Background: RAD51AP1 is a DNA-binding protein that enhances RAD51 recombinase activity.
Results: Our analyses revealed that RAD51AP1 possesses two DNA binding domains.
Conclusion: Both of the RAD51AP1 DNA binding domains are needed for protein function.
Significance: The results shed light on the mechanism of RAD51AP1 in the homology-directed repair of damaged DNA.
Keywords: DNA-binding protein, DNA Recombination, DNA Repair, DNA-Protein Interaction, Homologous Recombination, RAD51, RAD51AP1
Abstract
Homologous recombination catalyzed by the RAD51 recombinase is essential for maintaining genome integrity upon the induction of DNA double strand breaks and other DNA lesions. By enhancing the recombinase activity of RAD51, RAD51AP1 (RAD51-associated protein 1) serves a key role in homologous recombination-mediated chromosome damage repair. We show here that RAD51AP1 harbors two distinct DNA binding domains that are both needed for maximal protein activity under physiological conditions. We have finely mapped the two DNA binding domains in RAD51AP1 and generated mutant variants that are impaired in either or both of the DNA binding domains. Examination of these mutants reveals that both domains are indispensable for RAD51AP1 function in cells. These and other results illuminate the mechanistic basis of RAD51AP1 action in homologous DNA repair.
Introduction
Homologous recombination (HR)7 is a highly conserved DNA repair mechanism that rids chromosomes of double strand breaks and other deleterious lesions (1). As such, defects in HR lead to genome instability and tumorigenesis (2–4). In the HR reaction, the RAD51 recombinase polymerizes onto ssDNA derived from the primary lesion to form a helical protein filament known as the presynaptic filament. The presynaptic filament searches for and invades a homologous dsDNA donor, leading to the formation of a D-loop structure, which is resolved by one of several pathways to generate different recombinant types (5).
RAD51AP1 (RAD51-associated protein 1) interacts with RAD51 (6) and colocalizes with RAD51 to double strand break sites in cells (7). RAD51AP1 knockdown by RNA interference renders cells hypersensitive to DNA-damaging agents, such as mitomycin C (MMC), and prone to chromatid breaks (8, 9). RAD51AP1 binds DNA and stimulates the RAD51-mediated D-loop reaction (8, 9). Based on biochemical studies, it has been suggested that the DNA binding activity in the C-terminal portion of RAD51AP1 helps execute protein function (8). Surprisingly, as reported herein, RAD51AP1 possesses a second, distinct DNA binding domain that resides within the N-terminal portion of the protein. Characterization of point mutants impaired for the two DNA binding domains have provided evidence that both are needed for DNA damage resistance of cells. These results are quite unexpected and speak to the mechanism of action of RAD51AP1 in HR.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids for the Escherichia coli expression of the full-length human RAD51AP1 and the F1, F2, and F3 fragments with N-terminal maltose-binding protein and C-terminal (His)6 tags have been previously described (10). The C60 (residues 275–335) portion of RAD51AP1 was similarly cloned and expressed. All point mutations, truncations, and fusions of RAD51AP1 were generated with a site-directed mutagenesis kit (QuikChange XL, Stratagene) using the primers listed in supplemental Table 1.
Purification of RAD51AP1 and Mutants
RAD51AP1 and the F1, F2, and F3 fragments were purified using a procedure that encompasses cation exchange and affinity chromatographic steps, as described previously (10). The same procedure was employed in the purification of various RAD51AP1-derived polypeptides and mutants. RAD51 was expressed in E. coli and purified as described previously (11).
DNA Substrates
The DNA substrates used in the DNA binding and D-loop assays of this study have been described previously (12).
DNA Mobility Shift Assay
This assay was conducted as described previously (12).
