Background: NAD+ is required for nuclear enzymes that regulate chromatin and gene expression.
Results: The nuclear NAD+ synthase NMNAT-1 is required for PARP-1-dependent gene regulation.
Conclusion: The enzymatic activities of NMNAT-1 and PARP-1 are linked to the regulation of common target genes through functional interactions at gene promoters.
Significance: Our work reveals a new mechanism for the regulation of gene expression by NAD+.
Keywords: Chromatin, Gene Expression, NAD, Nucleus, Transcription, Chromatin Immunoprecipitation, Nicotinamide Mononucleotide Adenylyltransferase-1, Poly(ADP-ribose), Poly(ADP-ribose) Polymerase-1, Two-photon Microscopy
Abstract
NMNAT-1 and PARP-1, two key enzymes in the NAD+ metabolic pathway, localize to the nucleus where integration of their enzymatic activities has the potential to control a variety of nuclear processes. Using a variety of biochemical, molecular, cell-based, and genomic assays, we show that NMNAT-1 and PARP-1 physically and functionally interact at target gene promoters in MCF-7 cells. Specifically, we show that PARP-1 recruits NMNAT-1 to promoters where it produces NAD+ to support PARP-1 catalytic activity, but also enhances the enzymatic activity of PARP-1 independently of NAD+ production. Furthermore, using two-photon excitation microscopy, we show that NMNAT-1 catalyzes the production of NAD+ in a nuclear pool that may be distinct from other cellular compartments. In expression microarray experiments, depletion of NMNAT-1 or PARP-1 alters the expression of about 200 protein-coding genes each, with about 10% overlap between the two gene sets. NMNAT-1 enzymatic activity is required for PARP-1-dependent poly(ADP-ribosyl)ation at the promoters of commonly regulated target genes, as well as the expression of those target genes. Collectively, our studies link the enzymatic activities of NMNAT-1 and PARP-1 to the regulation of a set of common target genes through functional interactions at target gene promoters.
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1)9 is a nuclear enzyme that participates in a wide array of regulatory processes in cells. Originally characterized as a critical factor in DNA damage repair, PARP-1 has emerged as an important regulator of gene expression through multiple mechanisms, many of which involve its association with chromatin (1, 2). PARP-1 uses NAD+ as a substrate for poly(ADP-ribosyl)ation (PARylation), adding linear or branched poly(ADP-ribose) (PAR) chains to target proteins (3). The best studied target for PARylation is PARP-1 itself through an automodification reaction (3). However, other nuclear proteins, including histones and chromatin-associated proteins (e.g. transcription factors and histone-modifying enzymes), can also be modified by PARP-1 (1, 2, 4). PARylation of these nuclear target proteins regulates their binding to DNA and to their protein interaction partners, leading to changes in chromatin structure and transcription regulation (1, 2, 4).
PARylation by PARP-1 consumes NAD+ as a donor of ADP-ribose units, generating nicotinamide as a reaction byproduct (see Fig. 1A). In mammalian cells, an NAD+ salvage/recycling pathway uses nicotinamide for NAD+ synthesis (see Fig. 1A) (5). Nicotinamide phosphoribosyltransferase catalyzes the first step of the NAD+ salvage pathway, converting nicotinamide to nicotinamide mononucleotide (NMN). NMN is further converted into NAD+ by nicotinamide mononucleotide adenylyltransferase (NMNAT). The mammalian nicotinamide phosphoribosyltransferase enzyme is essential for early embryonic development (6) and has been found in both the cytosol and nucleus (7, 8).
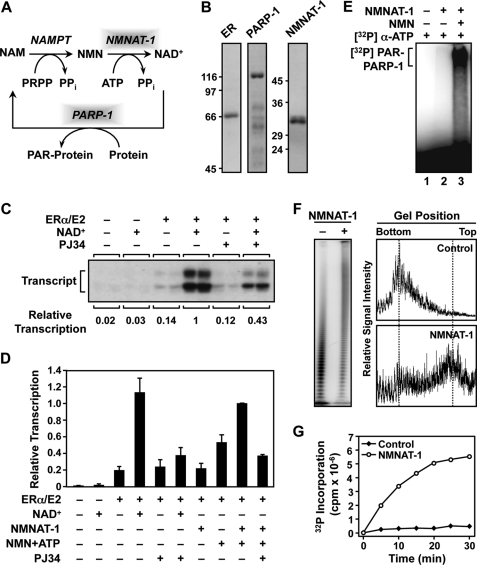
FIGURE 1.
NMNAT-1 produces NAD+ for PARP-1 automodification and PARP-1-dependent transcriptional regulation in vitro. A, the NAD+ salvage pathway produces NAD+ that is used by PARP-1 for PARylation of target proteins. NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyltransferase; PRPP, phosphoribosyl pyrophosphate. B, Coomassie Blue-stained gels of purified recombinant human ERα, PARP-1, and NMNAT-1 used in the in vitro studies. C and D, NMNAT-1 enhances ERα-dependent transcription with chromatin templates through a PARP-dependent mechanism. Chromatin was assembled using Drosophila S190 extract and transcribed using HeLa cell nuclear extract. The reactions were carried out in the presence or absence of either 1) NAD+ (300 μm) or 2) NMNAT-1 (100 nm) with its substrates NMN and ATP (both at 300 μm). PJ34, an inhibitor of PARP enzymatic activity, was added as indicated. The bars shown in D represent the mean of at least four independent experiments, and the error bars represent S.E. E, NAD+ synthesized in vitro by NMNAT-1 can be used by PARP-1 for automodification. PARP-1 auto(ADP-ribosyl)ation assays were carried out using purified PARP-1 and NMNAT-1. [α-32P]ATP and NMN (both at 200 μm) were used as the substrates for NMNAT-1-dependent NAD+ synthesis. All reactions included sheared salmon sperm DNA as an activator of PARP-1 enzymatic activity. Automodified PARP-1 was analyzed by SDS-PAGE and autoradiography. F, NMNAT-1 stimulates PAR chain elongation by PARP-1 independently of NAD+ production. [32P]NAD+ (0.67 μm) was used as the substrate for the PARP-1 ADP-ribosylation assay. The reactions were carried out in the presence of 200 μm ATP but without NMN so NMNAT-1 was not able to produce NAD+. Left, the reactions were subjected to digestion with proteinase K to release free PAR, run on a 10% denaturing polyacrylamide gel to resolve the PAR chains of different lengths, and analyzed by a PhosphorImager. Right, the relative signal intensities of the PAR chains of different lengths were quantified by scanning along the middle of each sample lane. G, the total amount of PAR synthesized by PARP-1 is stimulated by NMNAT-1. PARP-1 automodification reactions were carried out using [32P]NAD+ (0.67 μm) and [α-32P]ATP (20 μm) at the same specific activity. The amount of radiolabel incorporated by PARP-1 into PAR chains in the presence or absence of NMNAT-1 was quantified by a filter binding assay over the course of a 30-min reaction.
Three NMNAT enzymes exist in mammals (9–12). Among them, NMNAT-1 is exclusively nuclear (13). The unique nuclear localization of NMNAT-1 suggests that it may play a critical role in maintaining the functions of nuclear NAD+-dependent enzymes, such as PARP-1 and the protein deacetylase SIRT1. Several studies have suggested a role of NMNAT-1 in regulating NAD+-dependent nuclear functions. Overexpression of a chimeric protein with intact NMNAT-1 activity protects neurons against physical, chemical, and genetic insults (14–16). The results of some studies suggest that nuclear NAD+ production might be a key contributing factor for these protective effects (17, 18). We have recently shown that NMNAT-1 is recruited to target gene promoters through interactions with promoter-bound SIRT1, where it produces NAD+ that is used by SIRT1 to support its histone deacetylase activity (19). Furthermore, biochemical studies have shown that NMNAT-1 binds to automodified PARP-1 and stimulates its enzymatic activity in a phosphorylation-dependent manner (20), suggesting a signal-regulated role of NMNAT-1 in controlling PARP-1 function as well.
