Background: The role of ATM kinase activity in DNA replication was unknown.
Results: ATM kinase inhibition, but not ATM protein disruption, impedes DNA replication, and ATM physically and functionally interacts with proliferating cell nuclear antigen to regulate DNA synthesis.
Conclusion: ATM regulates DNA synthesis and binds to the replication machinery.
Significance: ATM kinase inhibition affects DNA replication in a different manner to ATM protein disruption.
Keywords: DNA polymerase, DNA repair, DNA replication, Protein kinases, Protein-protein interactions
Abstract
Ataxia telangiectasia (A-T) is a pleiotropic disease, with a characteristic hypersensitivity to ionizing radiation that is caused by biallelic mutations in A-T mutated (ATM), a gene encoding a protein kinase critical for the induction of cellular responses to DNA damage, particularly to DNA double strand breaks. A long known characteristic of A-T cells is their ability to synthesize DNA even in the presence of ionizing radiation-induced DNA damage, a phenomenon termed radioresistant DNA synthesis. We previously reported that ATM kinase inhibition, but not ATM protein disruption, blocks sister chromatid exchange following DNA damage. We now show that ATM kinase inhibition, but not ATM protein disruption, also inhibits DNA synthesis. Investigating a potential physical interaction of ATM with the DNA replication machinery, we found that ATM co-precipitates with proliferating cell nuclear antigen (PCNA) from cellular extracts. Using bacterially purified ATM truncation mutants and in vitro translated PCNA, we showed that the interaction is direct and mediated by the C terminus of ATM. Indeed, a 20-amino acid region close to the kinase domain is sufficient for strong binding to PCNA. This binding is specific to ATM, because the homologous regions of other PIKK members, including the closely related kinase A-T and Rad3-related (ATR), did not bind PCNA. ATM was found to bind two regions in PCNA. To examine the functional significance of the interaction between ATM and PCNA, we tested the ability of ATM to stimulate DNA synthesis by DNA polymerase δ, which is implicated in both DNA replication and DNA repair processes. ATM was observed to stimulate DNA polymerase activity in a PCNA-dependent manner.
Introduction
Ataxia telangiectasia (A-T)2 is a rare autosomal recessive human disorder characterized by neurodegeneration, immunodeficiency, an increased risk of cancer, and profound radiosensitivity (1). Cells derived from A-T patients are radiosensitive, contain increased chromosome aberrations, and exhibit cell-cycle checkpoint defects following exposure to ionizing radiation (1). These cell cycle checkpoint defects include a delayed arrest at G1/S and G2/M as well as an inability to arrest ongoing DNA replication in cells exposed to ionizing radiation, a phenomenon described as radioresistant DNA synthesis. Radioresistant DNA synthesis is caused by a defect in the inhibition of late origin firing in A-T cells following ionizing radiation (2). The contribution of DNA chain elongation arrest to the intra-S-phase checkpoint has been difficult to establish, because the DNA lesions that activate the checkpoint also directly arrest replication fork progression. However, early reports suggest that A-T cells have a slightly longer S-phase and contain an increase in replication intermediates relative to controls (3, 4).
Several observations suggest that mere checkpoint abrogation is unlikely to be the cause for the cellular radiosensitivity of A-T cells. For instance, holding irradiated A-T cells in prolonged periods of either G1 or G0, and thus giving them ample time to repair the damage (as would occur under wild-type conditions), still leads to increased levels of chromosome aberrations in A-T cells (5). The requirement of A-T mutated (ATM) for the repair of a small fraction of double strand breaks after γ-irradiation (6) or after cleavage by the endonuclease I-PpoI (7) directly implicate ATM in signaling to the DNA-repair machinery. It was proposed that ATM is particularly needed for the repair of double strand breaks in heterochromatin (8).
The gene defective in A-T patients, ATM, encodes a serine/threonine protein kinase that is critical for the induction of cellular responses to DNA damage, particularly to DNA double strand breaks (9). ATM kinase activity is increased following exposure to as little as 0.05 gray (Gy) of ionizing radiation and following the introduction of just two DNA double strand breaks (10). More than 1000 substrates of ATM and the related kinase A-T and Rad3-related (ATR) have been identified (11, 12). Although gene ontology analyses identified groups of substrates implicated in biological processes as diverse as immunity and defense, cell proliferation and differentiation, intracellular protein traffic, cell structure and motility, and the cell cycle, the largest group of 202 substrates was categorized as nucleoside, nucleotide, and nucleic acid metabolism (11). Of these 202 protein substrates, 46 are proteins implicated in DNA replication, recombination, and repair, and these include the large and fourth subunit of DNA polymerase ϵ as well as DNA polymerase θ and DNA polymerase λ (11). Neither the effects of a selective ATM kinase inhibitor on DNA replication nor the functional significance of these phosphorylations on DNA polymerase substrates have been reported. To our knowledge, whether small molecule ATM kinase inhibitors induce radioresistant DNA synthesis has not been investigated.
