Background: Most scorpion venom peptides adopt a single structural scaffold around four strictly conserved cysteines.
Results: Two K+ channel-blocking peptides from Tityus venoms share this cysteine spacing but fold into a distinct cystine-stabilized helix-loop-helix scaffold.
Conclusion: These peptides define a new structural group of scorpion venom peptides.
Significance: Cysteine spacing does not dictate the three-dimensional fold of small disulfide-rich proteins.
Keywords: NMR, Potassium Channels, Protein Folding, Protein Structure, Toxins, CSα/α fold, k-BUTX-Tt2b, Scorpion Toxin
Abstract
Scorpion venoms are a rich source of K+ channel-blocking peptides. For the most part, they are structurally related small disulfide-rich proteins containing a conserved pattern of six cysteines that is assumed to dictate their common three-dimensional folding. In the conventional pattern, two disulfide bridges connect an α-helical segment to the C-terminal strand of a double- or triple-stranded β-sheet, conforming a cystine-stabilized α/β scaffold (CSα/β). Here we show that two K+ channel-blocking peptides from Tityus scorpions conserve the cysteine spacing of common scorpion venom peptides but display an unconventional disulfide pattern, accompanied by a complete rearrangement of the secondary structure topology into a CS helix-loop-helix fold. Sequence and structural comparisons of the peptides adopting this novel fold suggest that it would be a new elaboration of the widespread CSα/β scaffold, thus revealing an unexpected structural versatility of these small disulfide-rich proteins. Acknowledgment of such versatility is important to understand how venom structural complexity emerged on a limited number of molecular scaffolds.
Introduction
Scorpion venoms are complex mixtures of biomolecules, products of millions years of evolution (1, 2). Venoms contain, among others components, peptide toxins capable of interacting specifically with potassium, sodium, and calcium channels (3–5). These toxic peptides have contributed considerably to the understanding of the structure and functional mechanism of the ion channels (6). Some of them are considered good candidates for the design and development of new drugs (7), although the number of structural and functional analyses of these peptides is still limited (8).
Potassium channels blocker toxins (KTx)6 can be classified into four subfamilies accordingly to the accepted nomenclature: α, β, γ, and κ (9, 10). α-KTx are peptides of 23–43 amino acids stabilized by three to four disulfide bonds, two of which are strictly conserved and link an α-helix and one strand of a β-sheet within the so-called cystine-stabilized α/β scaffold (CSα/β), the most common among scorpion toxins (10, 11). β-KTx toxins, known as “long chain,” are three disulfide-bridged peptides with ∼60 amino acids that also contain the cysteine pattern of peptides adopting the CSα/β scaffold, although no structure is available yet (12). γ-KTxs are blockers of the ether-á-go-go-related gene family of K+ channels; they are 36–47 amino acids long with three or four disulfide bridges, which also adopt the CSα/β scaffold (13). The last and smallest family, the κ-KTxs, displays purely helical structure stabilized by two disulfide bridges and fold into an α-hairpin fold known as CSα/α (cystine-stabilized helix-loop-helix) (11, 14–17). Peptides belonging to this family have been isolated from only two scorpion genera: Heterometrus (14, 15) and Opisthacanthus (16, 17). κ-KTxs are relatively poor blockers of K+ channels, despite presence of the typical functional dyad, Tyr and Lys, important for other activity of the toxin (3). It is still neither clear whether members of the κ-KTx family might all work through the same mechanism on potassium channels (16, 17), nor whether K+ channels are indeed their cognate receptors. In part this may be due to the lack of structural and functional characterization of peptides in this family.
This work describes the native isolation, recombinant production and structural characterization of a newly identified 28 amino acid K+ channel-blocking peptide from Tityus trivittatus venom, named κ-buthitoxin-Tt2b, κ-BUTX-Tt2b for short, following the nomenclature proposal of King et al. (18). The sequence shows high identity with peptides classified within the α-KTx subfamily 20 (19), including the six cysteines of the sequence signature of the CSα/β scaffold. Unexpectedly, the peptide adopts a CSα/α fold in solution. The same overall structure was determined for a recombinant version of another peptide from Tityus serrulatus classified within the same subfamily (UniProt P86271, Ts16 in Ref. 46). Peptides of the α-Ktx subfamily 20 were assumed to adopt the conventional CSα/β scaffold on the basis of their common cysteine spacing. However, both κ-BUTX-Tt2b and Ts16 adopt an unexpected fold even if sharing the cysteine pattern of conventional CSα/β peptides, thus offering new insights on the structural versatility of scorpion venom peptides. Acknowledging this versatility is important if the full potential of animal venoms as natural peptide libraries is to be realized.
EXPERIMENTAL PROCEDURES
Purification and Sequence Determination of the Novel Peptide
Venom from the scorpion Tityus trivitattus collected in the Province of Santa Fe, Argentina, was obtained by electrical stimulation and separated as earlier described (20). Only two steps of HPLC fractionation were necessary to obtain the pure peptide under study. An analytical C18 reverse phase column (Vydac, Hisperia) was used with a linear gradient from solvent A (water in 0.12% TFA, Sigma-Aldrich; all of the chemicals were purchased from this company, unless otherwise specified) to 60% solvent B (acetonitrile in 0.10% TFA), run for 60 min. The second separation used the same system with a gradient from 10 to 35% solvent B, run for 45 min. The amino acid sequence determination of the pure peptide was obtained using automatic Edman degradation with a Beckman LF 3000 protein sequencer, and MS measurements were obtained with an ion-trap equipment from Finnigan LCQDuo as earlier described (20). The peptide was named κ-buthitoxin-Tt2b.
Disulfide Bridge Determination
Pure peptide (18 μg) was digested with tosylphenylalanyl chloromethyl ketone-trypsin and endopeptidase V8 from Staphylococcus aureus (Roche Applied Science). Initially, 1 μg of trypsin was added in presence of an ammonium bicarbonate buffer (pH 8) and hydrolyzed for 5 h; then the endopeptidase V8 (1 μg) was added, and the reaction was incubated for 12 h at 36 °C. The peptides were separated using the same HPLC conditions used initially for the whole venom (see above) and sequenced.