A number of studies from our laboratory and others have revealed important roles for PARP-1 in regulating chromatin structure and gene transcription (21–32). For example, using genomic chromatin immunoprecipitation (ChIP) analyses, we have shown that PARP-1 is associated with transcriptionally active promoters across the genome in MCF-7 breast cancer cells (21). Several mechanisms for transcriptional regulation by chromatin-associated PARP-1 have emerged from gene-specific studies (24–26, 28–34). In many cases, these actions of PARP-1 require its enzymatic activity (22–25, 27–32). For example, using cell and molecular assays, we have shown that PARP-1 PARylates the histone demethylase KDM5B, which inhibits its association with chromatin and its ability to demethylate histone H3 trimethylated at lysine 4 in MCF-7 cells (32). The role of nuclear NAD+ production in transcriptional regulation by PARP-1, however, has not been explored in detail.
In this study, we used a variety of biochemical, molecular, cell-based, and genomic assays to examine the physical and functional interactions between NMNAT-1 and PARP-1. Our studies show that NMNAT-1 interacts with PARP-1 at target gene promoters and enhances PARP-1-dependent PARylation at these promoters by producing NAD+. Collectively, our results indicate a functional link between the enzymatic activities of NMNAT-1 and PARP-1 at gene promoters that regulates the expression of a set of common target genes.
EXPERIMENTAL PROCEDURES
Plasmids and Short Hairpin RNAs (shRNAs)
All human NMNAT-1 and PARP-1 constructs used in this study (i.e. pET15b and pGEX2TK for bacterial expression, pQCXIP for mammalian expression, and pSuper.Retro for shRNA-mediated knockdown) have been described previously (19, 28). Mutant versions of NMNAT-1 were generated by site-directed mutagenesis and confirmed by sequencing. For the human NMNAT-1 and PARP-1 shRNAs, multiple target sequences were tested for each gene, and the most effective sequences were used for the studies described herein: NMNAT-1, 5′-AACACAAGATTCTAGTCAA-3′ (shRNA 1); PARP-1, 5′-GGGCAAGCACAGTGTCAAA-3′ (shRNA 1) and 5′-ACACCTCTCTACTATATAA-3′ (shRNA 2) (19, 28). Potential off-target effects of the NMNAT-1 and PARP-1 shRNAs have been assessed in detail previously and were found to be of minimal concern (19, 28). An shRNA directed against firefly luciferase, 5′-GATATGGGCTGAATACAAA-3′ (35), was used as a control.
Antibodies
The custom rabbit polyclonal anti-PARP-1 and anti-NMNAT-1 antisera have been described previously (19, 22). Anti-FLAG M2 monoclonal antibody was from Sigma-Aldrich. Anti-PAR monoclonal antibody (clone 10H) was from Trevigen, Inc. (Gaithersburg, MD).
Expression and Purification of Recombinant Proteins
Recombinant NMNAT-1 was expressed in Escherichia coli and purified using nickel-nitrilotriacetic acid (NTA) affinity chromatography under standard non-denaturing conditions. Recombinant human PARP-1 was expressed in E. coli and purified as described previously (22, 23). Recombinant FLAG-tagged human estrogen receptor α (ERα) was expressed in Sf9 insect cells and purified as described previously (36, 37). The purified proteins were quantified by Coomassie Blue staining on SDS-PAGE gels using BSA as a standard.
Chromatin Assembly and in Vitro Transcription
pERE, an ∼3.2-kb DNA plasmid containing four tandem estrogen response elements (EREs) upstream of the adenovirus E4 core promoter (36), was assembled into chromatin using purified native Drosophila core histones and S190 extract derived from Drosophila embryos as described previously (36–38). Fifteen-microliter aliquots of S190-assembled chromatin containing 75 ng of DNA were incubated with or without 100 nm NMNAT-1, 300 μm NMN, 300 μm ATP, 300 μm NAD+, and 10 μm PJ34 (Alexis Biochemicals) as indicated for 20 min at room temperature. Next, 10 nm ERα and 100 nm 17β-estradiol were added where indicated and followed by a 20-min incubation at room temperature. These reactions were then preincubated for 15 min with HeLa cell nuclear extract (as a source of the RNA polymerase II transcriptional machinery) and transcribed for 30 min at 30 °C upon addition of rNTPs as described (36, 37). Synthesized RNA was detected using primer extension, followed by separation on a denaturing 8% urea-polyacrylamide gel with subsequent autoradiographic analysis. Data quantification was performed using a PhosphorImager (GE Healthcare) and ImageQuant software. All reactions were performed in duplicates, and at least three independent experiments were done to ensure reproducibility.
PARP-1 Auto(ADP-ribosyl)ation Assays
PARP-1 auto(ADP-ribosyl)ation assays were carried out as described previously (22, 23). All reactions contained 2.5 pmol of purified PARP-1 protein and sheared salmon sperm DNA as an allosteric activator of PARP-1 enzymatic activity. Purified NMNAT-1 (15 pmol) was included where indicated. The substrates for NMNAT-1, [α-32P]ATP and NMN, were supplied at a final concentration of 200 μm. For reactions where [32P]NAD+ was the sole source of radioactive label, the concentration of [32P]NAD+ was 0.67 μm, and 200 μm unlabeled ATP was included where indicated. For reactions where both [32P]NAD+ and [32P]ATP were sources of radioactive label, 20 μm [32P]ATP and 0.67 μm [32P]NAD+, both at a specific activity of 50 Ci/mmol, were included. Automodified PARP-1 was resolved by 6% SDS-PAGE and analyzed by autoradiography. For time course analyses, reactions were stopped with 1 ml of ice-cold 25% TCA. The precipitated protein was bound to filters and quantified in a scintillation counter. For determination of PAR chain length, the products of the PARP-1 automodification assay were deproteinized by incubation with proteinase K (39). The reactions were then extracted with phenol:chloroform (50%:50%) and precipitated with ethanol. Purified PAR was resolved on a 10% denaturing polyacrylamide gel (39).
Cell Culture, Ectopic Expression, and shRNA-mediated Knockdown
MCF-7 cells were kindly provided by Dr. Benita Katzenellenbogen (University of Illinois, Urbana-Champaign, IL) and maintained in Eagle's minimum essential medium (Sigma) supplemented with 5% calf serum and antibiotics. Phoenix-Ampho retrovirus producer cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) with 10% fetal bovine serum. Retroviruses were prepared following a standard transfection protocol using the Phoenix-Ampho cell line and the retroviral vectors described above (i.e. derivatives of pQCXIP and pSuper.Retro). The recombinant retroviruses were used to transduce MCF-7 cells, which were then selected with 1.0 μg/ml puromycin (Sigma-Aldrich) or 800 μg/ml G418 (Invitrogen) as appropriate for each vector. Ectopic expression and knockdown were screened by Western blotting.
Immunofluorescent Staining
MCF-7 cells were fixed in 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. After blocking in 5% goat serum, the cells were incubated for 1 h with primary antibodies (1:1000 dilution of PARP-1 antibody; 1:500 dilution of FLAG M2 antibody). FITC-conjugated goat anti-mouse and rhodamine-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories) were diluted 1:3000 for incubation. Images were taken with a Leica TCS SP2 confocal microscope at the Cornell University Life Sciences Core Laboratories Center.