We previously reported that ATM kinase inhibition, but not ATM protein disruption, blocks sister chromatid exchange, a phenotype attributed to the repair of damaged DNA replication forks, following DNA damage (13). Here we show that ATM kinase inhibition, but not ATM protein disruption, also inhibits DNA synthesis. Investigating a potential physical interaction of ATM with the DNA replication machinery, we found that ATM interacts with proliferating cell nuclear antigen (PCNA) both in vivo and in vitro. PCNA was originally characterized as an essential component of the eukaryotic DNA replication machinery wherein it functions as a DNA sliding clamp that enhances the processivity of replicative DNA polymerases (14). Subsequently PCNA was found to recruit other DNA-modifying enzymes, including repair proteins as diverse as DNA ligase I, XP-G, FenI, and MSH2 and -6 (15).
Using purified ATM truncation mutants we show that the interaction between ATM and PCNA is direct and mediated by the C terminus of ATM. A 20-amino acid region close to the ATM kinase domain was found to be sufficient for strong binding to PCNA. The peptide sequence mediating the interaction (ATM PBP) is distinct from previously identified PCNA-interacting motifs such as the PIP and KA boxes, and the observed binding is specific to ATM, because the homologous regions of other PIKK members, including the closely related kinase A-T and Rad3-related, do not bind PCNA. Because PCNA is known to stimulate DNA synthesis by DNA polymerase δ (polδ), in both DNA replication and several DNA repair processes, we tested the effect of ATM PBP in an in vitro DNA synthesis assay. We show that ATM stimulates DNA polymerase δ activity in a PCNA-dependent manner.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Expression Vectors
H460 large cell lung cancer cells were cultured in RPMI, and IMR90 lung fibroblasts, 293T embryonic kidney cells, and U2OS osteosarcoma cells were kept in DMEM, both supplemented with 10% fetal calf serum. Transfections were conducted according to the manufacturers' instructions using FuGENE6 (Roche Applied Science) for U2OS cells and Lipofectamine (Invitrogen) for 293T cells. Expression vectors for ATM without the 3′-untranslated region (UTR) were constructed by cutting a previously described ATM expression vector containing the 3′-UTR (16) with Bsu36I and XhoI and inserting an ATM C-terminal DNA sequence lacking the 3′-UTR, obtained by amplification with the appropriate primers.
In Vivo DNA Synthesis Assays
Cellular DNA synthesis was measured by subsequent incubation with medium containing 14C- or 3H-labeled thymidine as described (2). Incubation of cells with 14C was for 16 h, with 3H for 30 min. In the case of reconstitution experiments ATM knockdown cells were labeled with 14C before transfection with the indicated ATM expression vector. Tritium labeling was done 24 h after transfection.
Antibodies, Inhibitors, and Irradiation
Antibodies against ATM were purchased from Sigma; those against PCNA and heat shock cognate 70 (HSC70) were from Santa Cruz Biotechnology. KU60019 (Kudos Pharmaceuticals) was used at 1 μm concentration. Cells were γ-irradiated in a Shepherd Mark I Model 68 137Cs irradiator (J. L. Shepherd & Associates).