Design and Cloning of the Synthetic κ-BUTX-Tt2b Gene
The κ-BUTX-Tt2b synthetic gene coding for the peptide was assembled based on the amino acid sequence obtained and optimized for Escherichia coli codon usage. The κ-BUTX-Tt2b gene (see supplemental data Fig. SM1) was cloned into the expression vector pET32a (Novagen). The resulting plasmid pET32-κ-BUTX-Tt2b was confirmed by DNA sequencing. The Ts16 synthetic gene used was based on the κ-BUTX-Tt2b gene (sequence available in supplemental data Fig. SM1).
Protein Expression
Recombinant-κ-BUTX-Tt2b protein was overexpressed in tuner E. coli cells (Novagen) transformed with the expression vector pET32-κ-BUTX-Tt2b. LB medium supplemented with ampicillin was used to grown cells at 37 °C. Protein expression was induced at an A600 between 0.7–0.8 with isopropyl thio-β-galactopyranoside 0.5 mm. The cells were incubated for 6 h at 30 °C and harvested by centrifugation.
Recombinant Protein Purification
For κ-BUTX-Tt2b, the cell pellets were resuspended in lysis buffer (150 mm NaCl, 0.1 mg/ml lisozyme, 50 mm Tris/HCl, pH 8) and sonicated using (Misonix Sonicator 3000). The soluble fraction was separated by centrifugation at 32,000 × g for 30 min at 4 °C. Recombinant His6-tagged fusion toxin (20.3 kDa) was purified by metal-chelate affinity chromatography using a HiTrap Ni2+ column (GE Healthcare) and eluted with two column volumes of elution buffer (50 mm Tris/HCl, pH 8, 150 mm NaCl, 500 mm imidazole). Thrombin was used to release the κ-BUTX-Tt2b recombinant toxin from the N terminus thioredoxin fusion protein. Cleavage reaction was performed for 6 h at 18 °C according to the manufacturer's instructions. A second round of isolation by metal-chelate affinity chromatography was required to remove κ-BUTX-Tt2b from cleavage reaction subproducts. Final κ-BUTX-Tt2b purification was performed with reverse phase HPLC in a Pro-Star Varian instrument equipped with a binary gradient solvent system on a Jupiter C18 column (250 mm × 4.6 mm; Phenomenex) using a linear gradient from 12 to 30% of water-acetonitrile run for 30 min (containing 0.05% TFA) at a flow rate of 1 ml/min.
Protein Characterization
Mass Spectrometry
MALDI-TOF MS on a Bruker Daltonics Microflex LT equipment was used to determinate the molecular mass of the recombinant proteins. The data were acquired using the reflector operation mode acquiring 150 shots. The samples were prepared with α-cyano-4-hydroxycinnamic acid in a 1:10 ratio (21).
Electrophysiological Measurements
cRNAs for voltage-dependent potassium channels: Shaker-IR (Shaker channel with inactivation domain removed) and hKv1.2 subunits were transcribed using a mMESSAGE mMACHINE in vitro transcription kit (Ambion). cRNAs were injected into Xenopus laevis oocytes using 1–2 ng (in 50 nl) of K+-subunits. Potassium currents were recorded 2–4 days after cRNA injection.
Two-electrode Voltage Clamp
The bath solution was ND-96: 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH 7.4, adjusted with NaOH. Resistance for the voltage and current electrodes were 1.0–1.5 and 0.3–0.5 MΩ, respectively, when filled with 3 m KCl. Holding potential was −60 mV, and pulse protocol was applied from −60 mV to +60 mV with increments of 20 mV for 200 ms. Macroscopic currents were recorded using the CA1B high performance oocyte clamp amplifier (Dagan Co.), the Digidata 1440 digitizer, and the pClamp10 software (Molecular Devices). The currents were filtered at ⅕ of the sampling frequency (2 kHz). The volume of washing solution was ∼20 times greater than that of the chamber containing the oocytes. Inhibition was evaluated every minute until steady state was reached, usually from 8 to 10 min.
Nuclear Magnetic Resonance Spectroscopy
Sample Preparation
Native and recombinant κ-BUTX-Tt2b were prepared at 0.08 and 2.3 mm concentrations, respectively, by dissolving lyophilized protein in 95% H2O, 5% D2O (Cambridge Isotope Laboratories). The recombinant protein spectra were obtained at pH 3.5. No pH adjustment was done with the native sample. NMR data were recorded at 25 °C on an 800 MHz Varian spectrometer equipped with a HCN indirect detection probe. TOCSY spectra were recorded with isotropic mixing time of 75 and 20 ms. NOESY spectra were acquired using mixing times of 150 and 300 ms. The two-dimensional NMR spectra were conducted using a scheme of water suppression by gradient-tailored excitation for solvent suppression (22). All of the experiments were acquired using 2048 and 1024 complex points in direct and indirect dimensions, respectively. Ts16 recombinant protein (rTs16) spectra were recorded in a 500 MHz Varian instrument at 3.0 mm; experiments were obtained as before.
Data Processing and Analysis
NMRDraw and NMRpipe software were used for processing data (23). CARA1.5 software was used for NMR data analysis (24). Semi-automated assignment and distance geometry calculations were performed with CYANA 2.1 (25).
Molecular Dynamics Refinement
Structure refinement was performed using molecular dynamics AMBER 9 (26) suite with an explicit solvent model. Lowest CYANA target function values were used for further refinement. Molecular dynamics simulations and the energy minimizations were carried out with the topology and parameters of AMBER-99SB force field. Distance and dihedral angle constraints were used for AMBER calculations following the method proposed by Xia et al. (27). Geometric quality of structures was assessed using PROCHECK utility in the validation server of the Protein Data Bank.
RESULTS
κ-BUTX-Tt2b Isolation Sequence and Disulfide Determination
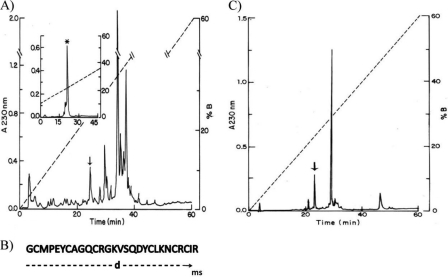
Fig. 1A shows HPLC separation of the soluble venom from T. trivitattus. The inset shows a second HPLC fractionation, after selecting only the component eluted at 24.7 min from the first run. The major component, labeled with an asterisk (see inset), was the peptide used for this study (∼3.1% of the soluble venom). The amino acid sequence determination of this pure peptide gave an unequivocal sequence assignment up to residue number 27 (Fig. 1B). The last amino acid, Arg in position 28, was inferred by mass spectrometry determination. The experimental mass determined for the native peptide was 3179.75 Da, and the theoretical mass expected for the first 27 residues directly determined was 3023.57, leaving a molecular mass of 156.18, which was identified as being that of arginine. This sequence was deposited in the UniProt Knowledgebase under the accession number B3A0L5. Sequence similarity searches revealed high identity with only two Tityus sp. venom peptides: Ts16 from T. serrulatus (P86271, 89%) and Tt28 from T. trivittatus (P0C183 (18), 65%). No disulfide pairing for any of these homologous peptides has been reported.
