Background: Acrolein is highly reactive and abundant in tobacco smoke.
Results: Acrolein induces DNA damage, inhibits excision repair and mismatch repair, causes repair protein degradation, and enhances mutagenesis.
Conclusion: Acrolein induces DNA damage and inhibits DNA repair that causes mutagenesis and initiates carcinogenesis.
Significance: This is the first demonstration that acrolein inhibits DNA repair pathways by induction of repair protein degradation.
Keywords: DNA Damage, DNA Mismatch Repair, DNA Nucleotide Excision Repair, DNA Repair, Lung Cancer, BPDE-DNA Damage, Acrolein, Mutagenesis
Abstract
Acrolein (Acr), a ubiquitous environmental contaminant, is a human carcinogen. Acr can react with DNA to form mutagenic α- and γ-hydroxy-1, N2-cyclic propano-2′-deoxyguanosine adducts (α-OH-Acr-dG and γ-OH-Acr-dG). We demonstrate here that Acr-dG adducts can be efficiently repaired by the nucleotide excision repair (NER) pathway in normal human bronchial epithelia (NHBE) and lung fibroblasts (NHLF). However, the same adducts were poorly processed in cell lysates isolated from Acr-treated NHBE and NHLF, suggesting that Acr inhibits NER. In addition, we show that Acr treatment also inhibits base excision repair and mismatch repair. Although Acr does not change the expression of XPA, XPC, hOGG1, PMS2 or MLH1 genes, it causes a reduction of XPA, XPC, hOGG1, PMS2, and MLH1 proteins; this effect, however, can be neutralized by the proteasome inhibitor MG132. Acr treatment further enhances both bulky and oxidative DNA damage-induced mutagenesis. These results indicate that Acr not only damages DNA but can also modify DNA repair proteins and further causes degradation of these modified repair proteins. We propose that these two detrimental effects contribute to Acr mutagenicity and carcinogenicity.
Introduction
Acrolein, an α,β-unsaturated aldehyde, is abundant in tobacco smoke, cooking fumes, and automobile exhaust fumes (1). Acr2 is also a by-product of lipid peroxidation generated endogenously in cells under oxidative stress (2). Inhaled Acr is extremely toxic in mouse models (3). In fact, the effects of Acr on lung carcinogenicity in mouse models have not been assessed due to excessive death of Acr-exposed mice (3). Nonetheless, it has been shown that intraperitoneal injection of Acr causes bladder tumors in rat models (4–7). Acr is a major metabolite of antitumor drugs cyclophosphamide and ifosfamide. Metabolically produced and inhaled Acr are excreted in urine and accumulated in the bladder (5, 6). It has been concluded that Acr is the culprit of bladder cancer in patients who have been administered cyclophosphamide and ifosfamide in long term treatment protocols (4–7).
Acr induces α- and γ-hydroxy-1,N2-cyclic propano-2′-deoxyguanosine (α-OH-Acr-dG and γ-OH-Acr-dG) adducts in human cells (8). It has been found that both types of Acr-dG adducts are mutagenic and that they induce mainly G to T and G to A mutations (9–18). By mapping Acr-dG adduct distribution at the nucleotide level in Acr-treated normal human bronchial epithelia (NHBE), we have found that the Acr-DNA binding spectrum in the p53 gene coincides with p53 mutational spectrum in lung cancer (19). Because Acr is abundant in tobacco smoke and its level is up to 10,000-fold that of benzo(a)pyrene (20, 21), we concluded that Acr is a major lung carcinogen. This conclusion is consistent with the finding that lung cancer is the number one cancer death in Taiwanese women, and yet only 5% of these women are tobacco smokers and that lung cancer incidence was greatly reduced in Taiwanese women in households that were equipped with fume extractors (22–26). These results strongly suggest that Acr-rich cooking fumes are involved in lung carcinogenesis.
To further understand the role of Acr in lung carcinogenicity, we examined how Acr-dG adducts are processed in NHBE and normal human lung fibroblasts (NHLF). We found that the Acr-dG adducts are poorly repaired in both Acr-treated NHBE and NHLF. However, cell lysates from untreated NHBE and NHLF are competent in repair of Acr-dG adducts, whereas cell lysates from Acr-treated cells are not. In searching for the mechanisms by which Acr inhibits DNA repair, we found that 1) Acr inhibits nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR); 2) Acr causes the reduction of XPA, XPC, hOGG1, MLH1, and PMS2 proteins and that this Acr-induced protein reduction can be prevented by a proteasome inhibitor; and that 3) Acr does not change the expression of these repair genes. Based on these results, we conclude that Acr modifications of DNA repair proteins, which trigger proteasome-mediated protein degradation, leading to reduced DNA repair activity. Consistent with this conclusion, we also found that Acr treatment enhances bulky DNA damage and oxidative DNA damage induced mutagenesis. These results indicate that Acr has two detrimental effects: it induces DNA damage and inhibits DNA repair. We propose that these effects contribute to Acr mutagenesis and carcinogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture, Acr Treatment, and Genomic DNA Isolation
NHBE were cultured in medium provided by Lonza (Basel, Switzerland). NHLF and lung adenocarcinoma cells (A549) (American Type Culture Collection, Manassas, VA) were grown in minimum essential medium supplemented with 10% FBS and DMEM supplemented with 10% FBS, respectively. Human NER-deficient XPA cells (GM05509) (National Institute of General Medical Sciences, Human Genetic Cell Repository) were grown in minimum essential medium supplemented with 15% FBS. Acr stock solutions (Sigma-Aldrich) were prepared freshly before use. Cells at 70% confluency were washed with PBS buffer (137 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4, 1.46 mm KH2PO4, pH 7.0) and treated with different concentrations of Acr (0–250 μm) in serum-free culture medium for 6 h at 37 °C in the dark. To determine Acr-dG adduct repair, cells were treated with 100 μm Acr for 6 h and then incubated at growth medium for different time. This treatment induced 50 to 90% of cytotoxicity in NHBE and NHLF, respectively. It should be noted that 50 μm Acr is equivalent to Acr concentration in lung tissue in an individual who has smoked 10–20 cigarettes (20). The Acr cytotoxicity was determined by WST-1 method as described previously (27). After treatment, the genomic DNA was isolated as described previously (19, 27).