In Vivo Interaction Assays
Whole cell lysates of H460 or U2OS cells were prepared by washing cells in PBS, lysing in TGN buffer (150 mm NaCl, 5 mm NaF, 1% Tween 20, 0.5% Nonidet P-40, 50 mm Tris-HCl, pH 7.5, protease inhibitors) on ice for 30 min and twice clearing by centrifugation. For immunoprecipitation of endogenous PCNA, lysates were incubated with antibodies against PCNA for 5 h and precipitated after four washes with TGN buffer. Anti-rabbit immunoglobulins served as the negative control. The immunoprecipitates with Protein A/G-agarose beads were tested for PCNA and ATM by immunoblots. Alternatively, in the case of exogenous PCNA, FLAG-tagged PCNA or hemagglutinin (HA)-tagged ATM was expressed in U2OS cells. 48 h after transfection the cells were washed, and the lysate was cleared by centrifugation and incubated with M2-agarose for 8 h. After washes with BC buffer (20 mm Tris-HCl 7.9, 20% glycerol, 0.2 mm EDTA, 0.5 mm PMSF, 1 mm DTT) with 150 mm KCl, the beads were boiled in reducing SDS buffer for elution. Inputs and eluates were examined by immunoblotting with antibodies against PCNA and ATM. In the case of the reciprocal immunoprecipitation, 293T cells were transfected with FLAG-tagged ATM and co-precipitation of ATM and PCNA was assessed in the same way. When investigating DNA dependence on the co-immunoprecipitations, lysates were incubated with M2-agarose in the presence or absence of 20 μg/ml ethidium bromide (Invitrogen) or 100 units of DNase I (Roche Applied Science).
In Vitro Interaction Assays
GST-fused proteins were expressed in Rosetta(DE3)pLysS cells at 30 °C and harvested 4–4.5 h after induction with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside in bacterial lysis buffer (20 mm HEPES (pH 7.9), 500 mm NaCl, 1 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF). Lysates were obtained by sonicating the bacterial resuspension and were cleared by ultracentrifugation (Sorvall T647.5, 35,000 rpm, 30 min, 4 °C). Appropriate amount of lysates were incubated with 10 μl of glutathione-Sepharose for at least 6 h and washed three times with BC buffer with 300 mm KCl and once with BC buffer with 100 mm KCl. Proteins were labeled with 35S-labeled methionine using TnT reticulate lysates, following the standard protocol provided by Promega (Madison, WI). In vitro translated proteins were incubated with resin-bound proteins (10 μg) by rotating at 4 °C for 4 h in BC buffer containing 100 mm KCl and 0.05 μg/μl BSA. After several washes with BC buffer containing 100 mm KCl and 300 mm KCl, the resin was boiled in SDS loading buffer before subjecting to SDS-PAGE. Gels were incubated in AmplifyTM (Amersham Biosciences) before drying and autoradiography to enhance the signal.
In Vitro DNA Synthesis Assays
DNA synthesis assays were carried out as described previously (17) with minor modifications. Briefly, a 5′-end-labeled primer (41 nucleotides: 0.05 pmol) annealed to a template (94 nucleotides) was extended by DNA polymerase δ (pol δ: 0.1 pmol) in the absence or presence of PCNA (0.25 pmol of trimer), replication factor C (RFC) (0.05 pmol), and indicated peptides for 15 min at 37 °C in a 40-μl reaction buffer containing 20 mm HEPES (pH 7.5), 100 mm NaCl, 10 mm MgCl2, 2 mm ATP, 40 μm dNTP mix, 0.1 mg/ml BSA, and 0.01% Triton X-100. All the peptides used in the DNA synthesis assays were diluted in DMSO, and all the reactions were adjusted to a final concentration of 2.5% DMSO, which did not interfere with DNA synthesis by pol δ. Reaction products were resolved by electrophoresis in denaturing 12.5% polyacrylamide gels and analyzed by using a PhosphorImager. ATM peptides consisted of a fluorescein-labeled tat-fused peptide from ATM, GRKKRRQRRRPPQLVTIQSFKAEFRLAGGVNLPK, and its mutant derivative, GRKKRRQRRRPPQLVEIQEFKAEFRLAGGVNLPK.