Affinity Pulldown Assay
The indicated RAD51AP1 species (5 μg) was incubated with RAD51 (5 μg) in 30 μl of buffer A (25 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, 50 mm KCl) at 4 °C for 30 min. Then, 20 μl of amylose resin (New England Biolabs) was added followed by gentle mixing at 4 °C for 30 min. After washing the resin, bound proteins were eluted with 20 μl of 2% SDS. The supernatant containing unbound proteins, wash, and the SDS eluate, 10 μl of each, was analyzed by 10% SDS-PAGE and Coomassie Blue staining.
D-loop Assay
Unless stated otherwise, all the steps were conducted at 37 °C. RAD51 (0.8 μm) was incubated with radiolabeled 90-mer Oligonucleotide (2.4 μm nucleotides; refer to Ref. 12 for sequence) in 9.5 μl of buffer R (25 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm MgCl2, 2 mm ATP, 1 mm DTT) for 5 min to assemble the presynaptic filament. Then, the indicated amount of RAD51AP1, RAD51AP1 polypeptide, or mutant was incorporated in 2 μl for 5 min, after which pBluescript replicative form I DNA (35 μm base pairs) was added in 1 μl. Following a 10-min incubation, reaction mixtures were treated with 0.5% SDS and 0.5 mg/ml proteinase K for 20 min and then subjected to agarose gel electrophoresis in TAE buffer (40 mm Tris, 20 mm NaOAc, pH 7.4, 2 mm EDTA) at 25 °C. Gels were dried and analyzed in a Personal Molecular Imager FX (Bio-Rad), with quantification done using the Quantity One software (Bio-Rad).
RAD51AP1 Expression Vectors, Transfection, and Selection of Stable Cell Lines
The eGFP-RAD51AP1 expression vector has been described (8). The eGFP-RAD51AP1res (resistant to siRNA) expression vector was generated by site-directed mutagenesis using the primer pair listed in supplemental Table 1. The N-K6RA or C-K7WA mutation8 was introduced as described for the E. coli protein expression vectors above (see “Results”). HeLa cells were transfected with the RAD51AP1res expression vectors using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Twenty-four hours after transfection, cells were trypsinized, diluted in normal growth medium containing 1.7 mg/ml G418, and plated in 96-well plates at 30–1000 cells/well. Plates were re-fed with G418 twice, and colonies arising from single cells were picked 12 days after transfection, expanded, and tested for expression of the ectopic proteins by Western blot analysis using anti-GFP (ab290, Abcam) and anti-RAD51AP1 antibody, as described previously (13). Down-regulation of endogenous RAD51AP1 in the stably transfected HeLa cell derivatives was mediated by the RAD51AP1-directed siRNA with the sense sequence CCUCAUAUCUCUAAUUGCAUU, as described previously (8). In parental HeLa cells, RAD51AP1 was depleted using RAD51AP1-directed siRNA with the sense sequence GCAGTGTAGCCAGTGATTA, as described previously (9).
MMC Cytotoxicity Test, Immunoprecipitation, and Western Blot Analysis
To test for MMC sensitivity, a colony formation assay was carried out using cell lines stably expressing wild type or mutant eGFP-RAD51AP1res and in which endogenous RAD51AP1 was depleted by siRNA, as described (8). Transiently transfected HeLa cells were used for immunoprecipitations. Briefly, 13 × 106 cells were seeded into 15-cm dishes and, 20 h later, transfected with 70 μg of RAD51AP1res plasmid and 35 μg of HA-RAD51 plasmid (expressed from pCMV-HA-RAD51) using Lipofectamine2000. Nuclear extract was prepared 24 h after transfection, and immunoprecipitation was carried out as described previously (9).