Acr Modification of Supercoiled Plasmids and UvrABC Incision Assay of Acr-dG Adducts
Supercoiled pGL3 and pUC18 plasmids, purified as described previously (14, 19), were modified with different concentrations of Acr (0–5 mm) for 24 h at 37 °C and purified by repeated phenol and diethyl ether extraction; the DNA was then precipitated with ethanol and dissolved in TE (10 mm Tris-HCl, pH 7.5, 1 mm EDTA) buffer. Methods for UvrA, UvrB, and UvrC protein purifications, UvrABC nuclease incision assays, and separations of the resultant DNA, were the same as described previously (14, 19, 27).
Acr-DNA Adduct Analysis by Two-dimensional Thin Layer Chromatography (TLC)/High Performance Liquid Chromatography
The two-dimensional TLC/HPLC method was the same as described previously (14, 19, 27). Genomic DNA was purified from Acr-treated cells (0–100 μm) and Acr-DNA adducts were analyzed by the 32P post-labeling and the two-dimensional TLC method on polyethyleneimine cellulose sheets (Anatech, Newark, DE) and followed by HPLC (14, 19, 27).
Host Cell Reactivation and in Vitro DNA Damage-dependent Repair Synthesis (DDR)
Methods for isolation of plasmid luciferase plasmid pGL3, pSV-β-galactosidase, pUC18, pBR322, the host cell reactivation assay, and DDR assay, were the same as described previously (19, 27).
MMR Assay
The effect of Acr treatment on MMR was analyzed in HeLa cells using a functional in vitro MMR assay (28, 29). Using HeLa cells is out of necessity as the current in vitro MMR assay can be only performed in nuclear extracts, which require a relative large quantity of cells. Exponentially growing HeLa cells (2 × 109) were treated with Acr (200 μm) for 3 h before harvesting for nuclear extract preparation as described (28, 29). MMR activity was determined by incubating the nuclear extract (100 μg) with a mismatch-containing circular plasmid as described (28, 29).
Mutation Assays
The methods used for supF mutation detection was the same as described previously (14, 19, 27). Briefly, shuttle vector pSP189 plasmid DNA was irradiated by UV (1500 J/m2), treated by H2O2 (100 mm, 37 °C for 30 min) or modified by benzo(a)pyrene diol epoxide (BPDE) (15 μm, at room temperature for 2 h) and transfected into Acr-treated NHLF for replication. Plasmids were recovered 72 h after transfection, and replicated plasmids were then transformed into MB7070 Escherichia coli indicator cells. The mutation frequency is determined by the number of mutant white colonies divided by the number of total colonies.
RT-PCR
Total RNAs were extracted using the PureLinkTM RNA Mini Kit (Invitrogen). Reverse transcription was performed on 1 μg of total RNAs using the SuperScriptTM III First-Strand Synthesis System (Invitrogen) at 50 °C. Quantification of mRNA levels was carried out by PCR on GeneAmp PCR System 9700 apparatus (Applied Biosystem). Briefly, 10 ng of cDNAs were amplified using 0.2 μm primers and 1X GoTaq Flexi DNA polymerase Mix (Promega).
Repair Protein Detection
The levels of repair proteins, XPA, XPC, hOGG1, Ref1, MLH1, MSH2, and PMS2 were detected by Western blotting. Briefly, following treatment with Acr, cells were washed in PBS and lysed by radioimmune precipitation assay buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) and a mixture of protease inhibitors (10 μm aprotinin, 10 μm pepstatin A, 10 μm leupeptin, and 1 mm phenylmethylsulfonyl fluoride (PMSF)). Proteins (50 μg) were quantified according to Bio-Rad protein assay kit (Bio-Rad) and separated by SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane and blocked for 1 h at room temperature in TBS-T with 5% nonfat milk. Western blots were probed with the primary antibodies: anti-XPA (1:500), anti-XPC (1:500) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-hOGG1 (1:500) (Novus Biologics), anti-MLH1 (1:500), anti-PMS2 (1:500) (BD Pharmingen, BD Biosciences), anti-MSH2 (1:500), anti-Ref-1 (1:500) (Santa Cruz Biotechnology, Santa Cruz, CA), and anti- α-tubulin (1:1000) (Calbiochem) in TBS-T with 5% nonfat milk overnight at 4 °C. Horseradish peroxidase-conjugated secondary antibodies (1:5000) (Santa Cruz Biotechnology, Santa Cruz, CA) were then added in 5% nonfat milk for 2 h at room temperature. Proteins were detected using the ECL plus chemiluminescence kit (PerkinElmer Life Sciences) and films (Fuji medical x-ray film, Düsserldorf, Germany) were scanned with a Laser Scanning Densitometer (CanoScan 8800F).