RESULTS
Acute ATM Kinase Inhibition Suppresses Cellular DNA Synthesis
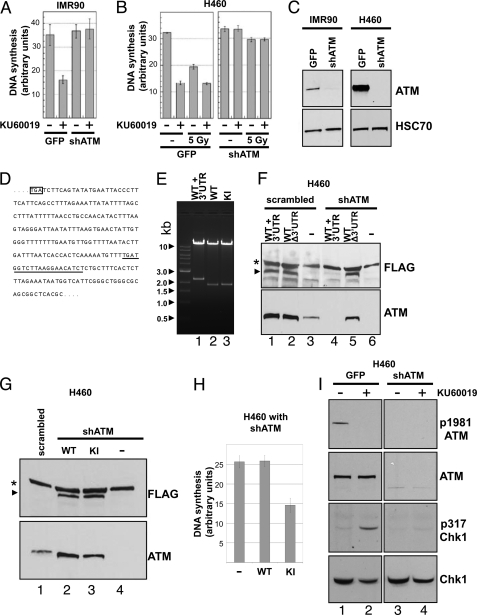
We recently showed that ATM kinase inhibition, but not ATM protein disruption, blocks sister chromatid exchange following DNA damage (13). To determine whether ATM kinase inhibition also influences DNA synthesis, we labeled the DNA of IMR90 human lung fibroblasts expressing shRNA, which disrupts ATM, or of a GFP-expressing control line (Fig. 1C, left panel) with 14C-labeled thymidine for one cycle. Cells where then divided and treated with the ATM kinase inhibitor KU60019 or vehicle from 30 min prior to a pulse of 30 min with 3H-labeled thymidine to the end of the pulse. At the end of the pulse the cells were harvested, and the incorporation of 3H and 14C into high molecular weight material was then measured. The ratio of 3H over 14C was used as a measure for relative DNA synthesis. Treatment with ATM inhibitor more than halved thymidine incorporation during the observed time interval in IMR90 cells, whereas DNA synthesis was not affected in IMR90 with disrupted ATM (Fig. 1A). The observed inhibitory effect of KU60019 on DNA synthesis is therefore ATM-dependent. Similarly, we tested the H460 large cell lung cancer cells with wild-type or knockdown levels of ATM (Fig. 1C, right panel). Again KU60019 inhibited DNA synthesis in an ATM-dependent manner (compare the first two lanes in the left and right panels in Fig. 1B). Irradiation of H460 with 5 Gy of ionizing radiation decreased DNA synthesis in an ATM-dependent manner (compare the first and third lanes in the left and right panels in Fig. 1B) as expected given the well established phenomenon of radioresistant DNA synthesis caused by ATM deficiency. Treatment with KU60019 decreased DNA synthesis even further in irradiated IMR90 with wild-type ATM levels. We next wanted to test whether a kinase-inactive ATM mutant showed a similar phenotype as observed with ATM inhibition. To do so we reconstituted ATM knockdown cells with wild-type or mutant ATM. The mutant ATM, ATM D2870A,N2875K, has been described previously (16) and lacks kinase activity. Fig. 1D shows part of the 3′-UTR of ATM mRNA. The underlined nucleotides are targeted by the shRNA vector used to make stable knockdown cell lines. A Bsu36I/XhoI double restriction of the expression vectors as well as sequencing was used to verify the lack of 3′-UTR in the wild-type and kinase activity-deficient ATM (Fig. 1E and under “Experimental Procedures”). An ATM vector lacking the 3′-UTR, but not an ATM vector containing the 3′-UTR, was able to reconstitute the ATM knockdown cell line (Fig. 1F). ATM knockdown cells expressing similar amounts of wild-type or kinase activity-deficient ATM (Fig. 1G) were used for DNA synthesis assays. Although wild-type ATM did not affect DNA synthesis, ATM lacking kinase activity strongly decreased DNA synthesis (Fig. 1H). Abolition of kinase by mutation and inhibition thus show the same phenomenon. The ATM inhibition-mediated stalling of DNA replication is associated with activation of Chk1 signaling as evidenced by phosphorylation of Chk1 at serine 317 (Fig. 1I) observed 1 h after ATM inhibition. Given that acute ATM inhibition, but not ATM disruption inhibited DNA synthesis, we speculated that ATM might interact with the replication machinery per se and interfere with DNA synthesis if kinase-inactivated.
FIGURE 1.