RESULTS
Two Distinct DNA Binding Domains in RAD51AP1
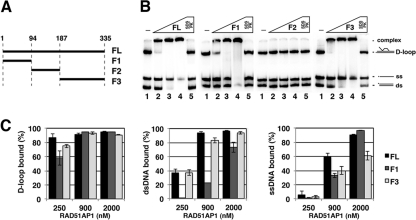
As reported before (8) and confirmed here with a D-loop substrate (supplemental Fig. 1, B and C), the C-terminal portion of RAD51AP1 harboring residues 188–335 (F3 fragment; Fig. 1A and supplemental Fig. 1A) binds DNA with a dissociation constant of ∼100 nm. However, the analysis showed that full-length RAD51AP1 (supplemental Fig. 1, B and C) has a higher affinity (dissociation constant ∼40 nm) for the substrate. This prompted us to address whether RAD51AP1 may have another DNA binding domain. Indeed, in another series of DNA binding experiments with a mixture of three substrates (ssDNA, dsDNA, and D-loop), a RAD51AP1 fragment harboring the N-terminal 94 residues (F1 fragment; Fig. 1A and supplemental Fig. 1A) could bind DNA (Fig. 1, B and C). In contrast, the middle portion of RAD51AP1 spanning residues 95–187 (F2 fragment; Fig. 1A and supplemental Fig. 1A) is clearly devoid of DNA binding activity (Fig. 1B). Thus, RAD51AP1 possesses two DNA binding domains residing within the N-terminal and C-terminal regions of the protein, respectively.
FIGURE 1.
Two distinct DNA binding domains in RAD51AP1. A, the RAD51AP1 fragments used in this study. FL, full-length RAD51AP1. B, purified full-length RAD51AP1 or RAD51AP1 fragments (0.25, 0.9, and 2.0 μm) were tested for the ability to bind radiolabeled D-loop, dsDNA, and ssDNA. C, quantification of results with error bars representing the mean ± S.D. from at least three independent experiments.
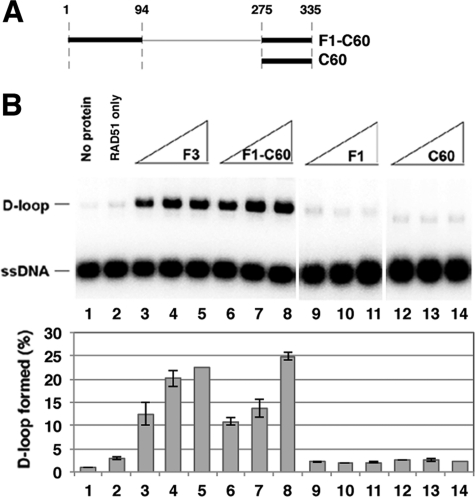
Consistent with previously published results (8), the F3 fragment could physically associate with RAD51 (supplemental Fig. 1D) and could also enhance the RAD51-mediated D-loop reaction (supplemental Fig. 1E). In contrast, neither F1 nor F2 could bind RAD51 (supplemental Fig. 1D) nor stimulate D-loop formation (supplemental Fig. 1E). Knowing that RAD51 interaction is indispensable for the HR function of RAD51AP1 (8, 9), we wondered whether endowing the F1 fragment with RAD51 binding activity would allow it to function in the D-loop reaction. We therefore appended the C-terminal 60 residues of RAD51AP1 that harbor the RAD51 binding domain (8, 9, 14) but lack DNA binding activity (supplemental Fig. 2C) to the F1 fragment and purified the F1-C60 polypeptide to test in biochemical systems (Fig. 2A and supplemental Fig. 2A). As expected, the F1-C60 polypeptide bound DNA and RAD51 (supplemental Fig. 2, B and C) and, importantly, exerted a strong stimulatory effect on D-loop formation to the level seen with the F3 fragment (Fig. 2B). The results also showed that C60 alone has no effect on D-loop formation (Fig. 2B). Thus, the N-terminal DNA binding activity of RAD51AP1, when endowed with a RAD51 interaction attribute, is functional in the RAD51-mediated D-loop reaction.
FIGURE 2.
