Background: G1/S transition is important for embryonic stem cell (ESC) proliferation; however, the transcriptional factor that regulates this remains largely unknown.
Results: CIBZ, a transcription factor, regulates ESC proliferation and G1/S transition.
Conclusion: CIBZ-associated ESC proliferation and G1/S transition is dependent on Nanog expression.
Significance: This study provides insights into the mechanism underlying the rapid proliferation and G1/S transition of ESCs.
Keywords: Cell Biology, Cell Cycle, Cell Death, Cell Growth, DNA-binding Protein, DNA Methylation, Embryonic Stem Cell, Proliferation, Regenerative Medicine
Abstract
Mouse embryonic stem cells (ESCs) require transcriptional regulation to ensure rapid proliferation that allows for self-renewal. However, the molecular mechanism by which transcriptional factors regulate this rapid proliferation remains largely unknown. Here we present data showing that CIBZ, a BTB domain zinc finger transcriptional factor, is a key transcriptional regulator for regulation of ESC proliferation. Here we show that deletion or siRNA knockdown of CIBZ inhibits ESC proliferation. Cell cycle analysis shows that loss of CIBZ delays the progression of ESCs through the G1 to S phase transition. Conversely, constitutive ectopic expression of exogenous CIBZ in ESCs promotes proliferation and accelerates G1/S transition. These findings suggest that regulation of the G1/S transition explains, in part, CIBZ-associated ESC proliferation. Our data suggest that CIBZ acts through the post-transcriptionally regulates the expression of Nanog, a positive regulator of ESC proliferation and G1/S transition, but does not affect Oct3/4 and Sox2 protein expression. Notably, constitutive overexpression of Nanog partially rescued the proliferation defect caused by CIBZ knockdown, indicating the role of CIBZ in ESC proliferation and G1/S transition at least in part depends on the Nanog protein level.
Introduction
Mouse embryonic stem cells (ESCs)2 are characterized by unlimited self-renewal pluripotency (1, 2). The ability of ESCs to self-renew is maintained through the combination of proliferation and the inhibition of differentiation. Although an increasing number of reports have documented the molecular mechanisms involved in ESC pluripotency and differentiation, little is known about their rapid proliferative properties. The core transcriptional factors Oct3/4, Sox2, and Nanog are involved in ESC pluripotency and proliferation through the regulation of gene expression of other factors as well as themselves (3). Genetic studies revealed that Oct3/4 and Sox2 are crucial for maintaining ESC pluripotency (4, 5), whereas Nanog-deficient ESCs mostly remain morphologically undifferentiated, mainly because of the normal Oct3/4 and Sox2 expression (6). Therefore, Nanog is not absolutely essential for the maintenance of ESC pluripotency. Loss-of-function of Nanog has been shown to result in the growth retardation of ESCs (7), while its overexpression promotes ESC proliferation (8, 9). Moreover, forced Nanog expression in Nanog-deficient hematopoietic stem cells and NIH3T3 fibroblasts facilitate proliferation of these cells (10, 11). These findings indicate that Nanog plays a critical role in the regulation of ESC proliferation.
One of the most striking features of ESC proliferation is the unique cell cycle structure that is characterized by a short G1 phase and a prolonged S phase. The shortening of the G1/S transition could account for the rapid proliferation of ESCs. A recent study indicated that Nanog knockdown induces cell cycle arrest and delays G1/S transition (7). Furthermore, the overexpression of Nanog in human ESCs promotes their proliferation and accelerates G1/S transition (9), indicating that the role of Nanog in ESC proliferation is dependent, at least partially, on G1/S transition. Considering the critical role of Nanog in ESC proliferation, elucidating the regulation of Nanog expression is essential. Transcriptional regulation of Nanog is known to require many transcriptional factors, including Oct3/4 and Sox2 as well as Nanog itself (12). In addition, Nanog is regulated by microRNA (miRNA)-134 through a post-transcriptional mechanism (13). However, it is unknown if other miRNA or transcriptional factors, apart from this miRNA-134, post-transcriptionally regulate Nanog expression.
Previously we identified a novel mouse BTB domain zinc finger transcription factor, CIBZ (14). CIBZ is ubiquitously expressed in adult mouse tissues (14), and various cell lines including ESCs and myoblast cells (15). We previously reported that CIBZ possesses transcriptional repression and activation domains, and that the repression activity of one repression domain depends on the interaction of CIBZ with CtBP protein (14). In mouse cells, except ESCs, CIBZ is involved in apoptosis regulation (15). Because CIBZ is expressed in mouse ESCs, we hypothesize that CIBZ may play a role in ESCs proliferation and G1/S transition.
To investigate the role of CIBZ in ESCs, knock-out/knockdown and overexpression strategies were employed. Here we show that ESC proliferation was regulated by the expression level of CIBZ. Our data indicated that CIBZ-dependent ESC proliferation is involved in the regulation of the G1/S transition. In addition, we present data showing that the phenotypes caused by altered CIBZ expression are dependent, at least partially, on the Nanog protein level.