RESULTS
Formation and Repair of Acr-dG Adducts in NHBE and NHLF
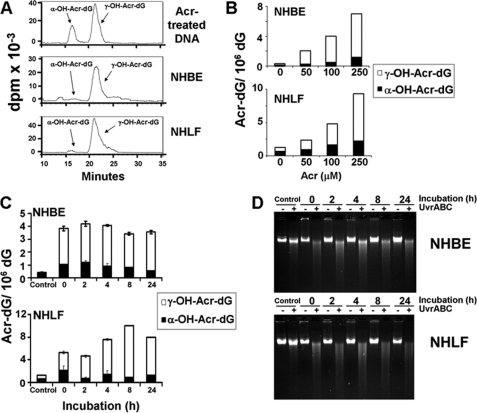
It has been long established that γ-Acr-dG adduct is the major adduct formed in DNA modified with Acr under neutral pH (8). However, Hecht and co-workers (30) recently reported that the ratio of α- and γ-Acr-dG adducts detected in human lung tissues varies dramatically among individuals. We, therefore, examined the types of Acr-dG adducts formed in primary cultured NHBE and NHLF by 32P post-labeling two-dimensional TLC and HPLC methods (14, 19, 27). Results in Fig. 1, A and B, show that α- and γ-Acr-dG adducts are the two isomeric Acr-dG adducts detected in Acr-treated NHBE and NHLF, that both types of adducts are formed in a dose-dependent manner and that the level of γ-Acr-dG adducts is much higher than the level of α-Acr-dG adducts. We then determined how α- and γ-Acr-dG adducts are processed in Acr-exposed human cells. Results in Fig. 1C show that both types of Acr-dG adduct were not significantly repaired in either NHBE or NHLF in 2–24 h. The lack of repair of Acr-dG adducts in Acr-treated cells was further confirmed by UvrABC-sensitive site determination. Previously, we found that E. coli nucleotide excision repair enzyme complex, UvrABC nuclease, is able to incise Acr-dG adducts formed in DNA fragments specifically and quantitatively (14, 19). Therefore, we determined the UvrABC sensitive sites formed in the genomic DNA of Acr-treated NHBE and NHLF after different incubation time. The results in Fig. 1D show that most, if not all, UvrABC-sensitive sites remain even after 24 h of incubation. It is worth noting that 50% of NHBE and 10% of NHLF are viable after Acr treatment (100 μm, 6 h). Therefore, these results indicate that both types of Acr-dG adducts are not repaired in Acr-treated NHBE and NHLF. We then determined the competency of NHBE and NHLF in repair of UV-induced DNA damage. NHBE and NHLF were irradiated with 20 J/m2, which induces a similar amount of UvrABC-sensitive sites as Acr treatment, and the UvrABC-sensitive sites formed in the genomic DNA after different incubation time were detected the same as in Fig. 1D. Results in supplemental Fig. S1 show that the majority of UV-induced UvrABC-sensitive sites are repaired after 8 h of incubation, indicating that NHBE and NHLF are NER-proficient.
FIGURE 1.
Formation and repair of Acr-induced DNA adducts in NHBE and NHLF. Exponentially growing cells were treated with different concentrations of Acr for 6 h at 37 °C. A and B, the genomic DNA was isolated, and the α-OH- and γ-OH-Acr-dG adducts were determined by 32P-post labeling/two-dimensional TLC/HPLC method as described previously (14, 19). C, cells treated with Acr (100 μm for 6 h) were incubated in normal culture medium for different time (0–24 h), and the unrepaired α- and γ-Acr-dG adducts in the genomic DNA were quantified as in B. D, Acr-DNA adducts formed in the genomic DNA of Acr-treated cells were detected by the UvrABC incision method as described previously (14, 19).
NER Is Pathway Responsible for Repair of α- and γ-Acr-dG Adducts
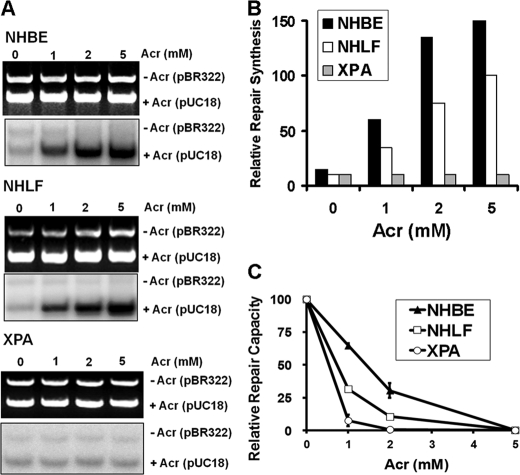
The results that Acr-treated NHBE and NHLF were unable to repair Acr-dG adducts seem counterintuitive because Acr-dG adducts are substrates of E. coli NER enzyme UvrABC nuclease and that NHBE and NHLF are NER-proficient (supplemental Fig. S1) (14, 19). Two possibilities could account for the lack of Acr-dG repair in Acr-treated NHBE and NHLF: 1) the NER system in human cells is incapable of repairing Acr-dG adducts; and 2) Acr treatment inhibits NER. To distinguish between these two possibilities, cell lysates isolated from NHBE and NHLF without Acr treatments were used to carry out in vitro DDR using Acr-modified DNA as substrates. As shown in Fig. 2, A and B, NHBE and NHLF cell lysates are competent at carrying out Acr-dG adduct-dependent repair synthesis. In contrast, cell lysates isolated from NER-deficient XPA cells are unable to carry out Acr-dG adduct-dependent repair synthesis. These results indicate that the NER system in NHBE and NHLF is able to recognize and repair Acr-dG adducts. This conclusion was further supported by the results in Fig. 2C, which demonstrate that NHBE cells and NHLF were able to repair more of Acr-modified luciferase gene than NER-deficient XPA cells. It is worth noting that NHBE are significantly more efficient in carrying out repair of Acr-dG adducts than NHLF.