Functionality of N-terminal DNA binding domain in D-loop reaction. A, the F1-C60 polypeptide and C60 fragment used. B, the F1-C60 polypeptide and the F1, F3, and C60 fragments (0.5, 0.75, and 1 μm) were tested with RAD51 in the D-loop reaction. The results were quantified and plotted; error bars are mean ± S.D. from at least three independent experiments.
Point Mutants Impaired for N-terminal DNA Binding Domain
To define the minimal DNA binding domain in the N-terminal region of RAD51AP1, we constructed and purified protein fragments that harbor residues 1–29, 1–49, or 1–69 appended to the C60 fragment (29-C60, 49-C60, and 69-C60, respectively) for biochemical testing (supplemental Fig. 3, A and B). As expected, all three polypeptides interacted with RAD51 (supplemental Fig. 3C). However, although 69-C60 and 49-C60 bound the DNA substrates as avidly as F1-C60 (compare supplemental Fig. 3D and supplemental Fig. 2C), 29-C60 had a much lower affinity for the substrates (supplemental Fig. 3D). Consistent with these results, a deletion of the first 53 residues in F1-C60 (F1Δ53-C60; supplemental Fig. 3, A and B) led to an impairment of DNA binding but had little or no effect on RAD51 interaction (supplemental Fig. 3, C and D). When tested in the D-loop assay, 69-C60 and 49-C60, but not 29-C60 or F1Δ53-C60, enhanced product formation (supplemental Fig. 3E).
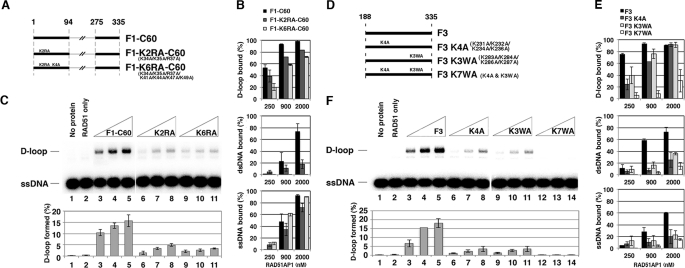
Altogether, the results above suggest that the region spanning residues 30 and 49 is important for DNA binding. This RAD51AP1 region harbors several highly conserved basic residues, Lys-34, Lys-35, Arg-37, Lys-41, Lys-44, Lys-47, and Lys-49, that could be involved in interaction with the negatively charged phosphodiester backbone of the DNA ligand (supplemental Fig. 4A). To test this idea, we constructed two compound mutants within the context of the F1-C60 polypeptide: the K2RA mutant with the first 3 (Lys-34, Lys-35, and Arg-37) basic residues changed to alanine, and the K6RA mutant with all 7 basic residues changed to alanine (Fig. 3A). When these two mutants, F1-K2RA-C60 and F1-K6RA-C60, were expressed in E. coli, they could be purified with similar yields as the wild type counterpart (supplemental Fig. 5A). Biochemical testing of these polypeptides revealed that although RAD51 interaction remains unaffected (supplemental Fig. 5B), both mutants are attenuated for binding D-loop and dsDNA, with the K6RA mutant exhibiting a more severe impairment (Fig. 3B). Importantly, we found that these mutants are largely devoid of the ability to stimulate the D-loop reaction (Fig. 3C).
FIGURE 3.
Effects of mutations in the N- and C-terminal DNA binding domains. A, mutants of the F1-C60 polypeptide. B, the F1-C60 polypeptide and mutants were tested for DNA binding, and the mean data ± S.D. from at least three independent experiments are shown. C, the F1-C60 polypeptide and mutants (0.5, 0.75, and 1 μm) were tested with RAD51 in the D-loop reaction. The results were quantified and plotted; error bars are mean ± S.D. from at least three independent experiments. D, mutants of the F3 fragment. E, the F3 fragment and mutants were tested for DNA binding, and the mean data ± S.D. from at least three independent experiments are shown. F, the F3 fragment and mutants (0.5, 0.75, and 1 μm) were tested with RAD51 in the D-loop reaction. The results were quantified and plotted; error bars are mean ± S.D. from at least three independent experiments.