FIGURE 1.
A, venom fractionation. Soluble venom of T. trivitattus was loaded into a C18 reverse phase column and eluted from the HPLC system with a linear gradient made of solvent A (0.12% TFA in water) to solvent B (acetonitrile in 0.10% TFA) run for 60 min. The component eluting at 24.7 min (arrow), which corresponds to 4.44% of the soluble venom applied, was recovered and loaded into the same system but resolved with a linear gradient from 8–35% solvent B run for 45 min. From the component marked with an arrow, ∼70% was the pure peptide (labeled with asterisk). B, a sample (∼2 nmol) of reduced and alkylated peptide was used for amino acid sequence determination using automatic sequencing and mass spectrometry, as described under “Experimental Procedures.” The first 27 residues were unequivocally determined by Edman degradation, and the last residue was inferred from the mass spectrometry analysis. C, separation of enzymatically digested peptide. A sample of native peptide (18 μg) was digested by trypsin and endopeptidase V8 in the conditions described under “Experimental Procedures” and applied to the C18 reverse phase column. All of the peptides were sequenced. The one labeled with an arrow eluted at 23.0 min was identified as corresponding to the disulfide between C1 and C5.
Fig. 1C shows the profile of HPLC separation after enzymatic hydrolysis. The peptide eluted at 23.0 min was compatible with a disulfide paring between the first (Cys2) and fifth cysteines (Cys24), C1 and C5. The automatic sequencer showed two amino acid residues for the first cycles: Gly and Asn. Position 2 was a blank, but Met was in position 3 and Pro was in position 4. It implies that endopeptidase V8 cleaved the peptide bond at the Glu5, and trypsin cleaved Lys22 and Arg25. The remaining peptides gave always more than two amino acid residues per cycle of the sequencer, because the core fragment of peptide after the enzymatic hydrolysis did not allow separating the other peptides containing the two remaining disulfide bridges. In this way, only one of the disulfide bridges of the toxin was directly determined.
Fusion Protein Expression and Purification
Most of the fusion protein was located in the soluble cellular lysate, and it was efficiently retained by the HiTrap Ni2+ column with purification yield of 36 mg/liter. Thrombin recognizes the sequence X3X2R/L—GSX1X2; therefore, the product of the hydrolysis reaction is the κ-BUTX-Tt2b sequence with additional glycine and serine at the N-terminal segment, rκ-BUTX-Tt2b. Optimal cleavage conditions were obtained at 18 °C after 6 h of reaction. Finally, another isolation by metal-chelate affinity chromatography purification step was performed.
Reverse Phase HPLC Purification and Protein Characterization
Under chromatographic conditions selected, rκ-BUTX-Tt2b elutes at 23.0 min with a MALDI-TOF mass of 3325.1 for [M+H]+, which is in good agreement with the theoretical mass of 3323.9 Da for an oxidized peptide forming three disulfide bridges, calculated with the ProtParam tool from ExPASy server (28). The purification procedures indicated above yielded 0.7 mg of rκ-BUTX-Tt2b per liter in LB culture medium.
Electrophysiological Measurements
Electrophysiological assays were done on Shaker-IR and hKv1.2 channels. Fig. 2 shows the effect of rκ-BUTX-Tt2b on both channels, using the experimental conditions described below. The protocol of test pulses was applied every 60 s to verify the current stability previous to the toxin addition. Only oocytes showing a minimum run-down were chosen for experimental assays. The control currents (black line) through Shaker-IR or hKv1.2 channels under whole cell recording are plotted. Fig. 2 shows representative traces for both channels only at +60 mV pulse. The addition of rκ-BUTX-Tt2b (200 μmol/liter) blocked partially outward currents (gray line). Recombinant toxin did not change basically the original shape of currents, suggesting that rκ-BUTX-Tt2b acts as a pore blocker, although, further experiments are needed to test this hypothesis. Current reduction was 88.7 ± 9.9 and 74.9 ± 7.1 (n = 3) for Shaker-IR and Kv1.2, respectively.
FIGURE 2.
Effect of κ-BUTX-Tt2b toxin on Shaker-IR (left panel) and Kv1.2 channels (right panel). Macroscopic K+ currents were recorded using two-electrode voltage-clamp technique in ND96 solution. Currents were elicited by depolarizing the membrane for 200 ms, from a holding potential of −60 mV to a pulse step of +60 mV. Pulse protocol was applied at 60 s each. Traces for control conditions are shown with a black line, and those in the presence of recombinant toxin are shown with a gray line (after 8 min of application at 200 μmol/liter). The current percentages of blockade for Shaker IR and Kv1.2 were 88.7 ± 9.9 and 74.9 ± 7.1 (mean ± S.E.), respectively. The inhibition of currents was not fully reversible (not shown).
Nuclear Magnetic Resonance Analysis
NMR experiments for the κ-BUTX-Tt2b native protein were obtained with poor signal to noise ratio at a very low concentration. The TOCSY experiment was obtained with sufficient sensitivity. However, the NOESY experiments displayed low sensitivity that made difficult to complete the sequential assignments. In contrast, NMR spectroscopic studies using higher concentration of rκ-BUTX-Tt2b revealed well resolved resonances with good chemical shift dispersion between 6.3 and 10.3 ppm. A comparison of the chemical shifts between the amide region signals of the recombinant (black) and the native (red) toxins shows just slight differences mainly caused by different pH levels and concentrations used for the experiments (see Fig. 3). Aliphatic region is almost identical. A NOESY HN region spectrum shows the HN-HN correlations consistent with the presence of helical structures (data not shown). The assignments were achieved following the standard protocol (29).