ATM inhibition affects DNA synthesis. A, acute ATM inhibition decreases DNA synthesis. IMR90 cells with wild-type or knockdown levels of ATM were treated with ATM kinase inhibitor KU60019 and assayed for DNA synthesis. B, H460 cells with wild-type (left panel) or knockdown levels of ATM (right panel) were treated with ATM kinase inhibitor KU60019 and/or 5 Gy of γ-irradiation before assaying for DNA synthesis. C, immunoblot of IMR90 and H460 with wild-type and knockdown levels of ATM. HSC70 served as loading control. D, partial sequence of the 3′-UTR of ATM. Underlined nucleotides were targeted by a shRNA vector to knockdown ATM. E, ATM vectors with and without 3′-UTR. Vectors for ATM mRNA with (lane 1) and without 3′-UTR (lanes 2 and 3; wild-type (WT), kinase-inactive mutant (KI)) were digested with Bsu36I and XhoI. F, efficiency of shRNA-mediated ATM knockdown. H460 cell lines with stable ATM knockdown or a scrambled control were transiently transfected with FLAG-tagged ATM expression vectors lacking or containing the 3′-UTR of ATM. Lysates were probed for ATM and FLAG by immunoblotting. The asterisk denotes a nonspecific band, the arrow FLAG-ATM. G, immunoblot of reconstituted ATM knockdown cells derived from H460. Lysates of knockdown cells transfected with shRNA-resistant expression vectors for wild-type (WT) or kinase-inactive mutant (KI) ATM were probed for ATM and FLAG. Untransfected knockdown cells and H460 with scrambled shRNA served as control. H, kinase-inactive mutant ATM inhibits DNA synthesis. Knockdown cells reconstituted with wild-type (WT) or kinase-inactive mutant ATM were assayed for DNA synthesis. I, ATM kinase inhibition activates Chk1 signaling. H460 with wild-type or knockdown levels of ATM were treated with the ATM inhibitor KU60019 for 1 h or not, and the lysates assayed by immunoblotting for Chk1 and ATM as well as their activating phosphorylation status. shATM, shRNA against ATM.
ATM Interacts with PCNA in Vivo
PCNA plays a key role in DNA replication and DNA repair by forming a sliding homotrimeric ring around DNA that serves as a docking platform for the recruitment of various DNA-modifying enzymes to the replication fork. Such enzymes include nucleases, ligases, helicases, and most importantly DNA polymerases (14). By tethering these enzymes to the DNA template, PCNA increases the processivity and catalytic activity of the machineries involved in DNA synthesis.
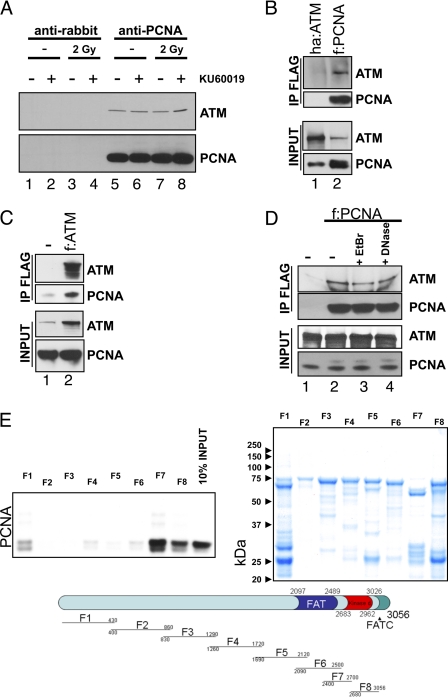
Because of the unique ubiquitous role of PCNA in DNA synthesis processes and to study the role of ATM in DNA synthesis, we tested whether ATM binds to PCNA. Whole cell lysates of H460 cells were incubated with antibodies against PCNA and precipitated. Anti-rabbit immunoglobulins served as negative control. The immunoprecipitates with Protein A/G-agarose beads were tested for PCNA and ATM by immunoblotting. ATM co-precipitated with PCNA independently of cell irradiation or treatment with the ATM-specific kinase inhibitor KU60019 (Fig. 2A, compare lanes 1 to 4 and lanes 5 to 8). To further corroborate the physical interaction of ATM with PCNA in vivo, FLAG-tagged PCNA or HA-tagged ATM were expressed in U2OS cells and the tagged PCNA precipitated with M2 (anti-FLAG)-agarose. After repeated washes the eluates were tested for the presence of PCNA and ATM. Endogenous ATM co-precipitated with PCNA, whereas no ATM was detected in the control cells transfected with HA-tagged ATM (Fig. 2B, compare lanes 1 and 2). Similarly, we expressed FLAG-tagged ATM in 293T cells (allowing better expression of ATM than U2OS) and found PCNA to co-precipitate with endogenous ATM (Fig. 2C). Lysate from untransfected cells served as control. To test whether the interaction of PCNA with ATM was mediated by DNA, we co-precipitated ATM and FLAG-tagged PCNA in the presence or absence of ethidium bromide or DNase. Neither treatment affected ATM co-precipitation with PCNA (Fig. 2D), indicating that DNA was neither bridging the interaction partners nor required for potential conformational changes associated with the interaction.