EXPERIMENTAL PROCEDURES
Targeted Disruption of the CIBZ Gene
A targeting vector containing both pgk-tk and pgk-neo cassettes was designed to replace the entire coding region of CIBZ gene. A 8.8 kb fragment and a 1.3 kb fragment, obtained by PCR, were used as the long and short arms for the targeting vector. The resulting targeting vector was linearized with XhoI digestion and introduced into RF8 ESCs by electroporation and selected with G418 (200 μg/ml) according to the protocol described (16). Genomic DNAs from G418-resistant colonies were screened for homologous recombination by Southern blotting and PCR. For Southern blot analysis, genomic DNA was digested with BamHI, separated on a 0.8% agarose gel and transferred to a positively charged nylon membrane (Roche). A 525 bp probe located outside the targeting construct was obtained by PCR and labeled with digoxigenin-11-dUTP (Roche). The hybridization was performed according to the manufacturer's instructions. For PCR analysis, three primers, one sense primer (5′- CCTGGGAGAATTTCCAACTAAGC-3′) and two antisense primers (5′-AAGTCGTCCTTGAGGTCCCTGGAGAGG-3′; 5′-AGAACCTGCGTGCAATCCATC-3′) were used. PCR with these three primers produces a 2.0 kb fragment from the wild-type locus and a 2.2 kb from the targeted locus. For generation of CIBZ knock-out ESC clone, the targeted clones were treated with high concentrations of G418 (1 mg/ml).
Cell Culture, siRNA, and Transient Transfection
Mouse RF8 ESCs were maintained on mitomycin-treated mouse embryonic fibroblasts (MEFs) in standard ESC culture medium (DMEM, 15% fetal bovine serum, 2 mm l-glutamine, 100 μm nonessential amino acids, 1% penicillin and streptomycin, and 0.1 mm β-mercaptoethanol), as previously described (16). ESCs were transfected with 50 nm CIBZ-specific or scrambled negative control Dicer substrate siRNA duplexes (Integrated DNA Technologies), as described previously (18). Transfection with siRNA was performed with INTERFERin reagent (Polyplus Transfection) according to the manufacturer's instructions. siRNA-transfected cells were cultured for 2 days after transfection and then analyzed by RT-PCR and Western blotting. Transient transfections of ESCs with CIBZ constructs were previously described (18). In brief, cells were transfected with Myc-tagged CIBZ or Myc alone using TransFast (Promega) according to the manufacturer's instructions. On day 2 after transfection, the cells were collected and analyzed by RT-PCR and Western blotting.
Cell Growth Assay
For cell growth assays, CIBZ−/− ESCs were seeded in duplicate, 1 × 105 cells/well into gelatin-coated 6-cm plates containing STO feeder cells. Every 24 h, two plates each of CIBZ−/− ESCs and control ESCs were trypsinized, and the dissociated cells were counted; non-viable cells were identified by performing the trypan blue exclusion assay. For evaluation of the cell growth of ESCs stably transfected with CIBZ, 2 × 105 cells were seeded into duplicate 6-well plates. The number of viable cells was estimated by trypan blue exclusion assay before and at 2 days after seeding. The average values of two plates are represented (±S.E.). Each experiment was performed three times.
BrdU Incorporation and Immunofluorescence Microscopy
5-Bromo-2′-deoxyuridine (BrdU) incorporation assay was performed according to a published protocol (19) with a slight modification. Transfected cells (4 × 105 cells/well) were seeded in 6-well plates; when cultures had reached 70% confluence, they were incubated with 10 μm BrdU (Sigma-Aldrich) for 45 min. The cells were fixed by the dropwise addition of ice-cold 70% ethanol while mixing, followed by incubation with 2N HCl/0.5% Triton X-100 for 30 min. The cells were then washed with phosphate-buffered saline (PBS) and incubated with anti-BrdU monoclonal antibody (Sigma-Aldrich) for 1 h followed by labeling with Alexa Fluor 488-conjugated secondary antibody (Invitrogen). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Data shown are representative of three independent experiments. The number of BrdU-positive cells per field of vision was determined. At least 10 different fields were counted per coverslip.
Annexin V-FITC/Propidium Iodide (PI) Assay
PI assays were performed as previously described (15). The annexin V kit (MBL) was used to detect phosphatidylserine translocation from the inner to the outer plasma membrane according to the manufacturer's protocol. Briefly, cells were washed once in PBS, followed by annexin V binding buffer containing annexin V and PI and incubated for 15 min at room temperature in the dark. The cells were analyzed by fluorescence-activated cell sorting (FACS; BD Biosciences) with acquisition of 20,000 events/sample to ensure adequate data.
Cell Cycle Analysis
For cell cycle analysis, cells were trypsinized and washed once in PBS and fixed by the dropwise addition into ice-cold 70% ethanol while mixing. These cells were washed twice in PBS, treated for 20 min with RNase A (250 μg/ml) at 37 °C and for 30 min with PI (50 μg/ml) at 4 °C. DNA content was analyzed by FACS, and cell number in each phase of the cell cycle was determined using ModFit LT software (BD Biosciences).