Two-photon Live Cell Imaging of Nuclear and Cytoplasmic Autofluorescence
MCF-7 cells were cultured in 35-mm-diameter glass bottom Petri dishes (Mattek Corp., Ashland, MA). Two-photon microscopy was performed with a laboratory-built Bio-Rad laser-scanning unit (MRC 1024) coupled to an Olympus IX-70 inverted microscope with an Olympus 100× objective (numerical aperture, 1.4; oil immersion) as described (40). The excitation was from a titanium-sapphire mode-locked laser (Tsunami, Spectra Physics) providing ∼100-fs pulses at an 80-Mhz repetition rate of 740-nm wavelength. The excitation power was ∼10 milliwatts. The cellular autofluorescence was detected by a bialkali photomultiplier tube through a 400–550-nm emission filter (40). Images taken on the same day were obtained with the same imaging conditions (e.g. detector gain, etc.) for signal comparison. Laser power fluctuation was minimal over the experimental period on the same day. Three-dimensional optical section series were acquired to ensure the correct assignment of the cell nucleus in the axial (z) direction. Nuclear and cytoplasmic regions were selected from the image based on intensity threshold, and the average intensities were calculated within these regions. Fluorescence signals from regions devoid of cells were subtracted as background.
Expression Microarray Analyses
To knock down target factors for expression microarray analyses, the following shRNA sequences were used: NMNAT-1, shRNA 1 (maintained under puromycin selection); PARP-1, shRNAs 1 and 2 (maintained under puromycin and G418 selection). For each factor studied, three independent isolates of MCF-7 cells recently (within 2–3 weeks) transduced with shRNA-expressing retroviruses were used. Studies on NMNAT-1 and PARP-1 were carried out independently, and each had its own matching luciferase control. Total RNA was isolated from the various MCF-7 cell lines using TRIzol Reagent (Invitrogen) followed by RNeasy columns (Qiagen).
Sample labeling and hybridization to Affymetrix Human Genome U133 Plus 2.0 and U133A 2.0 arrays were carried out under standard conditions at the Cornell Microarray Core Facility. The raw data were processed as described previously using Affymetrix GCOS software to obtain detection calls and signal values (19). Where applicable, the common probe sets of the two array platforms were normalized by scaling. The data sets were adjusted for batch effects using a parametric empirical Bayes method (41). Following this normalization, all values <0.01 were adjusted to 0.01, and the data were log2-transformed, median-centered for each array, and median-centered for each gene. Only those probe sets having “present” calls in at least two of the three replicates were included for further analysis. A two-tailed Student's t test was applied to the normalized data matrix to identify genes differentially expressed between each knockdown condition and the matching luciferase control. A p value cutoff of 0.05 was applied to define the differentially expressed gene set. A -fold change cutoff of log2 <−0.5 or >0.5 was applied to select significantly regulated gene sets. The expression microarray data sets can be accessed from the NCBI Gene Expression Omnibus (GEO) web site using accession numbers GSE13458 (NMNAT-1) and GSE12952 (PARP-1).
Reverse Transcription-Quantitative Real-time PCR (RT-qPCR)
Total RNA samples were purified using TRIzol Reagent (Invitrogen). cDNA samples were prepared by reverse transcription with an oligo(dT) primer and analyzed by SYBR Green real-time PCR under standard conditions (19). For data normalization, β-actin was used as an internal reference gene. The sequences of the primers used are listed in supplemental Table S1.
ChIP-qPCR and ChIP-chip
ChIP assays using MCF-7 cells were performed as described previously (19, 21, 42). Protein A-agarose beads were used for PARP-1 and PAR ChIP, and protein G-agarose beads were used for FLAG ChIP. For gene-specific analysis, qPCR was used to determine the enrichment of immunoprecipitated material relative to the input material using gene-specific primer sets for the specified regions. The sequences of the primers used are listed in supplemental Table S2. For ChIP-chip analysis, the immunoprecipitated genomic DNA was blunted, amplified by ligation-mediated PCR, and used to probe the NimbleGen HD2 human (HG18) promoter microarray (21). Additional details on the ChIP-chip experiments, which are shown in supplemental Fig. S2, can be found under supplemental “Experimental Procedures.”
ChIP-Western
ChIP assays were carried out as described above. After washing the samples in ChIP wash buffer, the protein A/G beads were resuspended in 2× SDS loading dye and boiled for 10 min. Samples were resolved by 10% SDS-PAGE for standard Western blot analysis.
GST-NMNAT-1 Interaction Assays
GST and GST-NMNAT-1 were expressed in bacteria using pGEX2TK-based constructs and purified using standard glutathione-agarose affinity chromatography. The purified proteins were quantified by Coomassie Blue staining on SDS-PAGE gels using BSA as a standard. HeLa cell nuclear extract was incubated with immobilized GST or GST-NMNAT-1, the samples were washed, and the specifically bound proteins were analyzed by Western blotting (19).
RESULTS
NMNAT-1 Produces NAD+ for PARP-1 Automodification and PARP-1-dependent Transcriptional Regulation in Vitro
As shown in the reaction diagram in Fig. 1A, PARP-1 catalyzes the PARylation of proteins using NAD+ as a donor of ADP-ribose units. To explore the NAD+-dependent activity of PARP-1 in the control of transcription, we used a variety of biochemical, molecular, cell-based, and genomic assays. We have shown previously that PARP-1 in the absence of NAD+ represses ERα-dependent transcription in an in vitro transcription assay with chromatin templates (22, 23, 43). The repression by PARP-1 is relieved following addition of NAD+ and subsequent automodification of PARP-1 (22, 23, 43). Using a similar in vitro transcription assay with chromatin templates, we examined whether the nuclear NAD+ synthase NMNAT-1 could produce NAD+ to support the activity of PARP-1 and relieve the PARP-1-mediated repression in these assays.
For the in vitro assays, we used recombinant His6-tagged NMNAT-1 expressed in E. coli and purified using nickel-NTA affinity chromatography as well as FLAG-tagged ERα expressed in Sf9 insect cells and purified using FLAG affinity chromatography (Fig. 1B). The ERα-responsive plasmid reporter construct pERE (36) was assembled into chromatin using Drosophila S190 chromatin assembly extract (38) and transcribed using HeLa cell nuclear extract as a source of the RNA polymerase II transcription machinery as described previously (37). As shown in Fig. 1C, addition of 300 μm NAD+ stimulated ERα-dependent transcription ∼7-fold. PJ34, an inhibitor of PARP enzymatic activity, significantly reduced the stimulatory effect of NAD+ (Fig. 1C), suggesting that a PARP family member from the chromatin assembly extract or the transcription extract, presumably PARP-1 as suggested by our previous results (22), was mediating the effect of NAD+ on ERα-dependent transcription. Importantly, adding NMNAT-1 and its substrates NMN and ATP to the transcription reaction had the same stimulatory effect as adding NAD+, and this effect was similarly diminished by PJ34 treatment (Fig. 1D). Together, these in vitro transcription assays demonstrate that NMNAT-1 can synthesize NAD+ for use by a PARP family member for transcriptional regulation.
To test directly whether NMNAT-1 can regulate PARP-1 activity through NAD+ synthesis, we examined NMNAT-1 function in a PARP-1 automodification assay. Recombinant His6-tagged PARP-1 expressed in E. coli and purified using nickel-NTA affinity chromatography was incubated with [α-32P]ATP with or without NMNAT-1 and NMN. Sheared DNA was added to all reactions as an activator of PARP-1 enzymatic activity. Under these conditions, PARP-1 was only automodified when NMNAT-1 and both of its substrates (i.e. ATP and NMN) were present (Fig. 1E). Collectively, the results from these in vitro assays show that NMNAT-1 produces NAD+ to support PARP-1 enzymatic activity and PARP-1-dependent transcriptional regulation with chromatin templates.