FIGURE 2.
Repair of Acr-dG adducts in NER-proficient lung cells (NHBE and NHLF) and NER-deficient XPA cells. The repair of Acr-dG adducts was determined by in vitro DNA DDR (A and B) and host cell reactivation (C). A, DDR was carried out by using cell lysates isolated from untreated NHBE, NHLF, and XPA cells and Acr-modified DNA as substrates. The methods for cell lysate preparation and DDR are the same as described previously (19, 27). Upper panels are DNA bands stained with ethidium bromide representing the input DNA, and lower panels are radioautograms of the same gel representing the extent of repair synthesis. The relative repair synthesis is shown in B. C, host cell reactivation was performed by the transfection of Acr-treated luciferase reporter plasmids and unmodified β-galactosidase plasmids into NHBE, NHLF, and XPA cells, and the relative repair capacity was measured the same as described previously (19, 27). Plasmid DNA was modified with different concentrations of Acr under pH 8 the same as described previously (14, 19, 27).
To test the possibility that deficiency in repair of Acr-dG adducts formed in NHBE and NHLF is due to the inhibition of NER by Acr, we examined cell lysates isolated from Acr-treated NHBE and NHLF for their capacity to repair the classical NER substrate, UV-irradiated DNA. Results in the upper panel of Fig. 3A (panel 1) show that the repair capacity in cell lysates isolated from Acr-treated cells has an inverse relationship to the concentrations of Acr, indicating that Acr treatment indeed causes inhibition of NER function. These results together indicate that Acr-dG adducts are substrates for human NER, but Acr treatment inhibits the repair activity. As a result, Acr-dG adducts are repaired poorly in Acr-treated NHBE and NHLF (Fig. 1, C and D).
FIGURE 3.
Effect of Acr treatment on NER and BER. A, NHBE, NHLF, and A549 (lung adenocarcinoma) were treated with different concentrations of Acr at 37 °C, cell extracts were prepared, and the in vitro DDR was carried out by the same method as described previously (14, 19, 27). Upper panels are DNA bands stained with ethidium bromide representing the input DNA, and lower panels are radioautograms of the same gel representing the extent of repair synthesis. B, control cell lysates were treated with different concentrations of Acr directly and DDR was carried out the same as in A. UV-irradiated (A, panel 1, and B, panel 3) and H2O2-modified (A, panel 2, and B, panel 4) pUC18 DNA was used as DNA substrates to detect NER and BER capacity, respectively. The relative repair efficiencies of cell lysates of different treatments were shown in the right. Note that this result shows that Acr treatment inhibits both NER and BER.
Acr Inhibits NER, BER, and MMR
The results shown in Figs. 1 and 2 that Acr treatment impairs NER raise two questions: 1) What is the mechanism by which Acr exerts the inhibition of NER? 2) Does Acr treatment affect other repair pathways such as BER and MMR? To address these questions, we performed in vitro DDR using UV-irradiated DNA, which is well known NER substrates, and H2O2-modified DNA as substrates for BER carrying out by cell lysates isolated from Acr-treated NHBE, NHLF and A549 cells. It has been well established that A549 cells are proficient in both NER and BER (19, 31, 32). Results in Fig. 3, A, panels 1 and 2, show that cell lysates isolated from NHBE, NHLF, and A549 cells treated with different concentrations of Acr show diminished activity in carrying out UV-induced- and oxidative-DNA-damage-dependent repair synthesis, indicating that Acr treatment inhibits BER as well as NER.
Because Acr can react with not only DNA but also proteins, two possible mechanisms can account for the rather general effect of Acr on NER and BER: 1) Acr treatment suppresses repair gene expression; and 2) Acr modifies repair proteins and causes repair protein dysfunction. To test the second possibility, we determined the effect of Acr on DNA repair by adding Acr directly to cell lysates. The results in Fig. 3, B, panels 3 and 4, show that adding Acr directly to the cell lysates inhibits the activity of cell lysates in carrying out NER and BER. These results suggest that the Acr inhibitory effect on NER and BER is via its direct interaction with the DNA repair proteins.