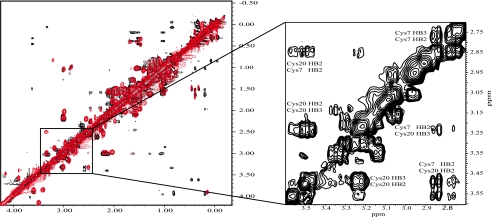
FIGURE 3.
NOESY spectra showing the aliphatic region of native and recombinant κ-BUTX-Tt2b spectra obtained with 0. 1 mm native (48 h of acquisition in red) and with 2.3 mm recombinant (in black). Differences are attributed to concentration and the glycine and serine in the N termini. The inset shows the correlations between Cys7 and Cys20 forming the cystine pair C2-C4 for the recombinant protein (Cys7 HB2–Cys20 HB2, Cys7 HB2–Cys20 HB3, Cys7 HB3–Cys20 HB2, and Cys7 HB3–Cys20 HB3 with distances found of 4.9, 4.0, 3.4, and 4.0 Å).
rκ-BUTX-Tt2b Disulfide Bond Connectivities and Three-dimensional Structure
All 30 rκ-BUTX-Tt2b spin systems were identified, and most proton resonance signals were unambiguously assigned (chemical shift completeness of 98.7%). The position of the disulfide bridges was established by analyzing the NMR experimental results independently. A NOE signal between Hβ-Hβ from Cys7 and Cys20 (Fig. 3, inset), which corresponds to C2-C4 cystine pair (residue numbers are indicated as in the native protein), was found. Also, there were observed NOE correlations between HN of Cys24 with Hβ of Cys2 and Hβ of Met3; all of these signals are in agreement with the formation of the C1-C5 pair. These results confirm a new cystine pairing for scorpion toxins when compared with the conventional pairs of CSα/β scorpion venom peptides (C1-C4, C2-C5, and C3-C6 (11)). To further support the assigned disulfide bonds, three CYANA calculations were performed with different input parameters following a protocol described by Fadel et al. (30). The first calculation included data without disulfide bond restrictions. This result provides the NOE constraint contributions for the structure, which was used as a reference. The second calculation was performed using as restrictions the most common disulfide pattern observed in KTxs toxins: C1-C4, C2-C5, and C3-C6. Finally, another set of structures were calculated considering the alternative C1-C5, C2-C4, and C3-C6 cystine pairing pattern, as suggested by the NOE NMR data assignment and mass spectrometry of a digested peptide. Table 1 shows the statistics and RMSD values of these three calculations. After comparison of backbone for the residues 3–30 of the lowest energy structure for each set of calculations, the RMSD against the reference structure gave 1.75 Å for the conventional disulfide bridges and 0.78 Å for the nonconventional disulfide bridges.
TABLE 1.
RMSD between 20 CYANA calculated structures for κ-BUTX-Tt2b varying the disulfide bridge patterns
The initial data set of NOE restraints for all calculations was the same. The first set was calculated using only NOE restrains, and no disulfide bridge constrains (No DSB) were used. A second structure calculation set was done considering conventional DSB (cDSB): C1–C4, C2–C5, and C3–C6 pairs. Last calculations were done including novel DSB (nDSB): C1–C5, C2–C4, and C3–C6 pairs. The reported numbers under the upper distance limit are the final set of NOEs that were included by CYANA, which automatically rejects or accepts NOEs to improve the target function value (25).
| κ-BUTX-Tt2b | No DSB | cDSB | nDSB |
|---|---|---|---|
| Upper distance limits | |||
| Total | 367 | 342 | 365 |
| Short range | 228 | 221 | 223 |
| Medium range | 64 | 55 | 64 |
| Long range | 75 | 66 | 78 |
| RMSD (3–30) | |||
| Average backbone (Å) | 0.43 | 0.52 | 0.39 |
| Average heavy atom (Å) | 1.09 | 1.19 | 1.01 |
| Target function value (Å) | 0.0 | 0.08 | 0.01 |
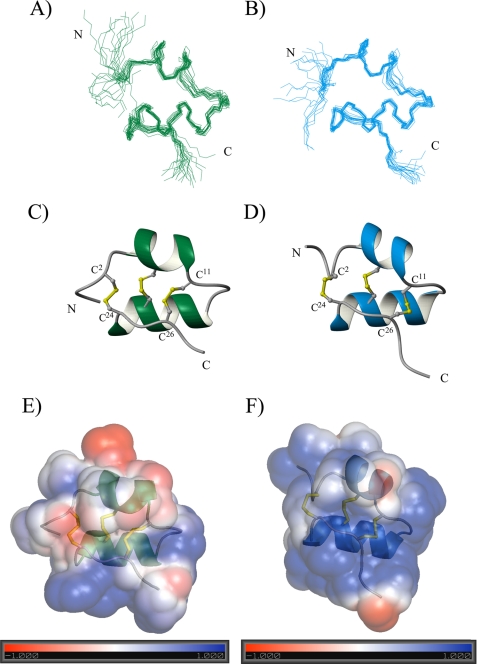
A total of 200 structures where generated with CYANA; only 20 structures (Fig. 4A) with the lowest target function are shown. The proton shift index is in good agreement with the secondary structure found (31). NOE correlations between HNi/HNi+1, Hαi/HNi+3, and Hαi/HNi+4 indicate that rκ-BUTX-Tt2b is mainly shaped by helical elements. The rκ-BUTX-Tt2b structure consists of an antiparallel helix-loop-helix topology with a new fold pattern stabilized by three nonconventional disulfide bonds between C1-C5, C2-C4, and C3-C6 for scorpion toxins. The helices are from residues Pro4–Gln10 and Val15–Asn23 and three disulfide bridges Cys2–Cys24, Cys7–Cys20, and Cys11–Cys26. A loop of four residues, Cys11–Lys14, connects the two helices. rκ-BUTX-Tt2b toxin adopts a cystine-stabilized helix-loop-helix fold (CSα/α). This is the first scorpion venom CSα/α peptide, which contains three disulfide bonds.
FIGURE 4.
NMR solution structure of rκ-BUTX-Tt2b and rTs16. A and B, superimposed 20 lowest energy structures, only backbone atoms are displayed (44). C and D, ribbon representation of both minimized structure; backbone trace and disulfide bridges are shown. E and F, electrostatic surface potential diagram showing the positive region around the Tyr19 is in blue, and negative charge is printed in red. The ribbon shows the orientation of the molecule. N and C termini are labeled in panels A–D. A, C, and E correspond to κ-BUTX-Tt2b, and B, D, and F correspond to Ts16.