NMNAT-1 Also Stimulates PARP-1 Enzymatic Activity Independently of NAD+ Production
Previous studies have shown that NMNAT-1 interacts with automodified PARP-1 and allosterically stimulates its enzymatic activity (20). To determine whether functional interplay between NMNAT-1 and PARP-1 occurs in our assay system, we carried out PARP-1 automodification reactions using [32P]NAD+ for isotopic labeling. In the presence of a low concentration of NAD+ (∼1 μm), the majority of PAR chains synthesized by PARP-1 were ∼10 ADP-ribose units in length (Fig. 1F). The addition of NMNAT-1 to the reaction in the presence of unlabeled ATP dramatically stimulated PARP-1 enzymatic activity, giving rise to much longer PAR chains (Fig. 1F). This effect of NMNAT-1 was also evident from the much slower migration of automodified PARP-1 on SDS-PAGE gels following auto(ADP-ribosyl)ation reactions in the presence of NMNAT-1 and ATP (supplemental Fig. S1A). Importantly, no NMN was added to these PARP-1 auto(ADP-ribosyl)ation reactions. Consequently, there was no net synthesis of NAD+ even though NMNAT-1 and ATP were both present in the reactions.
Due to the reversible enzymatic activity of NMNAT-1 (13), the presence of both NMNAT-1 and unlabeled ATP in the ADP-ribosylation reactions affects the specific activity of [32P]NAD+ (i.e. the [32P]phosphate groups are exchanged for cold phosphate groups). As a result, the amount of PAR synthesized in these reactions cannot be reliably determined by incorporation of radioactive label on PARP-1 from [32P]NAD+ (supplemental Fig. S1A). To bypass this issue, we carried out PARP-1 automodification reactions using both [32P]NAD+ and [α-32P]ATP at the same specific activity. Under these conditions, we clearly observed that NMNAT-1 increased the total amount of PAR produced by PARP-1 (Fig. 1G and supplemental Fig. S1B). Together, the results from our PARP-1 automodification assays demonstrate that NMNAT-1 can stimulate PAR synthesis and PAR chain elongation by PARP-1 independently of net NAD+ production.
NMNAT-1 Localizes to Nucleus and Mediates Nuclear NAD+ Synthesis
To study the function of NMNAT-1 in cells, we used the MCF-7 breast cancer cell as a model. We have shown previously that NMNAT-1 activity is required for the function of the NAD+-dependent protein deacetylase SIRT1 in MCF-7 cells (19). Stably expressed FLAG-NMNAT-1 localized to the nucleus of MCF-7 cells as assessed by immunofluorescent staining (Fig. 2, A and B), consistent with previous reports in other cell types (13). PARP-1 exhibited a nuclear distribution similar to that of NMNAT-1 (Fig. 2A). Thus, both NMNAT-1 and PARP-1 localize to the same cellular compartment where they can functionally interact.
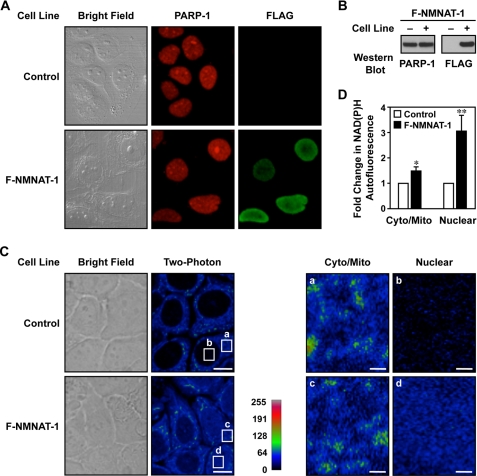
FIGURE 2.
NMNAT-1 localizes to nucleus and mediates nuclear NAD+ synthesis in MCF-7 cells. A, immunofluorescent staining of PARP-1 and NMNAT-1 in control and FLAG-NMNAT-1 (F-NMNAT-1)-expressing MCF-7 cells. B, Western blot analysis of PARP-1 and FLAG-NMNAT-1 protein levels in the MCF-7 cells used in A. C, ectopic expression of NMNAT-1 increases nuclear NAD(P)H autofluorescence in MCF-7 cells. Left, bright field and two-photon excitation microscopy images are shown for control and FLAG-NMNAT-1-expressing MCF-7 cells. Scale bars, 10 μm. Right, detailed images showing the two-photon microscopy signals from the cytoplasm/mitochondria (cyto/mito) and nucleus (nuclear) for control and FLAG-NMNAT-1-expressing MCF-7 cells. Scale bars, 1 μm. The white boxes labeled a–d in the left panels represent the regions labeled a–d magnified in the right panels. D, quantification of NAD(P)H autofluorescence levels from two-photon excitation images. Three to four different fields, each with seven to 10 cells, were analyzed for each condition. The bars shown represent the mean, and the error bars represent S.E. Statistical significance was determined using a two-tailed Student's t test. *, p value <0.1; **, p value <0.05.
To associate NMNAT-1 localization with its enzymatic activity, we used two-photon excitation microscopy (44). Two-photon excitation microscopy at the excitation and detection wavelengths that we used herein allows the detection of the intrinsic autofluorescence of NADH and NADPH, collectively referred to as NAD(P)H (40, 44). NAD+ does not exhibit this autofluorescence, but we can use the NAD(P)H signal as an indirect measurement of NAD+ levels in living cells (45) and an indicator for detecting NMNAT-1 enzymatic activity. This assay has the unique advantage of assaying relative NAD+ levels within specific intracellular compartments.
Using this approach, control MCF-7 cells (i.e. without ectopic expression of NMNAT-1) had very low levels of NAD(P)H autofluorescence in the nucleus (Fig. 2C). In contrast, MCF-7 cells with ectopic expression of NMNAT-1 showed dramatically elevated levels of NAD(P)H autofluorescence in the nucleus (3-fold increase; p value <0.05) with only a minor effect on NAD(P)H autofluorescence in the cytoplasm (1.5-fold; p value <0.1) (Fig. 2, C and D). One issue in interpreting these data is that protein-bound NAD(P)H has a longer fluorescence lifetime and contributes to a major fraction of NAD(P)H two-photon autofluorescence (40, 44, 45). Previous studies in mammalian cells, however, have observed very little change in the free NAD(P)H to protein-bound NAD(P)H ratio even under the extreme condition of hypoxia, which results in a detectable increase of two-photon autofluorescence (46). Thus, the dramatic increase of two-photon autofluorescence from our experiment is unlikely to be explained by changes in free NADH/protein-bound NADH ratios. Taken together, our results suggest that NMNAT-1 can specifically mediate nuclear NAD+ production.
Regulation of Gene Expression by NMNAT-1 and PARP-1
To examine the requirement for NMNAT-1 and PARP-1 in gene regulation, we knocked down NMNAT-1 and PARP-1 in MCF-7 cells. Stable expression of shRNAs targeting NMNAT-1 and PARP-1 reduced their protein levels to less than 10% of control cells expressing a functional shRNA targeting luciferase (Fig. 3A). We have previously characterized these shRNAs and have determined that off-target effects are not a major influence on gene regulation outcomes with their use (19, 28). Knockdown of NMNAT-1 reduces total cellular NAD+ levels in MCF-7 cells by about 35% without affecting total cellular nicotinamide levels as determined using a quantitative mass spectrometry assay (Ref. 19 and data not shown). Furthermore, knockdown of PARP-1 increases total cellular NAD+ levels in MCF-7 cells by about 45% while reducing total PAR levels by about 50% (28). These results highlight the potential functional interplay between NMNAT-1 and PARP-1 in cells at the level of NAD+ and PAR.
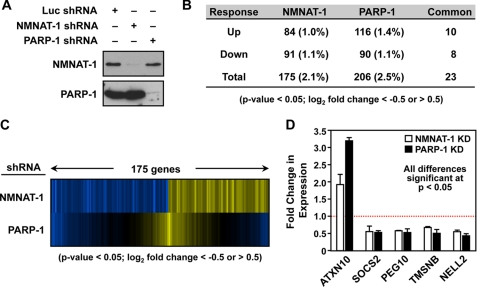
FIGURE 3.