FIGURE 2.
Direct interaction of ATM with PCNA. A, in vivo interaction of ATM with PCNA. PCNA from H460 cells treated in the indicated way was precipitated with an antibody against PCNA. Bound material was washed and analyzed by immunoblotting. Mock precipitates from identically treated cells served as negative controls. B, exogenous PCNA interacts with endogenous ATM. U2OS cells were transfected with a vector for either HA-tagged ATM or FLAG-tagged PCNA. Inputs (4%) and anti-FLAG (M2-agarose) immunoprecipitates were tested for ATM and PCNA. C, exogenous ATM interacts with endogenous PCNA. HEK293T cells were transfected with a vector for FLAG-tagged PCNA or not. Inputs (4%) and anti-FLAG (M2-agarose) immunoprecipitates were tested for ATM and PCNA. D, the interaction between PCNA and ATM is not mediated by DNA. Cells were transfected with FLAG-tagged PCNA. The inputs and anti-FLAG immunoprecipitates from lysates in the absence or presence of ethidium bromide (EtBr) or DNase were tested for ATM. Lysates from untransfected cells served as negative control. E, verification and mapping of binary ATM-PCNA interactions. GST alone or GST-fused truncation mutants of ATM were bound to glutathione-Sepharose and incubated with in vitro translated, 35S-labeled PCNA. Beads were washed, and eluted proteins were analyzed by autoradiography after SDS-PAGE. The lower panel shows a scheme, and the upper right panel shows a Coomassie stain of GST-fused ATM fragments. The interaction site of the ATM with PCNA lies within the C terminal regions F7 (2400–2700) and F8 (2680–3056) (upper left panel). f, FLAG tag; ha, HA tag; IP, immunoprecipitation; shATM, shRNA against ATM.
ATM Directly Interacts with PCNA in Vitro
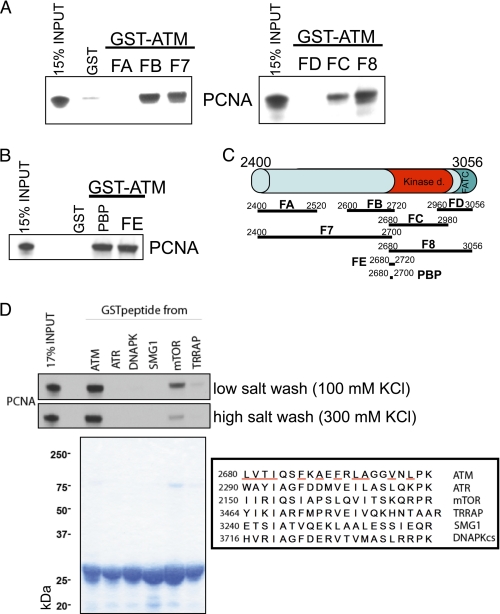
ATM is a protein of ∼350 kDa of 3056 amino acids. Despite its size, relatively few direct interaction partners have been identified to date. Examples of such binding partners include ATM itself (ATM forms a homodimer) (10), Tel2 (18), and the MRN complex (19). To establish whether PCNA binds ATM directly and to determine the regions involved in the interaction, bead-immobilized GST-fused ATM fragments purified from bacteria were incubated with in vitro translated 35S-labeled PCNA and washed. Eight overlapping fragments spanning the entire protein (scheme and Coomassie stain in Fig. 2E) were probed for PCNA binding. Labeled PCNA that bound GST-ATM fusion proteins was resolved by SDS-PAGE, and the gels were dried and exposed to film. PCNA binds directly to the C terminus of ATM, in particular to the fragments with amino acids 2400–2700 (F7) and 2680–3056 (F8) (Fig. 2E, upper left panel). The ATM C terminus encompasses the kinase domain (∼2680–2960), the PIKK-regulatory domain (∼2960–3025), and the FATC domain (∼3025–3056) (20). We next set out to fine map of the interaction domain of ATM and PCNA by truncating the C terminal bait even further (Fig. 3C). Using this iterative approach (Fig. 3, A and B), we found that the 21 ATM residues 2680–2700 are sufficient for the interaction with PCNA. This PCNA-binding peptide (PBP) is close to the catalytic site of ATM and is rich in hydrophobic residues.
FIGURE 3.