The structure with the lowest energy is shown in Fig. 4C, and its correspondent electrostatic potential surface diagram is shown in Fig. 4E. The rκ-BUTX-Tt2b atom coordinates and chemical shifts were deposited in the Protein Data Bank and Biological Magnetic Resonance Bank, with access codes 2LI3 and 17876, respectively.
Native versus Recombinant κ-BUTX-Tt2b
Using the NMR assignments of rκ-BUTX-Tt2b as a reference, it was possible to identify all 28 spin systems for native κ-BUTX-Tt2b. However, it was not possible to solve the tridimensional structure from the native sample because of the absence of a significant number of NOE signals. Chemical shift assignments were used to compare HN and aliphatic region between native and recombinant κ-BUTX-Tt2b. Little difference in chemical shift was observed between native and recombinant peptides. Similar overlapped NOE peaks were used as spectroscopic evidence that both peptides have the same fold (Fig. 3). Furthermore, secondary structure prediction based on proton chemical shifts of native κ-BUTX-Tt2b shows helical patterns as described for the recombinant protein.
rTs16 Three-dimensional Structure
The high identity between κ-BUTX-Tt2b and Ts16 strongly suggests that the latter would also adopt a CSα/α structure. To verify this hypothesis, the rκ-BUTX-Tt2b gene was used as a template for Ts16 cloning. Following the experimental procedures reported above, it was possible to obtain the NMR solution structure of rTs16.
All 31 rTs16 spin systems were identified (chemical shift completeness of 96.8%). The disulfide bridges positions for rκ-BUTX-Tt2b were determined based on the presence of Cys2–Cys24 and Cys7–Cys20 Hβ-Hβ NOE cross-peaks; neither Cys2–Cys20 nor Cys7–Cys24 showed any NOESY correlation between β protons.
Final CYANA calculations were performed as for rκ-BUTX-Tt2b considering 809 NOE distance restraints. rTs16 peptide has helical conformations between Lys4–Gln10 and Lys14–His23 NOE correlations between HNi/HNi+1, Hαi/HNi+3, and Hαi/HNi+4 indicate that Ts16 is mainly shaped by helical elements (see the supplemental data Fig. SM4). The rTs16 structure consists of an antiparallel helix-loop-helix topology with same fold pattern stabilized by three nonconventional disulfide bonds for scorpion toxins as for κ-BUTX-Tt2b. A loop of three residues, Cys11–Gly13, is connecting the two helices. Ts16 toxin thus also belongs to the CSα/α (Fig. 4, B, D, and F), which shows the prevalence of the three disulfide bridged CSα/α peptides in different scorpion venoms. Atom coordinates and chemical shifts were deposited in the Protein Data Bank (2LKA) and the Biological Magnetic Resonance Bank (17987). Table 2 shows the statistics and RMSD values for Ts16.
TABLE 2.
RMSD between 20 CYANA calculated structures for Ts16 using only NOE restrains considering the novel DSB (nDSB): C1–C5, C2–C4, and C3–C6 pairs.
| Ts16 | nDSBC |
|---|---|
| Upper distance limits | |
| Total | 421 |
| Short range | 251 |
| Medium range | 85 |
| Long range | 85 |
| RMSD (3–30) | |
| Average backbone (Å) | 0.35 |
| Average heavy atom (Å) | 1.00 |
DISCUSSION
New Structural Group of Scorpion Venom Peptides
The venom of the Argentinean scorpion T. trivitattus is quite toxic, having caused human fatalities (32). Although the interest on studying this venom started several years ago (33), very little is known about its components structure and function. There are only two publications describing components of this venom, both dealing with peptides that affect K+ ion permeability (19, 20). The former publication describes a new subfamily of α-KTx toxins. The reported peptide was called Tt28, with the systematic name α-KTx20.1. It has 29 amino acid residues with three disulfide bridges and is a fairly good blocker of K+ channels Kv1.2 (EC50 = ∼100 nm) and Kv1.3 (EC50 = ∼7 nm) but failed to have any effect on Kv1.1, Kv1.4, Shaker IR, or human ether-á-go-go related K+ channels. The three-dimensional structure of this toxin is not solved yet, and the positions of its disulfide bonds remain to be determined, but they were suggested to be similar to other α-KTx scorpion toxins based on the presence of the common cysteine spacing of the CSα/β proteins (19). The sequences of the two structures reported here share large identity with Tt28 (Fig. 5), so they would also be accordingly classified as α-KTx 20 peptides; however, enzymatic studies, NMR assignments, and molecular calculations with both peptides showed that they do not correspond to the typical CSα/β scaffold of most scorpion toxins known to date. Rather, both peptides adopt a CSα/α scaffold in solution.
FIGURE 5.
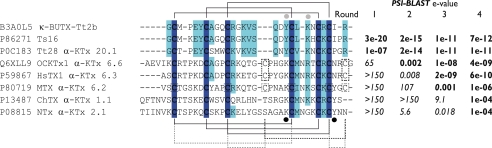
Multiple sequence alignment of scorpion venom peptides. Representative sequences retrieved during PSI-BLAST search (34) with κ-BUTX-Tt2b as query were aligned with COBALT (45). Conserved residues in κ-BUTX-Tt2b, Ts16, and Tt28 are highlighted (dark blue, complete conservation in all fourth round hits; light blue, partial conservation). PSI-BLAST columns indicate the expected values after a given number of rounds against SwissProt release of June 2011. The values in bold type indicate that the sequences were used for the next PSI-BLAST round. The values in italics indicate that the sequences were not used for the next round. Lines above the alignment connect the Cys residues according to the disulfides determined for κ-BUTX-Tt2b. The solid lines below the alignment connect the Cys residues according to the typical CSα/β scaffold. The dotted lines indicate the extra disulfide bridge of α-KTx subfamily 6 members. The gray dotted lines depict the unusual disulfide pattern of MTX (11).
All α Variation of CSα/β Scaffold
The CSα/α fold is very uncommon among scorpion venom peptides; all of the described examples are highly similar and have been collectively named κ-KTx (9, 11, 14–17). Disulfide pairing of κ-KTx, C1-C4, and C2-C3 is different from the one assigned to κ-BUTX-Tt2b and Ts16, C1-C5, C2-C4, and C3-C6; thus it is not possible to align the sequences based on Tytgat criteria (9), which establish that toxins must be aligned based on their cysteine position. Likewise, structure-based alignments of κ-BUTX-Tt2b and Ts16 with κ-KTx produce rather poor matches, thus indicating that their structure define a novel structural fold of scorpion venom peptides. The founding member of this group, although so far unnoticed (see below), would be Tt28 (19).