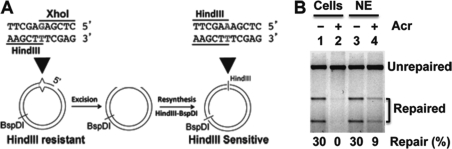
To examine the effect of Acr treatment on MMR, nuclear extracts were prepared from HeLa cells with and without Acr treatment and MMR assays were carried out using an in vitro MMR assay (Fig. 4A) (28, 29). Consistent with previous observations, nuclear extracts derived from untreated HeLa cells are competent in MMR (Fig. 4B, lane 1) (28, 29). However, nuclear extracts isolated from Acr-treated HeLa cells exhibited little MMR activity (Fig. 4B, lane 2), suggesting that Acr also impairs the MMR system. To determine the mechanism by which Acr inhibits MMR, we directly treated nuclear extracts isolated from untreated HeLa cells with Acr and measured the MMR activity of the treated extracts. The results revealed that the in vitro Acr treatment resulted in a significant (75%) reduction in MMR activity (Fig. 4B, lane 4), supporting the interpretation that the inhibitory effect on DNA repair activity by Acr is due to protein modifications.
FIGURE 4.
Acr treatment inhibits DNA MMR. A, schematic diagram of DNA substrate and in vitro MMR assay. B, MMR assay. HeLa nuclear extracts (75 μg) with the indicated treatments were incubated with 100 ng of heteroduplex DNA at 37 °C for 15 min in 15-μl reactions containing 10 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 1.5 mm ATP, and 0.1 mm dNTPs. DNA samples recovered were digested with restriction enzymes BspDI and HindIII (the scoring enzyme). Reaction products were analyzed through 1% agarose gel electrophoresis and visualized by UV illumination in the presence of ethidium bromide. Lanes 1 and 2 show MMR activity of nuclear extracts derived from HeLa cells (Cells) without and with Acr treatment, respectively; lanes 3 and 4 show repair activity of control HeLa nuclear extracts (NE) without and with Acr treatment, respectively.
To further test this possibility, we examined both steady state levels of mRNA and protein levels of key components required for NER (XPA and XPC), BER (hOGG1), and MMR (MLH1, MSH2, and PMS2) (33). We found that these repair proteins are highly expressed in A549 cells. However, Acr treatment causes a dose-dependent reduction of XPA, XPC, hOGG1, PMS2, and MLH1 proteins but has no effect on MSH2 and Ref1 (Fig. 5A). In contrast, Acr treatment does not affect the mRNA levels of these genes (Fig. 5B). These results further indicate that Acr impairs NER, BER, and MMR processes through its specific effects on repair proteins rather than their gene transcriptions. Similar results were also observed in Acr-treated NHBE and NHLF (Fig. 5, D and E). It has been long recognized that Acr can form Schiff base and carbonylate with amino acid such as lysine, cysteine, and histidine (1). It is possible that Acr modifications cause protein conformation change and that these Acr-modified proteins are subjected to degradation via proteasome similar to multiple ubiquitinated proteins (33). To test this possibility, cells were pretreated with the proteasome inhibitor MG132 (20 μm for 1 h) and then treated with Acr. Results in Fig. 5C show that Acr does not cause a reduction of repair proteins in cells pretreated with the proteasome inhibitor MG132, indicating that Acr-modified repair proteins are degraded by proteasomes. This Acr modification induced proteasome-dependent repair protein degradation, however, does not occur in cell lysates treated with Acr directly (Fig. 5F) even though the repair capacity in these cell lysates is greatly reduced (Fig. 3B). These results together suggest that Acr modification is sufficient to cause repair protein dysfunction.
FIGURE 5.
Effect of Acr treatment on protein and mRNA levels of XPA, XPC, hOGG1, MLH1, MSH2, PMS2, and Ref-1 genes in A549 cells, NHBE, and NHLF. A549 cells were treated with different concentrations of Acr for 3 h at 37 °C, the proteins were separated by SDS-PAGE and detected by Western blot (A), and the mRNA levels were detected by RT-PCR (B). In C, proteasome inhibitor MG132 (20 μm for 1 h) was added to A549 cells before Acr treatment, and the proteins were detected by the same method as described in (A). D and E, NHBE and NHLF were treated with different concentrations of Acr for 1 h at 37 °C. F, Acr was added directly to cell-free cell lysates (A549), which were used for in vitro DDR, incubated 3 h at 37 °C, and the proteins were detected as described in A. Note that 1) Acr treatment in vivo induces a dose-dependent reduction of XPA, XPC, hOGG1, MLH1, and PMS2 proteins but does not affect MSH2, Ref-1, and α-tubulin proteins; and 2) proteasome inhibitor MG132 inhibits the reduction of XPA, XPC, hOGG1, and MLH1 proteins induced by Acr treatments.
Acr Treatment Enhances Ultraviolet (UV) Light, BPDE, and Oxidative DNA Damage-induced Mutagenesis
Because Acr treatment causes XPA, XPC, and hOGG1 repair protein degradation and inhibits NER and BER, we expect Acr treatment will enhance bulky and oxidative DNA damage-induced mutagenesis. To examine this possibility, pSP189 shuttle vectors containing supF gene were irradiated with UV (254 nm), modified with BPDE, or modified with H2O2 and then transfected into NHLF with and without Acr treatment (10 μm for 1 h). After 72 h of incubation allowing DNA repair and subsequent replication of the transfected shuttle vectors to take place, the plasmid DNAs were recovered, and the supF mutations were detected by transforming the recovered plasmid DNAs into indicator E. coli cells. Results in Table 1 show that supF mutation frequencies were significantly higher in shuttle vectors that were transfected into Acr-treated cells than control cells. The same Acr treatment enhances H2O2- and UV-induced mutations by 2-fold (p < 0.05) and BPDE-induced mutations by 4-fold (p < 0.03). These results are consistent with the interpretation that Acr treatment inhibits DNA repair by NER, BER, and MMR.