Semiquantitative RT-PCR
Semiquantitative RT-PCR was performed as previously described (18). The primers were confirmed as unique using the non-redundant NCBI database, and are listed in supplemental Table S1. Primer annealing was performed at 58–60 °C for all primer sets. Reaction products were separated on 2% agarose gels and visualized by ethidium bromide staining. To identify the PCR products, single bands of the expected size were extracted from the gels and sequenced. GAPDH mRNA expression was used as internal control.
Western Blotting
Western blotting was performed as previously described (20). Briefly, cells were sonicated in lysis buffer. Proteins were separated on 8–12% SDS-PAGE, transferred onto PVDF membranes and probed with the following antibodies: anti-CIBZ (18), anti-Oct3/4 (MAB1759, R&D Systems), anti-Sox2 (S1451, Sigma-Aldrich), anti-Nanog (AB5731, Millipore), anti-α-tubulin (clone DM 1A, Sigma-Aldrich), anti-Cyclin E (sc-481, Santa Cruz Biotechnology), and anti-Cdk2 (sc-163, Santa Cruz Biotechnology). HRP-conjugated anti-mouse or anti-rabbit IgG (GE Healthcare) was used as the secondary antibody. Quantification of Western blots was performed with ImageJ software.
Plasmid Preparation and Stable Cell Line Generation
For cloning of EF1/Nanog, full-length Nanog cDNA including the Kozak consensus sequence was obtained by RT-PCR amplification of total RNA from RF8 ESCs. The PCR products were digested with NotI and SpeI and ligated into the corresponding sites of pRSV/EF1 vector (kindly provided by Dr. William J. Freed) (21) with the following modification: the pRSV/EF1 vector was digested with NotI and inserted with a linker oligonucleotides (5′-ACTAGTATCGATGCGGCCGC-3′) to generate an additional SpeI site. The EF1 promoter was subcloned into pcDNA3 following amplification by PCR from pRSV/EF1 using primers with containing a BglII site at the 5′-end and a KpnI site at the 3′-end to generate pcDNA3/EF1. pcDNA/EF1/Myc was created by ligation of the EF1 promoter from BglII-, KpnI-digested pRSV/EF1 into the corresponding sites of pcDNA3–6×Myc as previously described (14). For cloning of pcDNA3/EF1/Myc-CIBZ, the full length of CIBZ fragment digested with BamHI and ApaI from pcDNA3–6×Myc-CIBZ was ligated into the corresponding sites of pcDNA3/EF1/Myc. DNA sequences were verified by automated sequencing (ABI PRISM310).
For transfection, ESCs (5 × 105 cells in 10-cm plate) were transfected with 1 μg of the desired constructs using TransFast reagent (Promega) according to the manufacturer's protocol. Neomycin-resistant clones were picked after 12 days of G418 (200 μg/ml) selection and propagated with the same medium.
Statistical Analysis
Statistical analyses were performed using the Mann-Whitney U test. All data are expressed as means of ± S.E. Differences were considered significant if p < 0.05.
RESULTS
Generation of CIBZ Knock-out ESCs
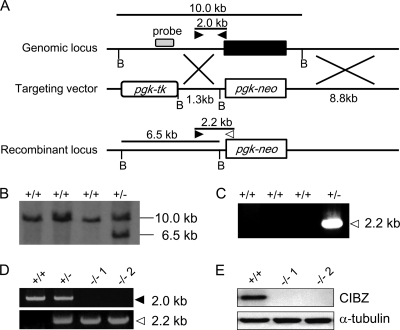
We have previously demonstrated that CIBZ is expressed in mouse ESCs (16). To investigate the role of CIBZ in ESCs, we replaced the entire coding region of CIBZ with the pgk-neomycin (pgk-neo) cassette (Fig. 1A). ESC clones that had undergone gene targeting were identified by Southern blotting and PCR (Fig. 1, B and C). The targeted clones were treated with high concentrations of G418 to select for homozygous clones (22). Two CIBZ homozygous-deficient clones (CIBZ−/−1 and CIBZ−/−2) were identified by PCR (Fig. 1D). The complete absence of CIBZ protein and CIBZ transcription was confirmed by Western blotting and RT-PCR (Figs. 1E and 2, A and B).
FIGURE 1.
Target disruption of CIBZ in mouse ESCs. A, WT genomic locus of CIBZ, targeting vector and the recombinant CIBZ locus. Bold box in the genomic locus represents the exon of the CIBZ gene. The open boxes in the targeting vector schematics represent pgk-tk and pgk-neo selectable marker genes. B, BamHI. B, Southern blot analysis of BamHI-digested genomic DNA from RF8 ESCs stably transfected with the targeting vector. DNA probe is indicated in A. C, PCR analysis of the recombinant locus using primers indicated by A (arrowheads). A 2.2 kb band results from the recombinant locus. D, generation of two CIBZ−/− ESC (CIBZ−/−1 and CIBZ−/−2) clones. CIBZ−/− homozygous cell lines were derived by selection using a high concentration of G418. Homologous recombination was confirmed by PCR using primers indicated in A. E, Western blotting showing CIBZ expression. α-Tubulin was used as a control for loading.