A and B, GST-fused truncation mutants of ATM were used to narrow down the interaction domain with PCNA. Interaction assays were done as described in Fig. 2E. ATM amino acids 2680–2700 (PBP) were sufficient to bind PCNA. C, scheme of GST-fused ATM truncations used. D, comparison of homologous regions in the PIKK family and their interaction with PCNA. GST-fused ATM PBP, but not the homologous peptides in other members of the human PIKK family interact with PCNA (upper left panel). The lower left panel shows a Coomassie stain of GST-PIKK fragments, and the right panel shows a sequence comparison of them. Hydrophobic residues in the ATM peptide are underlined.
Binding to PCNA Is Not Conserved among PIKK Members
ATM is a member of the phosphatidylinositol 3-kinase-related kinases (PIKKs), a family of serine/threonine protein kinases that includes A-T and Rad3-related, the catalytic subunit of DNA-dependent protein kinase, suppressor of morphogenesis in genitalia, mammalian target of rapamycin, and transformation/transcription domain-associated protein. To test whether PCNA binding is specific to ATM PBP or conserved in the analogous regions in other PIKK members (see Fig. 3D for a sequence alignment), we purified GST-tagged fusions of all corresponding PIKK peptides (Fig. 3D, lower left panel). Bead-immobilized GST-fused PIKK fragments were incubated with in vitro translated 35S-labeled PCNA and washed either with low salt or high salt buffers. PCNA binding was specific for ATM (Fig. 3D, upper left panel). Weak binding to mammalian target of rapamycin was observed under low (100 mm KCl), but not under higher salt (300 mm KCl) conditions. Of note, the fragment from the highly related A-T and Rad3-related kinase did not bind PCNA. Because PCNA binding to ATM PBP was not disrupted by increased salt concentrations, the interaction is probably mediated by hydrophobic residues.
The Interaction of PCNA with ATM Does Not Require the PCNA Interconnector Loop
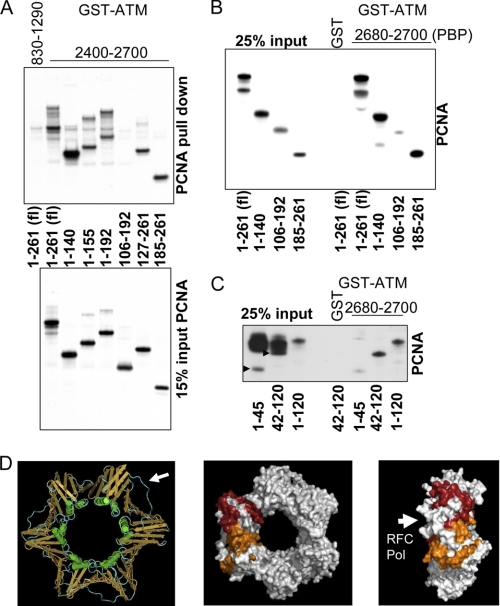
To determine where PCNA is bound by ATM we incubated bead-immobilized GST-fused ATM residues 2400–2700 or 830–1290 (as control) with in vitro translated 35S-labeled PCNA truncations. ATM interacted with both the N and C termini of PCNA, but not with a central fragment of residues 106–192 (Fig. 4A). The PCNA N-terminal residues 1–140 or the C-terminal residues 185–261 were sufficient for PCNA binding to ATM-(2400–2700). To confirm that ATM PBP (fragment 2680–2700) is sufficient for these interactions, we repeated the experiment with bead-bound GST-PBP. Again, the N and C termini of PCNA bound the shorter ATM fragment, but the central region of PCNA failed to do so (Fig. 4B). The crystal structure of PCNA revealed that it is a homotrimer forming a closed circular ring in which the C terminus of one monomer interacts tightly with the N terminus of the next one. Interestingly the C- and N-terminal domains of the monomer show structural similarity, each consisting of a 9-stranded antiparallel β sheet with two attached α helices, so that the ring is composed of six topologically identical domains (21). The major interaction sites of PCNA with partners are a hydrophobic pocket between the two domains of each monomer and the long loop connecting the two domains. Examples of proteins binding in that region are DNA pol δ, p21 (22), and flap endonuclease (Fen1) (23). These proteins contain a so-called PIP (PCNA-interaction protein) box with a QXX(M/L/I)XX(F/Y)(F/Y) consensus motif. The PIP boxes have been shown to bind to the interconnector loop in PCNA. Interestingly, ATM PBP does not bind to the PCNA interconnector loop (Fig. 4B, the interconnector loop spans PCNA amino acids 119–134), in agreement with the absence of a PIP consensus motif in the ATM PBP sequence.