Regulation of gene expression by NMNAT-1 and PARP-1 in MCF-7 cells. A, stable knockdown of NMNAT-1 and PARP-1 in MCF-7 cells using shRNA constructs with an shRNA targeting luciferase (Luc) as a control. NMNAT-1 and PARP-1 protein levels were determined by Western blot analyses. B, number of genes significantly affected by NMNAT-1 or PARP-1 knockdown in MCF-7 cells as determined by expression microarray analysis. The genes were selected using a two-tailed Student's t test (p value <0.05) and a -fold change cutoff (log2 -fold change <−0.5 or >0.5). Numbers in parentheses indicate the percentage of all expressed genes. Each gene set is divided into up- and down-regulated groups. Note that five genes in the overlapping set of 23 genes are differentially regulated by NMNAT-1 and PARP-1. C, expression profiles of genes affected by NMNAT-1 or PARP-1 knockdown in MCF-7 cells. The genes affected by NMNAT-1 knockdown were selected as described in B. The corresponding signals for the PARP-1 knockdown condition for the same genes are also shown. D, RT-qPCR confirmation of expression microarray data for a subset of target genes affected by NMNAT-1 and PARP-1 knockdown (KD) in a similar manner. The bars shown in D represent the mean of three or four independent experiments, and the error bars represent S.E. Statistical significance was determined by two-tailed Student's t test with a p value <0.05.
Next, we carried out expression microarray analyses using MCF-7 cells subjected to knockdown of NMNAT-1 or PARP-1. We identified ∼200 genes significantly affected by knockdown of either NMNAT-1 (175 genes) or PARP-1 (206 genes) (95% confidence by two-tailed Student's t test; log2 -fold change >0.5 or <−0.5) with about 10% overlap between the regulated gene sets using the thresholds and cutoffs reported here (Fig. 3B). Of the 175 genes significantly affected by NMNAT-1 knockdown, 23 (13%) were also significantly affected by PARP-1 knockdown (Fig. 3, B and C). For the purposes of this study, we focused on genes with potential co-regulation by NMNAT-1 and PARP-1 (i.e. overlapping or common genes; e.g. Fig. 3D). We have examined other types of NMNAT-1-dependent gene expression (e.g. interplay between NMNAT-1 and SIRT1) in a previous study (19). For genes in the overlapping set, knockdown of PARP-1 and NMNAT-1 generally had similar effects with respect to up-regulation or down-regulation (18 of 23 genes; Fig. 3, B–D). Overall, our expression analyses using the knockdown cells indicate that NMNAT-1 is required for the proper expression of a subset of PARP-1-responsive genes. We hypothesize that these are the genes that require PARP-1 enzymatic activity, which represent a subset of the total of PARP-1-regulated genes (28).
NMNAT-1 Is Recruited to Target Gene Promoters through Interactions with PARP-1
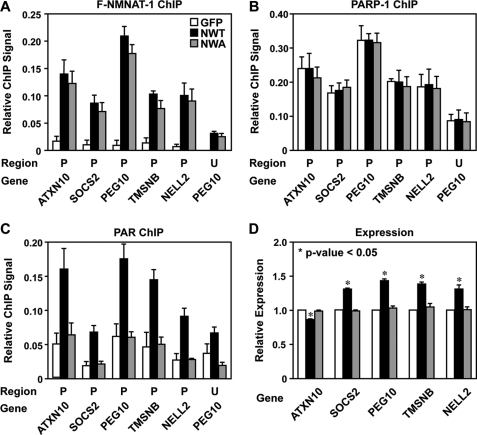
We have shown previously that PARP-1 localizes to the promoters of almost all expressed genes in MCF-7 cells (21, 28, 32). Furthermore, we have shown that NMNAT-1 also localizes to target gene promoters through interactions with chromatin-modulating enzymes, such as SIRT1 (19). Based on the functional interactions that we observed between NMNAT-1 and PARP-1 in our in vitro assays (Fig. 1), as well as a previously published report (20), we decided to investigate the possibility that NMNAT-1 might interact with PARP-1 at target gene promoters to regulate its activity in cells. To this end, we used ChIP assays to examine binding of endogenous PARP-1 and ectopically expressed FLAG-tagged NMNAT-1 to target gene promoters. We focused this analysis on genes regulated in common by NMNAT-1 and PARP-1 (Fig. 3D).
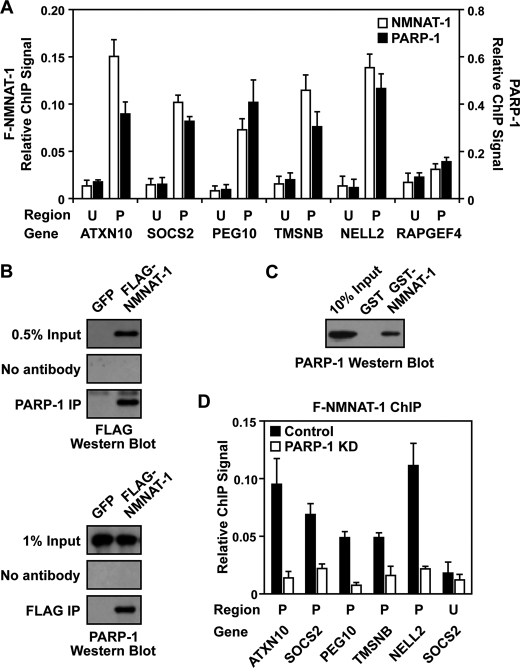
We observed that both NMNAT-1 and PARP-1 are enriched at the promoter regions of genes in the overlapping set (e.g. ATXN10, SOCS2, PEG10, TMSNB, and NELL2), but not at upstream regions (Fig. 4A). As a negative control, very low levels of NMNAT-1 and PARP-1 were found at the promoter of RAPGEF4, a gene unaffected by NMNAT-1 or PARP-1 knockdown, indicating that the ChIP signals were not due to nonspecific interactions with the antibodies (Fig. 4A). Overall, NMNAT-1 and PARP-1 exhibited very similar binding patterns at the genomic regions examined (Fig. 4A). This observation is supported by the results from global ChIP analyses (i.e. ChIP-chip), which show that significant peaks of NMNAT-1 and PARP-1 co-localize across the genome (supplemental Fig. S2). Together, these results suggest that NMNAT-1 may be recruited to chromatin through interactions with PARP-1 as we have observed previously for SIRT1 (19).
FIGURE 4.
NMNAT-1 and PARP-1 bind to promoter regions of commonly regulated target genes. A, ChIP-qPCR analysis of NMNAT-1 (FLAG) and PARP-1 at the promoter (P) and an upstream region (U; ∼10 kb from the transcription start site) of target genes in MCF-7 cells. B, ChIP-Western analysis of interactions between PARP-1 and NMNAT-1. MCF-7 cells expressing GFP or FLAG-NMNAT-1 (F-NMNAT-1) were used for PARP-1 and FLAG ChIP. The immunoprecipitated (IP) material was subjected to Western blotting for FLAG and PARP-1, respectively. The blots shown are representative of three independent experiments. C, GST-NMNAT-1 interaction assay with native PARP-1 from a nuclear extract. PARP-1 interaction with GST-NMNAT-1 bound to glutathione-agarose resin was detected by Western blot analysis. The blot shown is representative of three independent experiments. D, FLAG-based ChIP-qPCR analysis of NMNAT-1 localization to target gene promoters in control or PARP-1 knockdown (KD) cells. Each bar shown in A and D represents the mean of three or more independent experiments, and the error bars represent S.E.