Database searches with κ-BUTX-Tt2b and Ts16 sequences retrieved only Tt28 as a significant match. It was nonetheless possible to find other CSα/β peptides, mainly α-KTx, by remote homology searching within three rounds of PSI-BLAST against SwissProt database (34). The six cysteines could be satisfactorily aligned (Fig. 5). Similarly, Ts16 and Tt28 were retrieved within three PSI-BLAST rounds starting with typical α-KTx. Their low sequence similarity with α-KTx prevents an unambiguous homology assignment; thus further evidence, such as conserved gene structure, is needed to resolve whether or not there is an evolutionary link between these CSα/α peptides and the typical α-KTx.
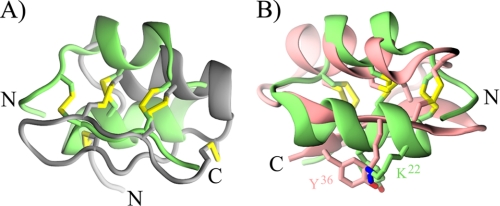
Structural comparison with TopMatch (35) revealed fairly good overlaps for two regions of κ-BUTX-Tt2b and Ts16 with the α-helix and second β-strand of several α-KTx retrieved during PSI-BLAST searches. Fig. 6A shows the structural alignment of κ-BUTX-Tt2b (green) and a chimeric peptide based on two α-KTx subfamily 6 peptides (Protein Data Bank code 1wpd (36) in gray); the structural alignment of main chain atoms over 15 residues has a RMSD of 1.8 Å. Detailed analysis of this structural superposition unveiled a tight overlap, 1.4 Å over the main chain atoms, between the four cysteines defining the sequence signature of the CSα/β scaffold in both structures (Cys7, Cys11, Cys24, and Cys26 in κ-BUTX-Tt2b and Cys9, Cys13, Cys29, and Cys31 for the chimera). Although we favor the noncanonical disulfide pattern for Ts16 on the basis of NOE cross-peaks, structure calculations with rTs16 suggest that it could form the typical CSα/β disulfide pattern, while maintaining the CSα/α scaffold. Moreover, it has been previously shown that another scorpion venom peptide, maurotoxin, could accommodate two different disulfide patterns without major changes in its CSα/β three-dimensional structure (37). It thus seems that the same cysteine pattern could accommodate more than one disulfide connectivity, as previously shown for the cyclic peptide kalata B1 (38).
FIGURE 6.
Structural comparison of κ-BUTX-Tt2b with α-KTx peptides. The structure of κ-BUTX-Tt2b (green) was aligned with the structures of a chimeric peptide based on HsTx1 and MTX (Protein Data Bank code 1wpd in gray) (A) and charybdotoxin (Protein Data Bank code 2crd in pink) (B); both panels have the same orientation. Cysteine residues are depicted as sticks in both panels; notice the almost perfect overlap of the four cysteines, which define the sequence signature of typical CSα/β peptides (RMSD of 0.76 Å over C-α atoms of the chimeric peptide), even if they are making different disulfide bridges. Functional dyad residues of charybdotoxin are shown as stick representation in B. In the superposition the distance between the side chain amino group of charybdotoxin Lys27 and κ-BUTX-Tt2b Lys21 is 1.43 Å, and the average distance of all side chain atoms of charybdotoxin Tyr36 and κ-BUTX-Tt2b Tyr19 is 1.3 Å.
From the structural comparison it is also evident that the main difference between κ-BUTX-Tt2b and Ts16 with respect to CSα/β peptides is the second α-helix of the newly described structures. It occupies the place of the first β-strand of CSα/β peptides; therefore the CSα/α fold of κ-BUTX-Tt2b and Ts16 seems to be an all α version of the CSα/β scaffold where the second β strand is conserved as an extended conformation of the C-terminal residues. A thought-provoking possibility is the existence of a structural switch between CSα/α and CSα/β conformations. Indeed, two chromatographic peaks were consistently found during the reverse phase chromatography of the recombinant peptides described here; each peak presents the same molecular mass in MALDI-TOF results (data not shown). Such structural transitions have been previously found in a number of proteins that undergo major shifts in secondary structure (39). Further characterization ought to be done to test this hypothesis.
Hint about Convergent Molecular Determinants for K+ Channel Blockade
Like many other peptides targeting K+ channels, most scorpion α-KTx blocks ion conduction through a common pharmacophore composed by a pore-plugging lysine and an aromatic residue located ∼7 Å apart, which has been dubbed as the “functional dyad” (11, 40). Neither the peptides described here nor the homologous Tt28 present the most common variant of the functional dyad among α-KTx (see dots above and below the alignment in Fig. 5). However, structural superposition showed that the functional dyad residues of charybdotoxin (ChTX), Lys27 and Tyr36 (Protein Data Bank code 2crd) (41), overlap with the nonhomologous residues Lys22 and Tyr19 of κ-BUTX-Tt2b (Fig. 6B), two residues conserved in Ts16 and Tt28. Thus, although mutagenesis studies would be needed to corroborate this hypothesis, it might be possible that convergent evolution has lead to the emergence of a novel variant of the functional dyad.
Note on Toxin Nomenclature and Classification
Here we followed the proposal of King et al. (18) about naming of novel toxins. Their classification scheme has the advantage of being systematic, although it intentionally let out the information about three-dimensional folding of the toxins. These authors stressed the fact that the knowledge about the three-dimensional scaffolds recruited into venoms is still “rudimentary” (18); the unexpected folding of peptides described here reinforces their cautionary remark. The newly described peptide was named κ-BUTX-Tt2b, acknowledging its activity against K+ channels (κ), the taxonomic classification of the original source (buthitoxin-Tt), and the fact that it is the second example of the second type of K+ channel blockers to be found in this species (2b, the first one would be Tt28).