Point Mutants Impaired for C-terminal DNA Binding Domain
According to the results of Modesti et al. (8), the RAD51AP1 C-terminal DNA binding domain resides within residues 226–290 of the protein. Sequence alignment of the deduced DNA binding domain in RAD51AP1 orthologs shows highly conserved basic and aromatic residues Lys-231, Lys-232, Lys-234, Lys-236, Lys-283, Lys-284, Lys-286, and Trp-287, that may be involved in ionic and stacking interactions with the phosphodiester backbone and bases in the DNA ligand, respectively (supplemental Fig. 4B). To test the relevance of these residues in DNA binding and functionality of RAD51AP1, we constructed and expressed three compound point mutants within the context of the F3 fragment, namely, K4A (K231A/K232A/K234A/K236A), K3WA (K283A/K284A/K286A/W287A), and K7WA, in which several or all (K7WA) of the aforementioned basic and hydrophobic residues have been changed to alanine (Fig. 3D). These mutant polypeptides could be purified with a similar yield as the wild type counterpart (supplemental Fig. 5C). In biochemical testing, all three mutants were competent for RAD51 interaction (supplemental Fig. 5D) but exhibited a varying degree of DNA binding deficiency, with the K7WA mutant being the most impaired in this regard (Fig. 3E). Importantly, the three mutants were nearly or completely devoid of stimulatory function in the D-loop reaction, in a manner that paralleled their DNA binding defect (Fig. 3F).
The Two DNA Binding Domains Are Indispensable for Protein Function
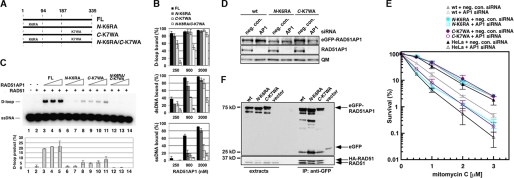
To ask whether both DNA binding domains are needed for RAD51AP1 function, we introduced the K6RA and K7WA mutations that impair the N-terminal and C-terminal DNA binding domains, singly (N-K6RA or C-K7WA) or in combination (N-K6RA/C-K7WA), into full-length RAD51AP1 (Fig. 4A). Following expression, the mutant proteins could be purified with a similar yield as the wild type counterpart (supplemental Fig. 5E). As expected, the three mutants could all associate with RAD51 with wild type affinity (supplemental Fig. 5F). Results from DNA binding experiments showed that the N-K6RA and C-K7WA mutants are both moderately impaired for DNA binding (Fig. 4B), which is not surprising given that each mutant still possesses a functional DNA binding domain. Importantly, the N-K6RA/C-K7WA double mutant is strongly impaired in this regard (Fig. 4B).
FIGURE 4.
Requirement for the two DNA binding domains in RAD51AP1 function. A, mutants of full-length RAD51AP1 (FL). B, binding of the D-loop, dsDNA, and ssDNA substrates by RAD51AP1 and mutants. Shown are the mean ± S.D. from at least three independent experiments. Note that the data for wild type protein are from Fig. 1C. C, the full-length RAD51AP1 and mutants (0.5, 0.75, and 1 μm) were tested with RAD51 in the D-loop reaction. The results were quantified and plotted; error bars are mean ± S.D. from at least three independent experiments. D, immunoblot analysis showing attenuation of endogenous RAD51AP1 but not of each eGFP-RAD51AP1res by siRNA treatment. QM indicates the loading control. wt, wild type; neg. con., negative control. E, results from colony formation assays to assess the fraction of surviving cells after MMC treatment. F, immunoprecipitation and Western blot analysis revealed that the wild type and eGFP-RAD51AP1res N-K6RA and C-K6WA mutants interact with both ectopically expressed HA-RAD51 and endogenous RAD51, whereas eGFP alone cannot.