To test this hypothesis, we examined interactions between NMNAT-1 and PARP-1 using ChIP-Western analyses and GST pulldown assays. FLAG-NMNAT-1 and PARP-1 co-immunoprecipitated with each other under our ChIP assay conditions (Fig. 4B). In addition, PARP-1 from nuclear extracts was specifically pulled down by GST-NMNAT-1, but not GST alone (Fig. 4C). These results suggest specific physical interactions between NMNAT-1 and PARP-1. Importantly, the binding of NMNAT-1 at target gene promoters was significantly reduced upon PARP-1 knockdown (Fig. 4D). In fact, NMNAT-1 was no longer enriched at promoter regions in PARP-1 knockdown cells (Fig. 4D, compare the promoter and upstream regions). Together, these results demonstrate that PARP-1 interacts with NMNAT-1 in vivo and plays an important role in recruiting NMNAT-1 to chromatin.
NMNAT-1 Regulates PARP-1 Activity at Target Gene Promoters
Co-localization of NMNAT-1 and PARP-1 in the ChIP assays suggests a functional interaction between the two enzymes at target gene promoters. We tested two aspects of this hypothesis: 1) regulation of PARP-1 localization by NMNAT-1 and 2) regulation of PARP-1 enzymatic activity by NMNAT-1. To do so, we used MCF-7 cells ectopically expressing FLAG-tagged wild-type NMNAT-1 (labeled “NWT”), catalytically inactive mutant NMNAT-1 (W169A; labeled “NWA”), or GFP (as a control). In ChIP assays, both versions of NMNAT-1 were recruited to target gene promoters at similar levels (Fig. 5A). Furthermore, the binding of PARP-1 to the same promoters was not affected by the ectopic expression of either version of NMNAT-1 (Fig. 5B) or by NMNAT-1 knockdown (supplemental Fig. S3). These results indicate that under the basal culture conditions used in our experiment NMNAT-1 does not regulate the association of PARP-1 with chromatin.
FIGURE 5.
NMNAT-1 regulates PARP-1 activity at target gene promoters. ChIP-qPCR and RT-qPCR analyses were carried out using MCF-7 cells ectopically expressing GFP, FLAG-NMNAT-1 (F-NMNAT-1) wild type (NWT), or FLAG-NMNAT-1 W169A enzymatic mutant (NMA), as indicated by shading as shown in A. For all panels, the bars shown represent the mean of three or more independent experiments, and the error bars represent S.E. A–C, ChIP for NMNAT-1 (FLAG) (A), PARP-1 (B), and PAR (using the PAR monoclonal antibody clone 10H) (C) at the promoter (P) and an upstream region (U; ∼10 kb from the transcription start site) of target genes. D, RT-qPCR analysis of target gene expression. β-Actin mRNA was used as an internal reference for data normalization. The results are presented as expression levels relative to those observed in the GFP control cells. Statistical significance was determined by two-tailed Student's t test. *, p value <0.05.
To explore the link between NMNAT-1 enzymatic activity and PARP-1 enzymatic activity, we used a PAR antibody in ChIP assays to detect PAR accumulation at target gene promoters. In contrast to the results with PARP-1 localization, we detected a 2–3-fold increase in PAR levels at target gene promoters in cells ectopically expressing wild-type NMNAT-1, but not in cells expressing GFP or the inactive mutant of NMNAT-1 (Fig. 5C). Together, our ChIP analyses indicate that NMNAT-1 regulates the enzymatic activity, but not the localization, of PARP-1 at target gene promoters.
In RT-qPCR assays, we observed modest but statistically significant and highly reproducible effects of ectopic expression of wild-type NMNAT-1 on the expression of the same set of target genes (Fig. 5D). As expected, these effects were opposite to the effect of NMNAT-1 knockdown (compare Figs. 5D and 3D). In addition, they were not observed upon ectopic expression of the catalytically inactive NMNAT-1 or GFP (Fig. 5D), indicating that the expression of these genes is dependent upon the enzymatic activity of NMNAT-1. These results are consistent with the results of the PAR ChIP assays (Fig. 5C), and together they suggest a direct role for NMNAT-1 in supporting the NAD+-dependent production of PAR by PARP-1 at target gene promoters.
DISCUSSION
Biochemical, Molecular, Cell-based, and Genomic Assays Link Activities of NMNAT-1 and PARP-1 in the Nucleus
NMNAT-1 and PARP-1, two key enzymes in the NAD+ metabolic pathway, localize to the nucleus where integration of their enzymatic activities has the potential to control a variety of nuclear processes. In the studies reported herein, we used a variety of biochemical, molecular, cell-based, and genomic assays to demonstrate that NMNAT-1 interacts with PARP-1 at target gene promoters in MCF-7 cells, where it enhances PARP-1-dependent PARylation. Specifically, using cell-free biochemical assays, we show that NMNAT-1 can produce NAD+ to support the catalytic activity of PARP-1 (Fig. 1, A–E), but can also enhance the PAR synthesis and PAR chain elongation activities of PARP-1 independently of NAD+ production (Fig. 1, F and G).
In addition, using genomic and gene-specific assays in MCF-7 cells, we show that 1) NMNAT-1 localizes to the nucleus where it catalyzes the production of NAD+ in a pool that may be distinct from other cellular compartments (Fig. 2); 2) depletion of NMNAT-1 or PARP-1 alters the expression of about 200 protein-coding genes each with about 10% overlap between the two gene sets (Fig. 3); 3) NMNAT-1 and PARP-1 interact in cells (Fig. 4, B and C); 4) NMNAT-1 and PARP-1 both bind to the promoters of commonly regulated target genes, and the binding of NMNAT-1 is dependent upon the presence of PARP-1 (Fig. 4, A and D); and 5) NMNAT-1 enzymatic activity is required for PARP-1-dependent PARylation at the promoters of commonly regulated target genes as well as the expression of those target genes (Fig. 5).
Model for Functional Interactions between NMNAT-1 and PARP-1 at Target Gene Promoters
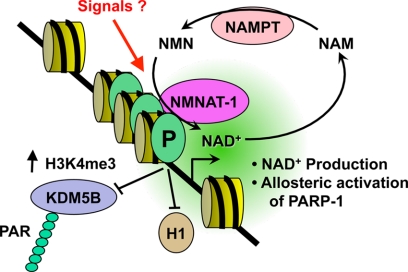
Collectively, our results support a model in which promoter-bound PARP-1 recruits NMNAT-1 to support PARP-1-dependent PARylation of target proteins (Fig. 6), such as the histone demethylase KDM5B as we have shown in a previous study (32) (compare for example the results for NELL2 between our current study and our previous study). NMNAT-1 supports PARP-1 catalytic activity in two ways: (1) by producing NAD+ for PARP-1-dependent PARylation of target proteins and (2) by stimulating PARP-1 catalytic activity independently of NAD+ production, possibly through an allosteric mechanism. Finally, the functional interactions of PARP-1 and NMNAT-1 with chromatin can be modulated as end points of cellular signaling pathways. This model functionally links the enzymatic activities of NMNAT-1 and PARP-1 to regulate the expression of a set of common target genes.
FIGURE 6.
Model for regulation of PARP-1 activity at target gene promoters by NMNAT-1. PARP-1 binds to target gene promoters where it blocks the binding of the linker histone H1 (21, 22) and PARylates chromatin-associated proteins, including transcription factors and histone-modifying enzymes, such as the histone H3 lysine 4 demethylase KDM5B (32) in an NAD+-dependent manner. PARP-1 interacts with and recruits NMNAT-1 to target gene promoters. NMNAT-1 supports PARP-1 activity in two ways: 1) by producing NAD+ for PARP-1 catalytic activity and 2) by stimulating PARP-1 catalytic activity in an NAD+ synthesis independent manner possibly through an allosteric mechanism. NMNAT-1 may also affect the gene regulatory activity of PARP-1 in a manner that is independent of the enzymatic activity of both PARP-1 and NMNAT-1. The functional interactions of PARP-1 and NMNAT-1 with chromatin may be modulated as end points of cellular signaling pathways (“Signals”) (20). P, PARP-1. H3K4me3, histone H3 lysine 4 trimethylation; NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyltransferase.