Alternatively we could have followed the “KTx” classification scheme for scorpion toxins active on K+ channels proposed by an international panel of experts (9). This nomenclature has the virtues of recapitulating what is known about the phylogeny of many scorpion toxins and of mirroring to a certain extent their pharmacology (10). With the exception of κ-KTx, all other scorpion KTx are supposed to fold according to the CSα/β scaffold, a conjecture largely supported by all of the structures known until now and the strict conservation of the cysteine pattern (11). Following this scheme, the amino acid sequence of the novel peptide described here would lead to classify it as another member of the α-KTx 20 subfamily, along with Tt28 (19) and Ts16. However, it was only until the rκ-BUTX-Tt2b structure was obtained that it was possible to realize that a new structural group of scorpion venom peptides has been found, a result corroborated by the solution structure of Ts16.
Assuming that Tt28 also folds into a CSα/α structure, the three similar peptides could be transferred to the only other group of scorpion venom peptides, which adopt the same structural pattern, the κ-KTx subfamily. However, we hesitate to do so because the helix-loop-helix topology of rκ-BUTX-Tt2b and Ts16 appear to be an elaboration of the most common CSα/β scaffold, rather than a variation of the κ-KTx α-hairpin. Such structural elaboration based on different cysteine patterns has been previously proposed to explain the emergence of the inhibitory cystine knot motif over an “ancestral” disulfide directed β-hairpin (42).
Concluding Remarks
The three-dimensional structures reported here highlight the structural versatility that could be attained by peptides sharing a common cysteine pattern, thus revealing a new trick of this old pattern. Further research would show whether or not they are unique examples of their kind, but providing that animal venoms are arsenal-like mixtures of bioactive compounds, among which a small number of disulfide rich scaffolds are highly prevalent (11, 43), we expect that other CSα/α peptides will be found in the future, thus helping to clarify their biological function in the venom and their relationships with other venom components.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance given to the project by Dr. Fernando Zamudio Zúñiga, Carmen Marquez, and Eréndira García. We are grateful to Professors E. Wanke (Università di Milano-Bicocca) and L. Toro (David Geffen School of Medicine at UCLA) for Kv1.2 and Shaker-IR clones, respectively.
This work was supported in part by Dirección General de Servicios de Cómputo Académico y Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México Projects IN205110 (to F. R. P.) and IN204110 (to L. D. P.) and Consejo Nacional de Ciencia y Tecnología Project 59297 (to F. R. P.).

This article contains supplemental Figs. SM1–SM4.
The atomic coordinates and structure factors (codes 2LI3 and 2LKA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- KTx
- K+ channel toxin
- Hα
- hydrogen in α position
- Hβ
- hydrogen in β position
- HN
- amide proton
- RMSD
- root mean square deviation
- CS
- cystine-stabilized.
REFERENCES
- 1. Froy O., Sagiv T., Poreh M., Urbach D., Zilberberg N., Gurevitz M. (1999) Dynamic diversification from a putative common ancestor of scorpion toxins affecting sodium, potassium, and chloride channels. J. Mol. Evol. 48, 187–196 [DOI] [PubMed] [Google Scholar]
- 2. Rodríguez de la Vega R. C., Schwartz E. F., Possani L. D. (2010) Mining on scorpion venom biodiversity. Toxicon 56, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 3. Mouhat S., Andreotti N., Jouirou B., Sabatier J. M. (2008) Animal toxins acting on voltage-gated potassium channels. Curr. Pharm. Des. 14, 2503–2518 [DOI] [PubMed] [Google Scholar]
- 4. Billen B., Bosmans F., Tytgat J. (2008) Animal peptides targeting voltage-activated sodium channels. Curr. Pharm. Des. 14, 2492–2502 [DOI] [PubMed] [Google Scholar]
- 5. Norton R. S., McDonough S. I. (2008) Peptides targeting voltage-gated calcium channels. Curr. Pharm. Des. 14, 2480–2491 [DOI] [PubMed] [Google Scholar]
- 6. Dutertre S., Lewis R. J. (2010) Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 285, 13315–13320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis R. J., Garcia M. L. (2003) Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2, 790–802 [DOI] [PubMed] [Google Scholar]
- 8. Jenkinson D. H. (2006) Potassium channels. Multiplicity and challenges. Br. J. Pharmacol. 147, S63–S71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tytgat J., Chandy K. G., Garcia M. L., Gutman G. A., Martin-Eauclaire M. F., van der Walt J. J., Possani L. D. (1999) A unified nomenclature for short-chain peptides isolated from scorpion venoms. α-KTx molecular subfamilies. Trends. Pharmacol. Sci. 20, 444–447 [DOI] [PubMed] [Google Scholar]
- 10. Rodríguez de la Vega R. C., Possani L. D. (2004) Current views on scorpion toxins specific for K+-channels. Toxicon 43, 865–875 [DOI] [PubMed] [Google Scholar]
- 11. Mouhat S., Jouirou B., Mosbah A., De Waard M., Sabatier J. M. (2004) Diversity of folds in animal toxins acting on ion channels. Biochem. J. 378, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diego-García E., Abdel-Mottaleb Y., Schwartz E. F., Rodríguez de la Vega R. C., Tytgat J., Possani L. D. (2008) Cytolytic and K+ channel blocking activities of β-KTx and scorpine-like peptides purified from scorpion venoms. Cell. Mol. Life Sci. 65, 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodríguez de la Vega R. C., Barraza O., Restano-Cassulini R., Possani L. D. (2009) Animal Toxins: State of the Art. Perspectives in Health and Biotechnology (Lima M. E., ed), pp. 193–204, Editora UFMG, Belo Horizonte, Brazil [Google Scholar]
- 14. Srinivasan K. N., Sivaraja V., Huys I., Sasaki T., Cheng B., Kumar T. K., Sato K., Tytgat J., Yu C., San B. C., Ranganathan S., Bowie H. J., Kini R. M., Gopalakrishnakone P. (2002) κ-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional diad in potassium channel selectivity. J. Biol. Chem. 277, 30040–30047 [DOI] [PubMed] [Google Scholar]