TABLE 1.
Effect of Acr treatment on UV-, BPDE-, and H2O2-induced mutations in supF gene
| pSP189a | Acrb | White colonies/total colonies | Mutation frequency | Fold changec | p valued |
|---|---|---|---|---|---|
| μm | × 104 | ||||
| Control | 0 | 19/119,440 | 1.6 | ||
| 10 | 20/92,560 | 2.2 | 1.1 | ||
| H2O2 | 0 | 28/11,920 | 23.5 | ||
| 10 | 29/6240 | 46.5 | 2.0 | 0.043004e | |
| UV | 0 | 28/7920 | 35.4 | ||
| 10 | 175/24,015 | 72.9 | 1.8 | 0.014156e | |
| BPDE | 0 | 74/46,080 | 16.1 | ||
| 10 | 111/17,680 | 62.8 | 3.9 | 0.027459e |
a H2O2, 100 μm, 37 °C for 30 min; UV, 1500 J/m2; BPDE, 15 μm, 25 °C for 2 h.
b Acr: 10 μm, 37 °C for 1 h.
c Fold change is between untreated and Acr-treated mutation frequency.
d p value, two independent experiments and statistical significance were tested by Student's t test.
e p value < 0.05.
DISCUSSION
It is quite intriguing that although Acr-dG adducts are clearly shown to be substrates for NER, Acr-dG adducts are not repaired in Acr-treated human lung cells, which are otherwise NER-proficient. We pursued a possible explanation to account for these puzzling results by looking for mechanism by which Acr treatment also inhibits DNA repair. Our results here show that Acr treatment indeed inhibits not only NER but also BER and MMR. The fact that Acr inhibits multiple DNA repair pathways, which are controlled by different repair proteins, raises the possibility that the effects of Acr on DNA repair occur mainly through mechanisms other than altered gene expression. We found indeed that adding Acr to the cell lysates also inhibits NER, BER, and MMR and that Acr treatment does not down-regulate the mRNA levels of DNA repair genes such as XPA, XPC, hOGG1, MLH1, and PMS2. Furthermore, we found that Acr causes a dose-dependent reduction of these repair proteins. These findings together, lead us to conclude that effect of Acr on DNA repair is primarily through its interaction with the DNA repair proteins.
Acr is rather reactive; it readily reacts with amino acids such as lysine, cysteine, and histidine and covalently bonds with these residues through Michael addition (1). It has been found that Acr is one of the most effective agents to deplete glutathione (1, 5, 6). We found that the NHBE are significantly more resistant than A549 cells and NHLF toward Acr-induced inhibition of NER and BER capacity. Because these three types of cells are competent in both NER and BER, it is intriguing that Acr causes different extent of inhibition in NER and BER capacity among these three types of cells (Fig. 3). This finding raises the possibility that the glutathione levels in these three types of cells could be different, therefore, the free Acr available to interact with NER and BER proteins in these cells treated with the same amount of Acr will be different. To test this possibility, the levels of glutathione and their sensitivity toward cell viability in these cells were determined. As shown in supplemental Fig. S2, NHBE have the relatively higher concentration of glutathione than A549 cells and NHLF, which is consistent with their high resistance to Acr-induced cytotoxicity. These results indicate that it is highly probable that Acr reacts with many cellular components as readily as to genomic DNA. Consequently, in Acr-exposed cells, many more Acr have already reacted with cellular proteins, including repair proteins, when Acr reacts with genomic DNA forming Acr-dG adducts. Because Acr modifications can cause protein dysfunction (1, 5, 6), this scenario may account for the lack of Acr-dG adduct repair in Acr-treated cells.
We observed significant reduction of XPA, XPC, hOGG1, PMS2, and MLH1 in cells treated with relatively low concentrations of Acr. This Acr treatment-dependent repair protein reduction appears to be mediated by proteasome degradation. On the other hand, the levels of α-tubulin, MSH2, and Ref1 were not affected in the same Acr-treated cells. Because Acr-sensitive amino acids such as Cys, His, and Lys are ubiquitous in these proteins, these results lead us to propose that only when the Acr modifications cause a significant change of protein conformation, then these modified proteins are subjected to proteasome-mediated degradation. It is possible that these extensively Acr-modified proteins are ubiquitinated, which are then subjected to proteasome degradation.
Based on the reactivity of Acr toward cellular components such as proteins and nucleic acid, it can be expected that the cytotoxicity of Acr may also act through its interaction with other organelles such as mitochondria, that activates death pathways as well as through the genotoxicity pathway. It has been found that activation of mitochondria apoptosis is a major cause of Acr cytotoxicity (34–40). In accord with the idea, we have found recently that cells with depleted mitochondria are more resistant to Acr (27).