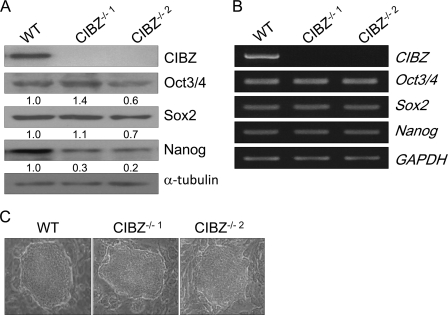
FIGURE 2.
Deletion of CIBZ in ESCs does not considerably affect their pluripotency. A and B, expression of the indicated proteins and genes in CIBZ−/− ESCs and control ESCs were detected by Western blotting (A) and semiquantitative RT-PCR (B), respectively. α-tubulin and GAPDH served as loading controls for immunoblotting and RT-PCR, respectively. C, representative morphology of CIBZ−/− ESCs and control ESCs cultured in the presence of STO feeder cells.
Undifferentiated ESCs Remain Largely Unaffected by CIBZ Loss
To determine whether CIBZ deletion affected undifferentiated ESCs, we carefully observed the morphology of ESCs under the same culture conditions. We observed that a vast majority of CIBZ−/− ESC colonies exhibited the morphological features of undifferentiated wild-type (WT) ESCs (Fig. 2C). Fig. 2, A and B shows that CIBZ−/− ESCs exhibited similar mRNA levels of pluripotency markers (Oct3/4, Sox2, and Nanog) and protein levels of Oct3/4 and Sox2 as compared with control ESCs. To further investigate whether transient knockdown of CIBZ affects the undifferentiated state of ESCs, two siRNA duplexes were used to specifically knockdown CIBZ expression. Both the CIBZ knockdown (CIBZkd) ESCs exhibited undifferentiated morphologies (supplemental Fig. S1C). Compared with control ESCs, the mRNA levels of the pluripotency markers and protein levels of Oct3/4 and Sox2 in CIBZkd ESCs were almost unchanged (supplemental Fig. S1, A and B). Compared with control ESCs, CIBZkd ESCs displayed no visible up-regulation of lineage-specific genes (supplemental Fig. S1D), including markers of ectoderm (Pax6 and Nestin), mesoderm (Brachyury and Tbx2), and endoderm (Gata4 and Gata6). These findings indicate that CIBZ is not essential to maintain ESCs in an undifferentiated state.
Loss of CIBZ in ESCs Show Reduced Proliferation
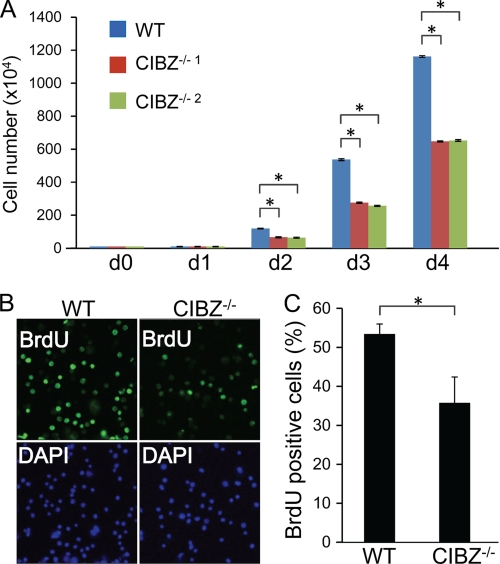
Loss of CIBZ expression in ESCs, either by gene deletion or by transient knockdown, resulted in decreased Nanog protein level (Fig. 2A, supplemental Fig. S1A), which is a positive regulator of proliferation. Moreover, CIBZ−/− ESCs grew slower than control ESCs under identical culture conditions, indicating that CIBZ−/− ESCs have reduced proliferation. To analyze this, cells were plated at low density and counted daily; it was found that cell number was significantly decreased in CIBZ−/− ESCs compared with control ESCs at 2–4 days (Fig. 3A). A considerable decrease in cell number was observed in CIBZkd ESCs compared with control ESCs (supplemental Fig. S2). These findings indicate that CIBZ is essential for rapid cell proliferation, potentially by regulation of the Nanog protein level.
FIGURE 3.
CIBZ deletion in ESCs resulted in reduced proliferation. A, analysis of cell growth in cultures of CIBZ−/− and control ESCs were seeded into 6-cm plate (1 × 105 cells/well). Cell number was counted daily after the seeding (d 0). Results are mean ± S.E. of three independent experiments (n = 6 in total, *, p < 0.05). B and C, knock-out of CIBZ resulted in reduced BrdU incorporation in CIBZ−/−1 ESCs at 3 days after seeding. B, representative fluorescence microscopy images of BrdU- or DAPI-positive cells. C, histogram showing percentages of BrdU- and DAPI-positive cells in each culture. The data shown are representative of 10 randomly chosen fields. Results are mean ± S.E. of three independent experiments. *, p < 0.05.