We next tested the proficiency of the three RAD51AP1 mutants in the D-loop reaction. Because the single mutants possess a functional DNA binding domain, the D-loop reaction was carried out under an increased level of KCl to the physiological level, i.e. 150 mm (versus 50 mm used in prior experiments) to reveal any deficiency that may accompany the DNA binding mutations. Under these conditions, either single mutant is less adept at RAD51 enhancement, whereas the double mutant is completely devoid of any RAD51 stimulatory activity (Fig. 4C). Thus, both RAD51AP1 DNA binding domains are required for protein function.
Phenotypic Analysis of DNA Binding Mutants
RAD51AP1-deficient cells are hypersensitive to genotoxic agents, such as the DNA cross-linker MMC. Here, we examined the relevance of the two DNA binding domains of RAD51AP1 in the repair of MMC-induced damage. Based on the eGFP-RAD51AP1 expression vector described earlier (8), we designed eGFP-RAD51AP1res mutants that harbor the N-K6RA or C-K7WA mutation. We generated HeLa cell lines stably expressing wild type or mutant eGFP-RAD51AP1res protein and selected clones with comparable levels of ectopically expressed protein (Fig. 4D). Although endogenous RAD51AP1 could be efficiently knocked down by siRNA, all three eGFP-RAD51AP1res variants were resistant to the treatment (Fig. 4D). Next, we tested the ability of wild type or mutant eGFP-RAD51AP1res to provide resistance to MMC-induced cell death upon depletion of endogenous RAD51AP1 (Fig. 4E). Importantly, neither the N-K6RA nor the C-K6WA mutant could protect cells from MMC-induced death as well as the wild type protein (Fig. 4E). By immunoprecipitation using anti-GFP antibody, we verified that both RAD51AP1 mutants retain the ability to associate with RAD51 (Fig. 4F). Taken together, our results suggested that both the N-terminal and the C-terminal DNA binding domains of RAD51AP1 are critical for its function in vivo. Nonetheless, both the N-K6RA and the C-K7WA DNA binding mutants appear to retain residual function because the expression of either mutant led to some protection from MMC-induced cell death when compared with parental HeLa cells depleted for endogenous RAD51AP1 (Fig. 4E). The retention of partial function by the single mutants is likely because of the presence of the other DNA binding domain endowing limited function.
DISCUSSION
A previous study by Modesti et al. (8) has provided clear evidence that the C-terminal region of RAD51AP1 possesses a DNA binding activity. In fact, as shown before and as reiterated in this study, a C-terminal RAD51AP1 fragment harboring the reported DNA binding domain (8) and RAD51 interaction activity (8, 9, 14) can function in the D-loop reaction. In this regard, our published studies have furnished evidence that RAD51 interaction is critical for RAD51AP1 function, as point and deletion mutations that compromise RAD51 interaction render RAD51AP1 ineffectual in the HR reaction (9, 10). By alignment against orthologs, we have identified residues that are conserved within the deduced DNA binding domain, and several compound point mutations of these residues weaken the DNA binding attribute of the C-terminal RAD51AP1 fragment and its ability to stimulate the RAD51-mediated D-loop reaction. Thus, our efforts have helped pinpoint RAD51AP1 residues that are important for the functionality of the C-terminal DNA binding domain.
Rather unexpectedly, we have found in this study a novel DNA binding domain that resides within the N-terminal region of RAD51AP1. Furthermore, we have shown that a polypeptide consisting of this DNA binding domain fused to the RAD51 interaction motif is capable of RAD51 enhancement in the D-loop reaction. Via biochemical mapping and sequence alignment of the deduced minimal domain against the equivalent region in orthologs, conserved residues that could be involved in DNA binding have been identified. Indeed, two compound point mutations that alter these residues compromise DNA binding activity and functionality of this domain.