The fact that we detected Acr-dG DNA adducts in the cells treated with Acr that causes 50–90% cytotoxicity suggests that Acr-dG adducts have potential to induce mutations. Indeed, it has been reported that Acr treatment induces significant mutations in human cells (41). Our present result also shows that Acr treatment enhances UV, BPDE, and oxidative DNA damage-induced mutagenesis. This suggests that Acr-dG adducts in these Acr treated cells are more likely to induce mutations because the DNA repair mechanisms are inhibited in these cells. If Acr-exposed cells are also co-exposed to other mutagens such as alkylating agents and agents that induce oxidative base damages, a scenario that exists in lung epithelial cells in tobacco smokers, then more than additive mutations can be expected because DNA repair is greatly suppressed by Acr.
In conclusion, we present evidence that Acr treatment induces not only DNA damage but also inhibits DNA repair. The mechanism by which Acr induced inhibition of DNA repair is through Acr protein modifications, which likely induce proteasome-dependent degradation of the modified proteins. As a result, Acr treatment also enhances both bulky- and oxidative-DNA-damage-induced mutagenesis. Therefore, we propose that the mutagenicity and carcinogenicity of Acr is via a combination of DNA damage and inhibition of DNA repair.
Supplementary Material
Acknowledgment
We thank Dr. Cathy Klein for reviewing this manuscript.
This work was supported by National Institutes of Health Grants CA114541, ES014641, CA99007, CA134892, GM089684, and ES00260. This work was also supported by a Kentucky Lung Cancer Research Grant (to G.-M. L.) and Chinese 111 project B06018.

This article contains supplemental Figs. S1 and S2.
- Acr
- acrolein
- BPDE
- benzo(a)pyrene diol epoxide
- BER
- base excision repair
- MMR
- mismatch repair
- NER
- nucleotide excision repair
- DDR
- DNA damage-dependent repair synthesis
- NHBE
- normal human bronchial epithelia
- NHLF
- normal human lung fibroblasts
- NP1
- nuclease P1
- XPA
- xeroderma pigmentosum complement group A
- XPC
- xeroderma pigmentosum complement group C
- α- and γ-OH-Acr-dG
- α- and γ-hydroxy-1,N2-propano-2′-deoxyguanosine.
REFERENCES
- 1. Stevens J. F., Maier C. S. (2008) Acrolein: Sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 52, 7–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chung F. L., Chen H. J., Nath R. G. (1996) Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis 17, 2105–2111 [DOI] [PubMed] [Google Scholar]
- 3. Bein K., Leikauf G. D. (2011) Acrolein, a pulmonary hazard. Mol. Nutr. Food Res. 55, 1342–1360 [DOI] [PubMed] [Google Scholar]
- 4. Comes R. M., Eggleton M. (2002) Concise International Chemical Assessment Document 43, World Health Organization, Geneva [Google Scholar]
- 5. Kehrer J. P., Biswal S. S. (2000) The molecular effects of acrolein. Toxicol. Sci. 57, 6–15 [DOI] [PubMed] [Google Scholar]
- 6. Ghilarducci D. P., Tjeerdema R. S. (1995) Fate and effects of acrolein. Rev. Environ. Contam. Toxicol. 144, 95–146 [DOI] [PubMed] [Google Scholar]
- 7. Cohen S. M., Garland E. M., St John M., Okamura T., Smith R. A. (1992) Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 52, 3577–3581 [PubMed] [Google Scholar]
- 8. Chung F. L., Young R., Hecht S. S. (1984) Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 44, 990–995 [PubMed] [Google Scholar]
- 9. Kanuri M., Minko I. G., Nechev L. V., Harris T. M., Harris C. M., Lloyd R. S. (2002) Error-prone translesion synthesis past γ-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 277, 18257–18265 [DOI] [PubMed] [Google Scholar]
- 10. Kawanishi M., Matsuda T., Nakayama A., Takebe H., Matsui S., Yagi T. (1998) Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 417, 65–73 [DOI] [PubMed] [Google Scholar]
- 11. Minko I. G., Washington M. T., Kanuri M., Prakash L., Prakash S., Lloyd R. S. (2003) Translesion synthesis past acrolein-derived DNA adduct, gamma -hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase η. J. Biol. Chem. 278, 784–790 [DOI] [PubMed] [Google Scholar]
- 12. Sanchez A. M., Minko I. G., Kurtz A. J., Kanuri M., Moriya M., Lloyd R. S. (2003) Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 16, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 13. VanderVeen L. A., Hashim M. F., Nechev L. V., Harris T. M., Harris C. M., Marnett L. J. (2001) Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 276, 9066–9070 [DOI] [PubMed] [Google Scholar]
- 14. Wang H. T., Zhang S., Hu Y., Tang M. S. (2009) Mutagenicity and sequence specificity of acrolein-DNA adducts. Chem. Res. Toxicol. 22, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang I. Y., Chan G., Miller H., Huang Y., Torres M. C., Johnson F., Moriya M. (2002) Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry 41, 13826–13832 [DOI] [PubMed] [Google Scholar]