To investigate the mechanism underlying CIBZ-associated proliferation defect, a BrdU (bromodeoxyuridine) incorporation assay was performed. Immunofluorescence microscopy revealed that CIBZ−/− ESCs and CIBZkd ESC exhibited decreased number of BrdU-positive cells compared with control ESCs cultures (Fig. 3, B and C, supplemental Fig. S3, A and B), indicating that fewer cells were entering S phase and that cells showed reduced proliferation. Apoptosis induction could be considered as a cause of cell number reduction. Compared with control ESCs, CIBZ−/− and CIBZkd ESCs expressed same levels of apoptosis markers (cleavage of PARP and caspase-3) (supplemental Figs. S4A and S5A) as well as similar apoptotic (annexin V-positive) cells (supplemental Figs. S4B and 5B). Taken together, these findings indicate that CIBZ deletion results in reduced ESC proliferation.
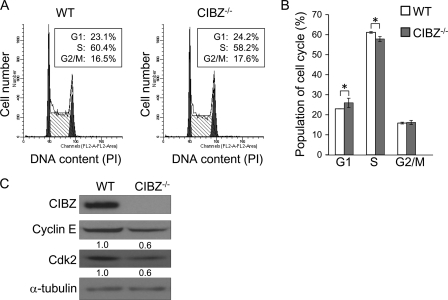
Loss of CIBZ in ESCs Results in Delayed G1/S Transition
To investigate the effect of CIBZ silencing on cell cycle progression, we performed cell cycle analysis by flow cytometry using PI staining. CIBZ−/− and CIBZkd ESC cultures exhibited proportion of cells increased in the G1 phase and concomitantly decreased in cells in the S phase, compared with that in control ESC cultures (Fig. 4, A and B, supplemental Fig. S6, A and B). To examine the role of CIBZ in regulating G1/S transition, we examined Cyclin E and cyclin-dependent kinase2 (Cdk2) expression, which regulators of the G1/S transition in mouse ESCs (23, 24). The Cyclin E and Cdk2 protein levels decreased in CIBZ−/− and CIBZkd ESCs compared with control ESCs (Fig. 4C, supplemental Fig. S7). These results collectively demonstrated the impaired proliferation caused by the effect of CIBZ silencing, at least partially, on the G1/S transition.
FIGURE 4.
Knock-out of CIBZ in ESCs delays G1/S transition. A and B, deletion of CIBZ delays G1/S transition. The cell cycle distribution of CIBZ−/−1 ESCs were analyzed by flow cytometry with PI staining. A, representative flow cytometry profiles show percentages in each stage of the cell cycle on day 3 after seeding cells. B, graphic representation of the percentage of cells in each phase. Results are mean ± S.E. of five independent experiments. *, p < 0.05. C, Western blotting analysis showed the expression of the indicated proteins in CIBZ−/−1 ESCs. α-Tubulin served as loading controls.
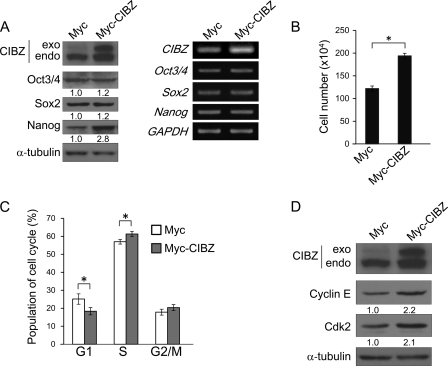
Constitutive Ectopic Expression of CIBZ Accelerated Proliferation and Promoted G1/S Transition
To examine further the mechanism through which CIBZ regulates ESC proliferation and G1/S transition, we established an ESC line stably expressing Myc-tagged CIBZ protein (Fig. 5A). This cell line exhibited morphological features of control ESCs (data not shown), and no measurable change in Oct3/4 and Sox2 mRNA and protein levels (Fig. 5A), thereby confirming that these cells remained undifferentiated. As seen in Fig. 5B, Myc-CIBZ ESC cultures exhibited increased cell number compared with control cells. Flow cytometry analysis revealed that ectopic expression of CIBZ decreased the proportion of cells in G1 and increased the proportion of cells in S phase (Fig. 5C). These findings suggest that ectopic CIBZ expression facilitates ESC proliferation dependent, at least partially, on promoting the G1/S transition.
FIGURE 5.
Constitutive ectopic expression of CIBZ promotes proliferation and G1/S transition. A, Western blotting (left panel) and semiquantitative RT-PCR (right panel) analysis of stable Myc tag and Myc-CIBZ transfected ESCs. Endo and exo indicate endogenously generated and exogenously added CIBZ, respectively. B, CIBZ stably expressing ESC line (2 × 105 cells) were seeded in a 6-well plate. The number of cells was counted before and after seeding at 2 days. Results are mean ± S.E. of three independent experiments (n = 6 in total, *, p < 0.05). C, graphic representation of the percentage of cells in each phase. Results are mean ± S.E. of five independent experiments. *, p < 0.05. D, Western blotting analysis showed the expression of the indicated proteins in Myc tag and Myc-CIBZ stable ESC line on Western blots.