- 15. Nirthanan S., Pil J., Abdel-Mottaleb Y., Sugahara Y., Gopalakrishnakone P., Joseph J. S., Sato K., Tytgat J. (2005) Assignment of voltage-gated potassium channel blocking activity to κ-KTx1.3, a non-toxic homologue of κ-hefutoxin-1, from Heterometrus spinifer venom. Biochem. Pharmacol. 69, 669–678 [DOI] [PubMed] [Google Scholar]
- 16. Chagot B., Pimentel C., Dai L., Pil J., Tytgat J., Nakajima T., Corzo G., Darbon H., Ferrat G. (2005) An unusual fold for potassium channel blockers. NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis. Biochem. J. 388, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Camargos T. S., Restano-Cassulini R., Possani L. D., Peigneur S., Tytgat J., Schwartz C. A., Alves E. M., de Freitas S. M., Schwartz E. F. (2011) The new κ-KTx 2.5 from the scorpion Opisthacanthus cayaporum. Peptides 32, 1509–1517 [DOI] [PubMed] [Google Scholar]
- 18. King G. F., Gentz M. C., Escoubas P., Nicholson G. M. (2008) A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 52, 264–276 [DOI] [PubMed] [Google Scholar]
- 19. Abdel-Mottaleb Y., Coronas F. V., de Roodt A. R., Possani L. D., Tytgat J. (2006) A novel toxin from the venom of the scorpion Tityus trivittatus, is the first member of a new alpha-KTX subfamily. FEBS Lett. 580, 592–596 [DOI] [PubMed] [Google Scholar]
- 20. Coronas F. V., de Roodt A. R., Portugal T. O., Zamudio F. Z., Batista C. V., Gómez-Lagunas F., Possani L. D. (2003) Disulfide bridges and blockage of Shaker B K+-channels by another butantoxin peptide purified from the Argentinean scorpion Tityus trivittatus. Toxicon 41, 173–179 [DOI] [PubMed] [Google Scholar]
- 21. Gobom J., Schuerenberg M., Mueller M., Theiss D., Lehrach H., Nordhoff E. (2001) α-Cyano-4-hydroxycinnamic acid affinity sample preparation. A protocol for MALDI-MS peptide analysis in proteomics. Anal. Chem. 73, 434–438 [DOI] [PubMed] [Google Scholar]
- 22. Piotto M., Saudek V., Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 23. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24. Bartels C., Xia T., Billeter M., Güntert P., Wüthrich K. (1995) J. Biomol. NMR 6, 1–10 [DOI] [PubMed] [Google Scholar]
- 25. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 26. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The AMBER biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia B., Tsui V., Case D. A., Dyson H. J., Wright P. E. (2002) Comparison of protein solution structures refined by molecular dynamics simulation in vacuum, with a generalized Born model, and with explicit water. J. Biomol. NMR 22, 317–331 [DOI] [PubMed] [Google Scholar]
- 28. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) “Protein Identification and Analysis Tools on the ExPASy Server,” in The Proteomics Protocols Handbook (Walker John M., ed) 2nd Edition, pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 29. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley, New York [Google Scholar]
- 30. Fadel V., Bettendorff P., Herrmann T., de Azevedo W. F., Jr., Oliveira E. B., Yamane T., Wüthrich K. (2005) Automated NMR structure determination and disulfide bond identification of the myotoxin crotamine from Crotalus durissus terrificus. Toxicon 46, 759–767 [DOI] [PubMed] [Google Scholar]
- 31. Wishart D. S., Sykes B. D., Richards F. M. (1992) The chemical shift index. A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 32. de Roodt A. R., Coronas F. I., Lago N., González M. E., Laskowicz R. D., Beltramino J. C., Saavedra S., López R. A., Reati G. J., Vucharchuk M. G., Bazán E., Varni L., Salomón O. D., Possani L. D. (2010) General biochemical and immunological characterization of the venom from the scorpion Tityus trivittatus of Argentina. Toxicon 55, 307–319 [DOI] [PubMed] [Google Scholar]
- 33. de Roodt A. R., Gimeno E., Portiansky E., Varni L., Dolab J. A., Segre L., Litwin S., Vidal J. C. (2001) A study on the experimental envenomation in mice with the venom of Tityus trivitattus Kraepelin 1898 (Scorpiones, Buthidae) captured in Argentina. J. Nat. Toxins 10, 99–109 [PubMed] [Google Scholar]
- 34. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sippl M. J., Wiederstein M. (2008) A note on difficult structure alignment problems. Bioinformatics 24, 426–427 [DOI] [PubMed] [Google Scholar]
- 36. Regaya I., Beeton C., Ferrat G., Andreotti N., Darbon H., De Waard M., Sabatier J. M. (2004) Evidence for domain-specific recognition of SK and Kv channels by MTX and HsTx1 scorpion toxins. J. Biol. Chem. 279, 55690–55696 [DOI] [PubMed] [Google Scholar]
- 37. Fajloun Z., Mosbah A., Carlier E., Mansuelle P., Sandoz G., Fathallah M., di Luccio E., Devaux C., Rochat H., Darbon H., De Waard M., Sabatier J. M. (2000) Maurotoxin versus Pi1/HsTx1 scorpion toxins. Toward new insights in the understanding of their distinct disulfide bridge patterns. J. Biol. Chem. 275, 39394–39402 [DOI] [PubMed] [Google Scholar]
- 38. Bryan P. N., Orban J. (2010) Proteins that switch folds. Curr. Opin. Struct. Biol. 20, 482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. (2003) Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 278, 8606–8616 [DOI] [PubMed] [Google Scholar]
- 40. Dauplais M., Lecoq A., Song J., Cotton J., Jamin N., Gilquin B., Roumestand C., Vita C., de Medeiros C. L., Rowan E. G., Harvey A. L., Ménez A. (1997) On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 272, 4302–4309 [DOI] [PubMed] [Google Scholar]
- 41. Bontems F., Roumestand C., Gilquin B., Ménez A., Toma F. (1991) Refined structure of charybdotoxin. Common motifs in scorpion toxins and insect defensins. Science 254, 1521–1523 [DOI] [PubMed] [Google Scholar]
- 42. Smith J. J., Hill J. M., Little M. J., Nicholson G. M., King G. F., Alewood P. F. (2011) Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc. Natl. Acad. Sci. U.S.A. 108, 10478–10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., Rodríguez de la Vega R. C. (2009) The toxicogenomic multiverse. Convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 44. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 29–32 [DOI] [PubMed] [Google Scholar]
- 45. Papadopoulos J. S., Agarwala R. (2007) COBALT. Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 46. Bordon K. C. F., Cologna C. T., Tytgat J., Arantes E. C. (2011) 17th Congress of the European Section of the International Society of Toxinology, Valencia, September 11–15, 2011 (Calvete J. J., chairman), Poster 128, SOM Ciencia, Valencia, Spain [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.