To address whether the two DNA binding domains are redundant or act in concert in the promotion of RAD51AP1 function, we have introduced mutations that inactivate either of the domains, singly or in combination, into the full-length protein, purified the mutants, and tested them in the D-loop reaction. Under conditions of physiological ionic strength, the RAD51AP1 single and double mutants all exhibit a functional defect that mirrors their DNA binding deficiency. Importantly, by genetic complementation, we have furnished evidence that the two DNA binding domains are both required for full biological activity of RAD51AP1.
Recent studies have provided evidence for functional synergy between RAD51AP1 and the tumor suppressor PALB2 in the RAD51-mediated homologous DNA pairing reaction (13). We note that PALB2 itself also possesses a DNA binding activity (13). It will be of particular interest to test whether maximal enhancement of RAD51 by the RAD51AP1-PALB2 pair requires both the DNA binding domains of RAD51AP1 as well as the DNA binding activity of PALB2.
Supplementary Material
Acknowledgments
We are grateful to Mauro Modesti and Zhiyuan Shen for providing the eGFP-RAD51AP1 and pCMV-HA-RAD51 plasmids, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1ES015252, RO1ES015632, RO1ES07061, PO1CA129186 (to P. S.), R01CA120315 (to D. S.), and P01CA092584 (to D. S. and P. S.).

This article contains supplemental Figs. 1–5 and Table 1.
The multiple mutation designations used are: N-K6RA, K34A/K35A/R37A/K41A/K44A/K47A/K49A C-K7WA, K231A/K232A/K234A/K236A/K283A/K284A/K286A/W287A.
- HR
- homologous recombination
- RAD51AP1
- RAD51-associated protein 1
- MMC
- mitomycin C
- eGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Sung P., Klein H. (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 2. Hartlerode A. J., Scully R. (2009) Mechanisms of double strand break repair in somatic mammalian cells. Biochem. J. 423, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lim D. S., Hasty P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 6. Kovalenko O. V., Golub E. I., Bray-Ward P., Ward D. C., Radding C. M. (1997) A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res. 25, 4946–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mizuta R., LaSalle J. M., Cheng H. L., Shinohara A., Ogawa H., Copeland N., Jenkins N. A., Lalande M., Alt F. W. (1997) RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. U.S.A. 94, 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Modesti M., Budzowska M., Baldeyron C., Demmers J. A., Ghirlando R., Kanaar R. (2007) RAD51AP1 is a structure-specific DNA-binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell 28, 468–481 [DOI] [PubMed] [Google Scholar]
- 9. Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D. W., Tsai M. S., Williams G. J., Rydberg B., Sung P., Schild D. (2007) Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell 28, 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunlop M. H., Dray E., Zhao W., Tsai M. S., Wiese C., Schild D., Sung P. (2011) RAD51-associated protein 1 (RAD51AP1) interacts with the meiotic recombinase DMC1 through a conserved motif. J. Biol. Chem. 286, 37328–37334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chi P., Van Komen S., Sehorn M. G., Sigurdsson S., Sung P. (2006) Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair 5, 381–391 [DOI] [PubMed] [Google Scholar]
- 12. Dray E., Dunlop M. H., Kauppi L., San Filippo J., Wiese C., Tsai M. S., Begovic S., Schild D., Jasin M., Keeney S., Sung P. (2011) Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51-associated protein 1 (RAD51AP1). Proc. Natl. Acad. Sci. U.S.A. 108, 3560–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dray E., Etchin J., Wiese C., Saro D., Williams G. J., Hammel M., Yu X., Galkin V. E., Liu D., Tsai M. S., Sy S. M., Schild D., Egelman E., Chen J., Sung P. (2010) Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17, 1255–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovalenko O. V., Wiese C., Schild D. (2006) RAD51AP2, a novel vertebrate- and meiotic-specific protein, shares a conserved RAD51-interacting C-terminal domain with RAD51AP1/PIR51. Nucleic Acids Res. 34, 5081–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.