CIBZ-dependent ESC Proliferation Is Mediated by Nanog Protein
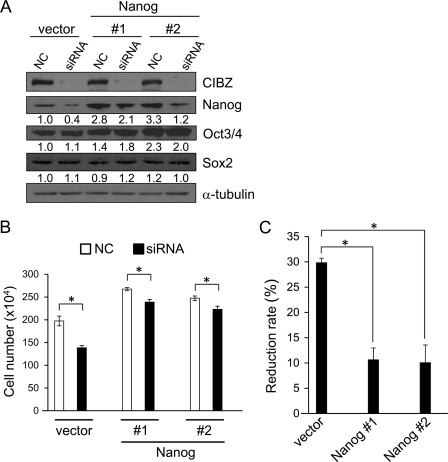
The aforementioned data showed that CIBZ expression regulates the Nanog protein level without affecting Nanog mRNA expression (Figs. 2, A and B and 5A, supplemental Fig. S1, A and B). These findings suggests that CIBZ post-transcriptionally regulates Nanog expression. Because Nanog is a positive regulator of ESC proliferation, we hypothesized that CIBZ-dependent proliferation depends on Nanog protein level, and that the overexpression of Nanog in CIBZkd ESCs might rescue the decreased proliferation due to CIBZ knockdown. To address this possibility, we generated two ESC lines stably expressing Nanog, and the overexpression level of Nanog was confirmed by Western blotting (Fig. 6A). Nanog overexpression displayed no visible change in CIBZ expression (Fig. 6A, lines 1, 3, and 5). These observations, together with CIBZ loss- and gain-of-function data that CIBZ regulates the expression of Nanog protein (Figs. 2A and 5A), suggest that Nanog is a downstream target of CIBZ. The cell number of Nanog-overexpressing ESC cultures was ∼30% higher than that of a vector cell culture (Fig. 6B), indicating that forced Nanog expression in ESCs promotes proliferation. Notably, CIBZ knockdown in the vector cells resulted in a 29.8% reduction in cell number compared with control knockdown ESCs, whereas CIBZ knockdown in Nanog-overexpressing ESCs resulted in ∼10.1% reduction (3-fold decrease) in cell number compared with control knockdown ESCs (Fig. 6, B and C). These data demonstrated that CIBZ-associated ESC proliferation is dependent, at least in part, on Nanog protein expression.
FIGURE 6.
Constitutive overexpression of Nanog in ESCs rescues the compromised proliferation by siRNA-mediated CIBZ knockdown. A, Western blotting analysis of CIBZ knockdown in vector ESC line and Nanog-overexpressing ESC lines (#1 and #2). CIBZ siRNA or scrambled negative siRNA (NC) was transiently transfected into the vector and Nanog ESC lines. At 2 days after transfection, cells were collected and analyzed by immunoblotting with the indicated antibodies. B and C, analysis of proliferation of ESCs (4 × 105) transfected with CIBZ siRNA or scrambled siRNA and plated in a 6-well plate. B, cell number was counted after seeding at 2 days. Results are mean ± S.E. of three independent experiments (n = 6 in total, *, p < 0.05). C, graphic representation of cell number reduction rate, which was calculated by decreased number of scrambled siRNA cells relative to CIBZ siRNA cells/number of scrambled siRNA cells. *, p < 0.05.
DISCUSSION
We showed that loss-of-function of CIBZ in ESCs, either by gene deletion or by transient knockdown, results in cell number reduction. The following data indicate that this reduction is due to impaired proliferation but not apoptosis: (i) reduced BrdU incorporation indicated that loss of CIBZ in ESCs leads to a reduction in the number of proliferating cells (Fig. 3, B and C, supplemental Fig. S3, A and B); (ii) Immunoblotting and immunofluorescence analysis revealed that loss of CIBZ does not induce apoptosis (supplemental Figs. S4, A and B and S5, A and B); (iii) WST-8 assay, a highly sensitive and reproducible method for the evaluation of proliferation, revealed that CIBZkd ESCs proliferated slower than control ESCs (data not shown). Moreover, we found that constitutive ectopic expression of CIBZ in ESCs resulted in increased cell number (Fig. 5B). These data indicate that CIBZ positively regulates ESC proliferation.
Cell cycle analysis using flow cytometry revealed that loss of CIBZ in ESCs delayed the cell cycle at G1/S transition (Fig. 4, A and B, supplemental Fig. S6, A and B), whereas its overexpression promoted G1/S transition (Fig. 5C). In somatic cells, Cyclin D-Cdk4 and Cyclin E-Cdk2 complexes are crucial for G1/S transition (23). In ESCs, however, Cyclin D-Cdk4 complex is barely detectable, while Cyclin E-Cdk2 complex is present and constitutively active (23, 25, 26). Cyclin E and Cdk2 expression is essential for G1/S transition and contributes to rapid ESC proliferation (23). Our data showed that loss of CIBZ expression in ESCs reduced Cyclin E and Cdk2 protein levels (Fig. 4C, supplemental Fig. S7), whereas constitutive ectopic expression of CIBZ increased these levels (Fig. 5D). These data indicate that CIBZ regulates G1/S transition by affecting Cyclin E and Cdk2 protein levels.