A previous study investigating functional interactions between NMNAT-1 and PARP-1 has shown that NMNAT-1 can bind to automodified PARP-1 and stimulate its enzymatic activity (20). Our results agree with and further extend this conclusion with the observation that both PAR chain elongation and PAR synthesis activities of PARP-1 are allosterically regulated by NMNAT-1. This conclusion is based on the observation that NMNAT-1 can stimulate PARP-1 enzymatic activity in the absence of ongoing NAD+ biosynthesis (Fig. 1, F and G, and supplemental Fig. S1), suggesting that NAD+ synthesis and PARP-1 stimulation are separable biochemical functions of NMNAT-1. Importantly, the results of our cell-based assays indicate that NMNAT-1 regulates PARP-1 activity at target gene promoters. We have previously described a similar functional interplay between NMNAT-1 and the NAD+-dependent protein deacetylase SIRT1 (19), which suggests some common features for functional interplay between NMNAT-1 and the NAD+-dependent enzymes that it regulates.
Why might PARP-1 recruit NMNAT-1 to target gene promoters for localized actions? As we have noted previously with respect to NMNAT-1/SIRT1 interactions at promoters, the need for localized NAD+ production seems unnecessary given the very rapid diffusion rate of small molecules in the nucleus. Even localized NAD+ production within the nuclear compartment may not lead to an accumulation of promoter-localized pools of elevated NAD+ accessible to PARP-1. In this regard, interactions between NMNAT-1 and PARP-1 at target gene promoters may facilitate more efficient NAD+ utilization by PARP-1, perhaps through a substrate channeling mechanism (47). We and others have proposed a model in which NAD+ biosynthetic enzymes, such as NMNAT-1, form a complex with SIRT1 and channel NAD+ directly to it, creating a microdomain of high NAD+ concentration (19, 48). Our results provide further evidence in support of this hypothesis.
A prediction from our model is that PARP-1 enzymatic activity should be required for genes regulated by both PARP-1 and NMNAT-1. Our results indicate that this is indeed the case, but it depends on the PARP-1-dependent end point that is examined. We have shown previously that the expression of genes, such as NELL2 and TMSNB (also known as TMSL8), as well as other PARP-1- and NMNAT-1-dependent genes that we examined in the current study, is not inhibited by the PARP inhibitor PJ34 at concentrations that inhibit total cellular production of PAR (see supplemental Fig. S2A in Ref. 32). Importantly, however, for these same genes, PARP-1 enzymatic activity is required for a number of key molecular end points, such as the inhibition of KDM5B binding (KDM5B is a histone demethylase) and the maintenance of histone H3 lysine 4 trimethylation (H3K4me3) levels at the promoters (see supplemental Fig. S2, G and H, in Ref. 32). Our results suggest that NAD+ synthesis by NMNAT-1 is required to support these chromatin-modulating activities of PARP-1. Given the likely allosteric interactions between PARP-1 and NMNAT-1, it is also possible that NMNAT-1 is affecting the gene regulatory activity of PARP-1 in a manner that is independent of the enzymatic activity of both PARP-1 and NMNAT-1. These possibilities will be explored in future studies.
Possible NMNAT-1-dependent Functional Interplay between PARP-1 and SIRT1?
Previous studies have shown that in response to stress and other physiological stimuli PARP-1 and SIRT1 can regulate the activity of each other through several different mechanisms (e.g. competition for NAD+ and reciprocal covalent modification) (49–51). The NAD+ competition model may be relevant to our studies with NMNAT-1. Based on our microarray expression data from MCF-7 cells using NMNAT-1, PARP-1, and SIRT1 knockdown (Refs. 19 and 28 and data herein), we find that 75% of the genes requiring both PARP-1 and NMNAT-1 for expression also require SIRT1, and 54% of the genes requiring both NMNAT-1 and SIRT1 also require PARP-1. Furthermore, our current and published data indicate that NMNAT-1 associates with both PARP-1 and SIRT1 at the same promoter regions (19). Thus, PARP-1 and SIRT1 are functioning at the same promoters in an NMNAT-1-dependent manner. Perhaps competition for the regulatory effects of NMNAT-1 may underlie the inhibitory cross-talk that has been observed between PARP-1 and SIRT1.
Compartmentalization of Distinct Pool of Nuclear NAD+?
Among mammalian NMNAT enzymes, NMNAT-1 is exclusively located in the nucleus, suggesting a unique role for this enzyme in nuclear NAD+ synthesis and the regulation of NAD+-dependent functions in the nucleus. Previous studies have failed, however, to detect any effect of NMNAT-1 on total cellular NAD+ production (14, 18). Using a sensitive mass spectrometry-based assay with 18O-labeled standards, we have previously observed about a 35% reduction in total cellular NAD+ levels upon knockdown of NMNAT-1 in MCF-7 cells (19). As we discuss below, most of this effect may be limited to the nuclear compartment. The discrepancy between our observations and the previous reports may result from differences in cell types or the relative expression levels of NMNAT-1. Because NAD+ levels in the cytosol are higher than those in the nucleus, it is possible that under some conditions total cellular NAD+ measurements may underestimate the decrease in nuclear NAD+ levels that we observed upon knockdown of NMNAT-1.
To complement and extend our spectrometry-based NAD+ measurements, we used two-photon excitation microscopy. We observed that ectopic expression of NMNAT-1 dramatically increases nuclear NAD(P)H autofluorescence in MCF-7 cells without major alterations in the levels of cytoplasmic/mitochondrial NAD(P)H autofluorescence (Fig. 2, C and D). In this assay, we measured NAD(P)H autofluorescence as an indicator for NAD+ (45). Interestingly, the increase in NAD(P)H autofluorescence observed upon ectopic expression of NMNAT-1 was largely confined to the nucleus, indicating that the outcomes of NMNAT-1 catalytic activity are largely confined to the nucleus. These results suggest the presence of a distinct nuclear pool of NAD+ that is functionally compartmentalized from the cytoplasmic and mitochondrial pools of NAD+. The integrity of such a pool might break down under conditions of cellular stress (e.g. under conditions of caloric restriction; for a review, see Ref. 51), allowing for global integration of NAD+ levels across cellular compartments. These intriguing possibilities will have to be explored in more detail in future studies using even more sophisticated approaches. If verified, a distinct nuclear pool of NAD+ would point to a critically important role for NMNAT-1 in controlling nuclear events that depend on NAD+.
Supplementary Material
Acknowledgments
We thank Dr. Watt Webb for generous use of the two-photon microscope; Dr. Kristine M. Frizzell Hussey for supplying PARP-1 reagents and data sets; Gabor Bethlendy, Leo Iniguez, and Heidi Rosenbaum from Roche NimbleGen for promoter microarrays, reagents, and technical assistance with the ChIP-chip experiments; and the students of the 2009 Cold Spring Harbor Laboratory Eukaryotic Gene Expression course for assistance with the ChIP-chip experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK069710 from the NIDDK (to W. L. K.) and a National Research Service award postdoctoral fellowship from the NIDDK (to M. J. G.). This work was also supported by a grant from the Endocrine Society (to W. L. K.); postdoctoral fellowships from the New York State Health Research Science Board (to T. Z.), the American Heart Association (AHA) (to M. J. G.), and the Susan G. Komen Breast Cancer Foundation (to D. D. R.).; and predoctoral fellowships from the Alfred P. Sloan Foundation (to J. G. B.) and the AHA (to R. K.).

This article contains supplemental “Experimental Procedures,” Figs. S1–S3, and Tables S1 and S2.