- 16. Yang I. Y., Hossain M., Miller H., Khullar S., Johnson F., Grollman A., Moriya M. (2001) Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 276, 9071–9076 [DOI] [PubMed] [Google Scholar]
- 17. Yang I. Y., Johnson F., Grollman A. P., Moriya M. (2002) Genotoxic mechanism for the major acrolein-derived deoxyguanosine adduct in human cells. Chem. Res. Toxicol. 15, 160–164 [DOI] [PubMed] [Google Scholar]
- 18. Yang I. Y., Miller H., Wang Z., Frank E. G., Ohmori H., Hanaoka F., Moriya M. (2003) Mammalian translesion DNA synthesis across an acrolein-derived deoxyguanosine adduct. Participation of DNA polymerase η in error-prone synthesis in human cells. J. Biol. Chem. 278, 13989–13994 [DOI] [PubMed] [Google Scholar]
- 19. Feng Z., Hu W., Hu Y., Tang M. S. (2006) Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U.S.A. 103, 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujioka K., Shibamoto T. (2006) Determination of toxic carbonyl compounds in cigarette smoke. Environ. Toxicol. 21, 47–54 [DOI] [PubMed] [Google Scholar]
- 21. Hoffmann D., Hecht S. S. (1990) Handbook of Experimental Pharmacology (Cooper C. S., Grover P. A., eds) pp. 70–74, Springer-Verlag Berlin, Germany [Google Scholar]
- 22. Gao Y. T., Blot W. J., Zheng W., Ershow A. G., Hsu C. W., Levin L. I., Zhang R., Fraumeni J. F., Jr. (1987) Lung cancer among Chinese women. Int. J. Cancer 40, 604–609 [DOI] [PubMed] [Google Scholar]
- 23. Ko Y. C., Lee C. H., Chen M. J., Huang C. C., Chang W. Y., Lin H. J., Wang H. Z., Chang P. Y. (1997) Risk factors for primary lung cancer among non-smoking women in Taiwan. Int. J. Epidemiol. 26, 24–31 [DOI] [PubMed] [Google Scholar]
- 24. Seow A., Poh W. T., Teh M., Eng P., Wang Y. T., Tan W. C., Yu M. C., Lee H. P. (2000) Fumes from meat cooking and lung cancer risk in Chinese women. Cancer Epidemiol. Biomarkers Prev. 9, 1215–1221 [PubMed] [Google Scholar]
- 25. Koo L. C., Ho J. H. (1990) Worldwide epidemiological patterns of lung cancer in nonsmokers. Int. J. Epidemiol. 19, S14–23 [DOI] [PubMed] [Google Scholar]
- 26. Siegfried J. M. (2001) Women and lung cancer: Does oestrogen play a role? Lancet Oncol. 2, 506–513 [DOI] [PubMed] [Google Scholar]
- 27. Tang M. S., Wang H. T., Hu Y., Chen W. S., Akao M., Feng Z., Hu W. (2011) Acrolein induced DNA damage, mutagenicity, and effect on DNA repair. Mol. Nutr. Food Res. 55, 1291–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holmes J., Jr., Clark S., Modrich P. (1990) Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl. Acad. Sci. U.S.A. 87, 5837–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 30. Zhang S., Villalta P. W., Wang M., Hecht S. S. (2007) Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 20, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mehta M., Chen L. C., Gordon T., Rom W., Tang M. S. (2008) Particulate matter inhibits DNA repair and enhances mutagenesis. Mutat. Res. 657, 116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng Z., Hu W., Tang M. S. (2004) Trans-4-hydroxy-2-nonenal inhibits nucleotide excision repair in human cells: A possible mechanism for lipid peroxidation-induced carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 8598–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kern J. C., Kehrer J. P. (2002) Acrolein-induced cell death: A caspase-influenced decision between apoptosis and oncosis/necrosis. Chem. Biol. Interact. 139, 79–95 [DOI] [PubMed] [Google Scholar]
- 35. Li X., Liu Z., Luo C., Jia H., Sun L., Hou B., Shen W., Packer L., Cotman C. W., Liu J. (2008) Lipoamide protects retinal pigment epithelial cells from oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 44, 1465–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo J., Robinson J. P., Shi R. (2005) Acrolein-induced cell death in PC12 cells: Role of mitochondria-mediated oxidative stress. Neurochem. Int. 47, 449–457 [DOI] [PubMed] [Google Scholar]
- 37. Roy J., Pallepati P., Bettaieb A., Averill-Bates D. A. (2010) Acrolein induces apoptosis through the death receptor pathway in A549 lung cells: Role of p53. Can. J. Physiol. Pharmacol. 88, 353–368 [DOI] [PubMed] [Google Scholar]
- 38. Roy J., Pallepati P., Bettaieb A., Tanel A., Averill-Bates D. A. (2009) Acrolein induces a cellular stress response and triggers mitochondrial apoptosis in A549 cells. Chem. Biol. Interact 181, 154–167 [DOI] [PubMed] [Google Scholar]
- 39. Sun L., Luo C., Long J., Wei D., Liu J. (2006) Acrolein is a mitochondrial toxin: effects on respiratory function and enzyme activities in isolated rat liver mitochondria. Mitochondrion 6, 136–142 [DOI] [PubMed] [Google Scholar]
- 40. Tanel A., Averill-Bates D. A. (2005) The aldehyde acrolein induces apoptosis via activation of the mitochondrial pathway. Biochim. Biophys. Acta 1743, 255–267 [DOI] [PubMed] [Google Scholar]
- 41. Curren R. D., Yang L. L., Conklin P. M., Grafstrom R. C., Harris C. C. (1988) Mutagenesis of xeroderma pigmentosum fibroblasts by acrolein. Mutat. Res. 209, 17–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.