In addition to the effects on the cell cycle, loss or gain of CIBZ had no discernible effect on Oct3/4 and Sox2 mRNA or their protein levels (Figs. 2, A and B and 5A, supplemental Fig. S1, A and B). Oct3/4 and Sox2 expression is consistent with the observation that CIBZ−/− and CIBZkd ESCs remained largely undifferentiated (Fig. 2C, supplemental Fig. S1C). The effects of CIBZ loss- and gain-of-function on ESC proliferation and the G1/S transition are similar to those of Nanog, which plays crucial roles in the regulation of these processes (7, 9–11). These findings strongly indicate that the role of CIBZ in the regulation of ESC proliferation may be dependent on the Nanog protein level. Since the overexpression of Nanog largely rescued the proliferation defect caused by CIBZ knockdown in ESCs (Fig. 6, B and C), we conclude that the regulation of ESC proliferation by CIBZ is mediated, at least in part, on the Nanog protein level.
Phosphoinositide 3-kinase (PI3K) signaling plays a crucial role in the G1/S transition (27). Interestingly, inhibition of PI3K activity in ESCs leads to reduced Nanog protein level, suggesting a functional link between PI3K signaling and Nanog expression (28). Treatment of ESCs with PI3K inhibitor results in reduced CIBZ and Nanog protein levels (data not shown), suggesting that CIBZ-mediated regulation of Nanog occurs via the PI3K pathway, and consequently, promotes G1/S transition. In human ESCs, Nanog is a direct transcriptional regulator of CDC25A and CDK6, which are S-phase regulators, and promotes G1/S transition (9). In our experiments, the mRNA levels of CDC25A and CDK6, did not change in CIBZ−/− ESCs and CIBZkd ESCs compared with control ESCs (data not shown), suggesting that CIBZ-associated regulation of cell cycle is not dependent on these genes. One possible explanation for this discrepancy is that different mechanisms may exist in human and mouse ESCs, because constitutively overexpressing NANOG in human ESCs fails to induce Cyclin E or CDK2 protein expression (9). Further experiments are required to determine the distinct pathways between human and mouse ESCs.
Our data indicated that CIBZ is involved in regulating the level of Nanog protein, without discernible effect on Nanog transcription. Compared with control ESCs, the almost equal mRNA level of Nanog in the CIBZ-deficient/knockdown ESCs and CIBZ-overexpressing ESCs were also confirmed by RT-PCR using two different primer pairs specific to Nanog gene (data not shown). Moreover, the transient overexpression of CIBZ in ESCs resulted in up-regulation of Nanog protein, but failed to induce the level of Nanog mRNA (data not shown). These observations indicate that CIBZ modulates Nanog expression post-transcriptionally; whether this is a translational or post-translational mechanism remains to be determined. One possibility is that CIBZ regulates Nanog protein expression by suppressing its ubiquitin degradation, because studies have demonstrated that Nanog protein is regulated by ubiquitin-proteasomal pathway (29, 30). However, the observation that the overexpression of CIBZ in ESCs failed to inhibit Nanog degradation suggests that CIBZ is not required for the stabilization of Nanog protein (data not shown). Tay et al. (32) reported that two miRNAs (miRNA-296 and -470) regulate the translation of Nanog via targeting sites in the CDS of Nanog gene without affecting its mRNA level. It is possible that CIBZ may regulate the expression of such kind of miRNAs, and thereby modulating the expression of Nanog protein. In addition, CIBZ may regulate Nanog expression by regulating miRNA expression including that of the reported miRNA-134. Recently, we reported that CIBZ, a methyl-CpG-binding protein, suppresses myogenin in a methylation-dependent manner (18). However, the report that mouse ESCs without DNA methylation still maintain normal ESC proliferation (31), together with the findings that CIBZ deletion in ESCs has no discernible effect on the genome-wide DNA methylation (data not shown), indicate that the regulation of Nanog expression by CIBZ may not rely on DNA methylation. Further experiments should be undertaken to elucidate how CIBZ regulates Nanog expression at the molecular level.
Supplementary Material
Acknowledgments
We thank Drs. Manabu Sugai and Pin Lin for reading of the manuscript. We thank members of the Kawaichi laboratory for technical advice.
This work was supported by a grant-in-aid for Scientific Research (C), and the Global COE program, from Japan's Ministry of Education, Culture, Sports, Science, and Technology.

This article contains supplemental Figs. S1–S7 and Table S1.
- ESCs
- embryonic stem cells
- siRNA
- small interfering RNA
- CIBZ
- CtBP-interacting BTB zinc finger protein
- CDK
- cyclin-dependent kinase
- PARP
- poly (ADP-ribose) polymerase.