- PARP
- poly(ADP-ribose) polymerase
- ERα
- estrogen receptor α
- KDM5B
- lysine-specific demethylase 5B
- NTA
- nitrilotriacetic acid
- NMNAT
- nicotinamide mononucleotide adenylyltransferase
- PAR
- poly(ADP-ribose)
- PARylation
- poly(ADP-ribosyl)ation
- qPCR
- quantitative real-time PCR
- NMN
- nicotinamide mononucleotide.
REFERENCES
- 1. Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krishnakumar R., Kraus W. L. (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 4. Kim M. Y., Zhang T., Kraus W. L. (2005) Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 5. Rongvaux A., Andris F., Van Gool F., Leo O. (2003) Reconstructing eukaryotic NAD metabolism. BioEssays 25, 683–690 [DOI] [PubMed] [Google Scholar]
- 6. Revollo J. R., Körner A., Mills K. F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R. R., Milbrandt J., Kiess W., Imai S. (2007) Nampt/PBEF/Visfatin regulates insulin secretion in β cells as a systemic NAD biosynthetic enzyme. Cell Metab. 6, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rongvaux A., Shea R. J., Mulks M. H., Gigot D., Urbain J., Leo O., Andris F. (2002) Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32, 3225–3234 [DOI] [PubMed] [Google Scholar]
- 8. Kitani T., Okuno S., Fujisawa H. (2003) Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett. 544, 74–78 [DOI] [PubMed] [Google Scholar]
- 9. Yalowitz J. A., Xiao S., Biju M. P., Antony A. C., Cummings O. W., Deeg M. A., Jayaram H. N. (2004) Characterization of human brain nicotinamide 5′-mononucleotide adenylyltransferase-2 and expression in human pancreas. Biochem. J. 377, 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raffaelli N., Sorci L., Amici A., Emanuelli M., Mazzola F., Magni G. (2002) Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem. Biophys. Res. Commun. 297, 835–840 [DOI] [PubMed] [Google Scholar]
- 11. Emanuelli M., Carnevali F., Saccucci F., Pierella F., Amici A., Raffaelli N., Magni G. (2001) Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J. Biol. Chem. 276, 406–412 [DOI] [PubMed] [Google Scholar]
- 12. Schweiger M., Hennig K., Lerner F., Niere M., Hirsch-Kauffmann M., Specht T., Weise C., Oei S. L., Ziegler M. (2001) Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett. 492, 95–100 [DOI] [PubMed] [Google Scholar]
- 13. Berger F., Lau C., Dahlmann M., Ziegler M. (2005) Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 [DOI] [PubMed] [Google Scholar]
- 14. Mack T. G., Reiner M., Beirowski B., Mi W., Emanuelli M., Wagner D., Thomson D., Gillingwater T., Court F., Conforti L., Fernando F. S., Tarlton A., Andressen C., Addicks K., Magni G., Ribchester R. R., Perry V. H., Coleman M. P. (2001) Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 4, 1199–1206 [DOI] [PubMed] [Google Scholar]
- 15. Conforti L., Tarlton A., Mack T. G., Mi W., Buckmaster E. A., Wagner D., Perry V. H., Coleman M. P. (2000) A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci. U.S.A. 97, 11377–11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coleman M. (2005) Axon degeneration mechanisms: commonality amid diversity. Nat. Rev. Neurosci. 6, 889–898 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Zhai Q., Chen Y., Lin E., Gu W., McBurney M. W., He Z. (2005) A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 170, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Araki T., Sasaki Y., Milbrandt J. (2004) Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 19. Zhang T., Berrocal J. G., Frizzell K. M., Gamble M. J., DuMond M. E., Krishnakumar R., Yang T., Sauve A. A., Kraus W. L. (2009) Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J. Biol. Chem. 284, 20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berger F., Lau C., Ziegler M. (2007) Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyltransferase 1. Proc. Natl. Acad. Sci. U.S.A. 104, 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 22. Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 23. Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 25. Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. (2004) Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 119, 815–829 [DOI] [PubMed] [Google Scholar]
- 26. Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. (2005) PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18, 83–96 [DOI] [PubMed] [Google Scholar]
- 27. Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Oshimura M., Feinberg A. P., Lobanenkov V., Klenova E., Ohlsson R. (2004) Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 36, 1105–1110 [DOI] [PubMed] [Google Scholar]
- 28. Frizzell K. M., Gamble M. J., Berrocal J. G., Zhang T., Krishnakumar R., Cen Y., Sauve A. A., Kraus W. L. (2009) Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 284, 33926–33938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petesch S. J., Lis J. T. (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tulin A., Spradling A. (2003) Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562 [DOI] [PubMed] [Google Scholar]
- 31. Tulin A., Stewart D., Spradling A. C. (2002) The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16, 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krishnakumar R., Kraus W. L. (2010) PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simbulan-Rosenthal C. M., Rosenthal D. S., Luo R., Samara R., Espinoza L. A., Hassa P. O., Hottiger M. O., Smulson M. E. (2003) PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene 22, 8460–8471 [DOI] [PubMed] [Google Scholar]
- 34. Hassa P. O., Covic M., Hasan S., Imhof R., Hottiger M. O. (2001) The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J. Biol. Chem. 276, 45588–45597 [DOI] [PubMed] [Google Scholar]
- 35. Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. (2004) Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330 [DOI] [PubMed] [Google Scholar]
- 36. Kraus W. L., Kadonaga J. T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kraus W. L., Kadonaga J. T. (1999) in Steroid/Nuclear Receptor Superfamily: a Practical Approach (Picard D., ed) pp. 167–189, Oxford University Press, New York [Google Scholar]
- 38. Bulger M., Kadonaga J. (1994) Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5, 241–262 [Google Scholar]
- 39. Keith G., Desgrès J., de Murcia G. (1990) Use of two-dimensional thin-layer chromatography for the components study of poly(adenosine diphosphate ribose). Anal. Biochem. 191, 309–313 [DOI] [PubMed] [Google Scholar]
- 40. Kasischke K. A., Vishwasrao H. D., Fisher P. J., Zipfel W. R., Webb W. W. (2004) Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science 305, 99–103 [DOI] [PubMed] [Google Scholar]
- 41. Johnson W. E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 [DOI] [PubMed] [Google Scholar]
- 42. Kininis M., Chen B. S., Diehl A. G., Isaacs G. D., Zhang T., Siepel A. C., Clark A. G., Kraus W. L. (2007) Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 27, 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langelier M. F., Ruhl D. D., Planck J. L., Kraus W. L., Pascal J. M. (2010) The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285, 18877–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams R. M., Piston D. W., Webb W. W. (1994) Two-photon molecular excitation provides intrinsic 3-dimensional resolution for laser-based microscopy and microphotochemistry. FASEB J. 8, 804–813 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Q., Piston D. W., Goodman R. H. (2002) Regulation of corepressor function by nuclear NADH. Science 295, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 46. Vishwasrao H. D., Heikal A. A., Kasischke K. A., Webb W. W. (2005) Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J. Biol. Chem. 280, 25119–25126 [DOI] [PubMed] [Google Scholar]
- 47. Srere P. A. (1987) Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124 [DOI] [PubMed] [Google Scholar]
- 48. Grubisha O., Smith B. C., Denu J. M. (2005) Small molecule regulation of Sir2 protein deacetylases. FEBS J. 272, 4607–4616 [DOI] [PubMed] [Google Scholar]
- 49. Pillai J. B., Isbatan A., Imai S., Gupta M. P. (2005) Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2α deacetylase activity. J. Biol. Chem. 280, 43121–43130 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J. (2003) Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? BioEssays 25, 808–814 [DOI] [PubMed] [Google Scholar]
- 51. Zhang T., Kraus W. L. (2010) SIRT1-dependent regulation of chromatin and transcription: linking NAD+ metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta 1804, 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.