic acetylcholine receptor. J. Biol. Chem. 281, 35381–35396 [DOI] [PubMed] [Google Scholar]
- 48. Adkins E. M., Samuvel D. J., Fog J. U., Eriksen J., Jayanthi L. D., Vaegter C. B., Ramamoorthy S., Gether U. (2007) Membrane mobility and microdomain association of the dopamine transporter studied with fluorescence correlation spectroscopy and fluorescence recovery after photobleaching. Biochemistry 46, 10484–10497 [DOI] [PubMed] [Google Scholar]
- 49. Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Granas C., Ferrer J., Loland C. J., Javitch J. A., Gether U. (2003) N-terminal truncation of the dopamine transporter abolishes phorbol ester- and substance P receptor-stimulated phosphorylation without impairing transporter internalization. J. Biol. Chem. 278, 4990–5000 [DOI] [PubMed] [Google Scholar]
- 51. Daniels G. M., Amara S. G. (1999) Regulated trafficking of the human dopamine transporter. Clathrin-mediated internalization and lysosomal degradation in response to phorbol esters. J. Biol. Chem. 274, 35794–35801 [DOI] [PubMed] [Google Scholar]
- 52. Miranda M., Wu C. C., Sorkina T., Korstjens D. R., Sorkin A. (2005) Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C. J. Biol. Chem. 280, 35617–35624 [DOI] [PubMed] [Google Scholar]
- 53. Marchese A., Heiber M., Nguyen T., Heng H. H., Saldivia V. R., Cheng R., Murphy P. M., Tsui L. C., Shi X., Gregor P., George S. R., O'Dowd B. F., Docherty J. M. (1995) Cloning and chromosomal mapping of three novel genes, GPR9, GPR10, and GPR14, encoding receptors related to interleukin 8, neuropeptide Y, and somatostatin receptors. Genomics 29, 335–344 [DOI] [PubMed] [Google Scholar]
- 54. Seachrist J. L., Anborgh P. H., Ferguson S. S. (2000) β2-Adrenergic receptor internalization, endosomal sorting, and plasma membrane recycling are regulated by rab GTPases. J. Biol. Chem. 275, 27221–27228 [DOI] [PubMed] [Google Scholar]
- 55. Moore R. H., Millman E. E., Alpizar-Foster E., Dai W., Knoll B. J. (2004) Rab11 regulates the recycling and lysosome targeting of β2-adrenergic receptors. J. Cell Sci. 117, 3107–3117 [DOI] [PubMed] [Google Scholar]
- 56. Tsao P. I., von Zastrow M. (2000) Type-specific sorting of G protein-coupled receptors after endocytosis. J. Biol. Chem. 275, 11130–11140 [DOI] [PubMed] [Google Scholar]
- 57. Hanyaloglu A. C., McCullagh E., von Zastrow M. (2005) Essential role of Hrs in a recycling mechanism mediating functional resensitization of cell signaling. EMBO J. 24, 2265–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hanyaloglu A. C., von Zastrow M. (2007) A novel sorting sequence in the β2-adrenergic receptor switches recycling from default to the Hrs-dependent mechanism. J. Biol. Chem. 282, 3095–3104 [DOI] [PubMed] [Google Scholar]
- 59. Cong M., Perry S. J., Hu L. A., Hanson P. I., Claing A., Lefkowitz R. J. (2001) Binding of the β2 adrenergic receptor to N-ethylmaleimide-sensitive factor regulates receptor recycling. J. Biol. Chem. 276, 45145–45152 [DOI] [PubMed] [Google Scholar]
- 60. Wang Y., Lauffer B., Von Zastrow M., Kobilka B. K., Xiang Y. (2007) N-ethylmaleimide-sensitive factor regulates β2 adrenoceptor trafficking and signaling in cardiomyocytes. Mol. Pharmacol. 72, 429–439 [DOI] [PubMed] [Google Scholar]
- 61. Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 62. Gage R. M., Kim K. A., Cao T. T., von Zastrow M. (2001) A transplantable sorting signal that is sufficient to mediate rapid recycling of G protein-coupled receptors. J. Biol. Chem. 276, 44712–44720 [DOI] [PubMed] [Google Scholar]
- 63. Mollenhauer H. H., Morré D. J., Rowe L. D. (1990) Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim. Biophys. Acta 1031, 225–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sorkina T., Hoover B. R., Zahniser N. R., Sorkin A. (2005) Constitutive and protein kinase C-induced internalization of the dopamine transporter is mediated by a clathrin-dependent mechanism. Traffic 6, 157–170 [DOI] [PubMed] [Google Scholar]
- 65. Karthikeyan S., Leung T., Birrane G., Webster G., Ladias J. A. (2001) Crystal structure of the PDZ1 domain of human Na+/H+ exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains. J. Mol. Biol. 308, 963–973 [DOI] [PubMed] [Google Scholar]
- 66. Hanley J. G. (2008) PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol. Ther. 118, 152–160 [DOI] [PubMed] [Google Scholar]
- 67. Thorsen T. S., Madsen K. L., Rebola N., Rathje M., Anggono V., Bach A., Moreira I. S., Stuhr-Hansen N., Dyhring T., Peters D., Beuming T., Huganir R., Weinstein H., Mulle C., Strømgaard K., Rønn L. C., Gether U. (2010) Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc. Natl. Acad. Sci. U.S.A. 107, 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Park M., Penick E. C., Edwards J. G., Kauer J. A., Ehlers M. D. (2004) Recycling endosomes supply AMPA receptors for LTP. Science 305, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 69. Pan L., Wu H., Shen C., Shi Y., Jin W., Xia J., Zhang M. (2007) Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J. 26, 4576–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P., Unger V. M. (2008) Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Habermann B. (2004) The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 5, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jeyifous O., Waites C. L., Specht C. G., Fujisawa S., Schubert M., Lin E. I., Marshall J., Aoki C., de Silva T., Montgomery J. M., Garner C. C., Green W. N. (2009) SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat. Neurosci. 12, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lauffer B. E., Melero C., Temkin P., Lei C., Hong W., Kortemme T., von Zastrow M. (2010) SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lauffer B. E., Chen S., Melero C., Kortemme T., von Zastrow M., Vargas G. A. (2009) Engineered protein connectivity to actin mimics PDZ-dependent recycling of G protein-coupled receptors but not its regulation by Hrs. J. Biol. Chem. 284, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. van Weering J. R., Verkade P., Cullen P. J. (2010) SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin. Cell Dev. Biol. 21, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hoogenraad C. C., Popa I., Futai K., Martinez-Sanchez E., Wulf P. S., van Vlijmen T., Dortland B. R., Oorschot V., Govers R., Monti M., Heck A. J., Sheng M., Klumperman J., Rehmann H., Jaarsma D., Kapitein L. C., van der Sluijs P. (2010) Neuron specific Rab4 effector GRASP-1 coordinates membrane specialization and maturation of recycling endosomes. PLoS Biol. 8, e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.