REFERENCES
- 1. Evans M. J., Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2. Martin G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chambers I., Smith A. (2004) Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23, 7150–7160 [DOI] [PubMed] [Google Scholar]
- 4. Niwa H., Miyazaki J., Smith A. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 5. Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- 7. Chen T., Du J., Lu G. (2012) Cell growth arrest and apoptosis induced by Oct4 or Nanog knockdown in mouse embryonic stem cells: a possible role of Trp53. Mol. Biol. Rep. 39, 1855–1861 [DOI] [PubMed] [Google Scholar]
- 8. Liu N., Feng X., Fang Z., Ma F., Lu S., Lu M., Han Z. (2008) Identification of genes regulated by nanog, which is involved in ES cells pluripotency and early differentiation. J. Cell. Biochem. 104, 2348–2362 [DOI] [PubMed] [Google Scholar]
- 9. Zhang X., Neganova I., Przyborski S., Yang C., Cooke M., Atkinson S. P., Anyfantis G., Fenyk S., Keith W. N., Hoare S. F., Hughes O., Strachan T., Stojkovic M., Hinds P. W., Armstrong L., Lako M. (2009) A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J. Cell Biol. 184, 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka Y., Era T., Nishikawa S., Kawamata S. (2007) Forced expression of Nanog in hematopoietic stem cells results in a γδT-cell disorder. Blood 110, 107–115 [DOI] [PubMed] [Google Scholar]
- 11. Zhang J., Wang X., Chen B., Suo G., Zhao Y., Duan Z., Dai J. (2005) Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem. Biophys. Res. Commun. 338, 1098–1102 [DOI] [PubMed] [Google Scholar]
- 12. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tay Y. M., Tam W. L., Ang Y. S., Gaughwin P. M., Yang H., Wang W., Liu R., George J., Ng H. H., Perera R. J., Lufkin T., Rigoutsos I., Thomson A. M., Lim B. (2008) MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells 26, 17–29 [DOI] [PubMed] [Google Scholar]
- 14. Sasai N., Matsuda E., Sarashina E., Ishida Y., Kawaichi M. (2005) Identification of a novel BTB-zinc finger transcriptional repressor, CIBZ, that interacts with CtBP corepressor. Genes Cells 10, 871–885 [DOI] [PubMed] [Google Scholar]
- 15. Oikawa Y., Matsuda E., Nishii T., Ishida Y., Kawaichi M. (2008) Down-regulation of CIBZ, a novel substrate of caspase-3, induces apoptosis. J. Biol. Chem. 283, 14242–14247 [DOI] [PubMed] [Google Scholar]
- 16. Matsuda E., Shigeoka T., Iida R., Yamanaka S., Kawaichi M., Ishida Y. (2004) Expression profiling with arrays of randomly disrupted genes in mouse embryonic stem cells leads to in vivo functional analysis. Proc. Natl. Acad. Sci. U.S.A. 101, 4170–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deleted in proof
- 18. Oikawa Y., Omori R., Nishii T., Ishida Y., Kawaichi M., Matsuda E. (2011) The methyl-CpG-binding protein CIBZ suppresses myogenic differentiation by directly inhibiting myogenin expression. Cell Res. 21, 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du J. X., Yun C. C., Bialkowska A., Yang V. W. (2007) Protein inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor Krüppel-like factor 5. J. Biol. Chem. 282, 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuda E., Agata Y., Sugai M., Katakai T., Gonda H., Shimizu A. (2001) Targeting of Krüppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J. Biol. Chem. 276, 14222–14229 [DOI] [PubMed] [Google Scholar]
- 21. Zeng X., Chen J., Sanchez J. F., Coggiano M., Dillon-Carter O., Petersen J., Freed W. J. (2003) Stable expression of hrGFP by mouse embryonic stem cells: promoter activity in the undifferentiated state and during dopaminergic neural differentiation. Stem Cells 21, 647–653 [DOI] [PubMed] [Google Scholar]
- 22. Mortensen R. M., Conner D. A., Chao S., Geisterfer-Lowrance A. A., Seidman J. G. (1992) Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12, 2391–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White J., Dalton S. (2005) Cell cycle control of embryonic stem cells. Stem Cell Reviews 1, 131–138 [DOI] [PubMed] [Google Scholar]
- 24. Koledova Z., Kafkova L. R., Calabkova L., Krystof V., Dolezel P., Divoky V. (2010) Cdk2 inhibition prolongs G1 phase progression in mouse embryonic stem cells. Stem Cells Dev 19, 181–194 [DOI] [PubMed] [Google Scholar]
- 25. Neganova I., Lako M. (2008) G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 213, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neganova I., Zhang X., Atkinson S., Lako M. (2009) Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 28, 20–30 [DOI] [PubMed] [Google Scholar]
- 27. Paling N. R., Wheadon H., Bone H. K., Welham M. J. (2004) Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 279, 48063–48070 [DOI] [PubMed] [Google Scholar]
- 28. Storm M. P., Bone H. K., Beck C. G., Bourillot P Y., Schreiber V., Damiano T., Nelson A., Savatier P., Welham M. J. (2007) Regulation of Nanog expression by phosphoinositide 3-kinase-dependent signaling in murine embryonic stem cells. J. Biol. Chem. 282, 6265–6273 [DOI] [PubMed] [Google Scholar]
- 29. Moretto-Zita M., Jin H., Shen Z., Zhao T., Briggs S. P., Xu Y. (2010) Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc. Natl. Acad. Sci. U.S.A. 107, 13312–13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramakrishna S., Suresh B., Lim K. H., Cha B. H., Lee S. H., Kim K. S., Baek K. H. (2011) PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev 20, 1511–1519 [DOI] [PubMed] [Google Scholar]
- 31. Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H. R., Nakayama J., Okano M. (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a, and Dnmt3b. Genes cells 11, 805–814 [DOI] [PubMed] [Google Scholar]
- 32. Tay Y., Zhang J., Thomson A. M., Lim B., Rigoutsos I. (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.