FIGURE 4.
A, in vitro translated truncation mutants of PCNA were used to narrow the domains of PCNA mediating the interaction with ATM. ATM-(2400–2700) binds to both the C and N termini of PCNA, as evidenced by the interaction of PCNA truncation mutants with ATM. ATM-(830–1290) served as negative control. B, GST-ATM-(2680–2700) (PBP) is sufficient to bind PCNA. Of note, the interconnecting loop in fragment 106–192 did not bind ATM. C, ATM PBP binds to amino acids 41–120 of PCNA. Arrows indicate PCNA fragments 1–45 and 42–120 in the input. D, model of PCNA. The left panel shows the trimeric structure of PCNA with the DNA binding helices in green, the β sheets in orange, and the interconnecting loop in blue indicated by an arrow. The structure of human PCNA was from the Molecular Modeling Database, MMDB ID 31339 (24). The central panel shows a planar view of PCNA with the regions of a monomeric subunit sufficient to bind the ATM-(2680–2700) peptide colored in orange (45–106) and red (185–261). The right panel shows a side view of PCNA. The face bound by DNA polymerases and RFC is indicated by an arrow. fl, full length.
To further define the PCNA interaction sites for ATM PBP, we attempted to generate shorter PCNA truncations but found that several translated very poorly. Nevertheless, we were able to show with two reasonably expressing fragments that PCNA region 42–120 bound very well to ATM PBP, whereas PCNA-(1–45) did not (Fig. 4C). Fig. 4D shows a model of the structure of human PCNA obtained from the Molecular Modeling Database, MMDB ID 31339 (24). The left panel shows the trimeric structure of PCNA with the DNA binding helices colored in green, the β sheets in orange, and the interconnecting loop in blue, indicated by an arrow. The middle panel shows PCNA with the N-terminal (45–106) and C terminal (185–261) regions of one monomer that are sufficient for ATM PBP binding colored in orange and red, respectively. The interaction sites of PCNA with ATM determined by deletion mutagenesis would indicate that ATM-(2680–2700) binds the grove between the domains of a PCNA monomer (Fig. 4D). The interconnecting loop is neither needed for the interaction nor does it bind on its own (Fig. 4, B and C). The side view of PCNA in Fig. 4D, right panel, shows that the interaction of the ATM peptide is likely on the opposite face of where RFC and DNA pol δ bind (indicated by an arrow in the left panel), as deduced from the crystal structure of yeast PCNA with yeast RFC (25) and human PCNA with a peptide derived from the pol p66 subunit (23). Because crystal structures suggest that, as expected, DNA polymerases in general bind to the same side of the ring (26), the binding of ATM is unlikely to interfere with PCNA-DNA polymerase interactions.
ATM Stimulates PCNA-dependent DNA pol δ Activity
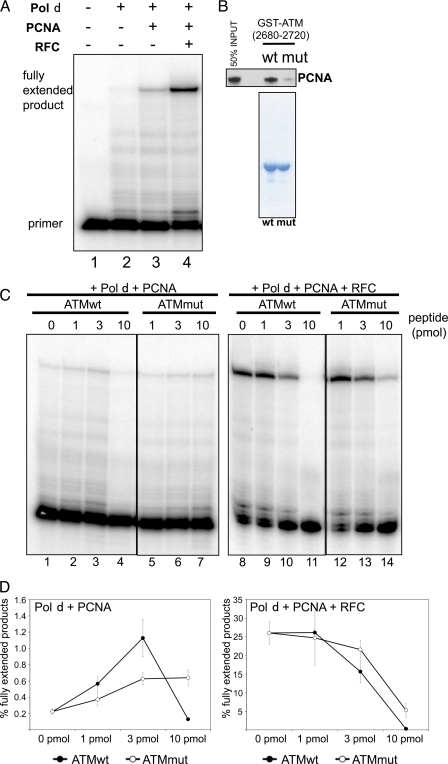
DNA pol δ is one of the three replicative polymerases and is believed to be the enzyme primarily responsible for lagging strand synthesis (27). Pol δ is also involved in DNA repair by filling the gap during mismatch repair and nucleotide excision repair and extending the invading strand in homologous recombination (28, 29). The activity of pol δ is strongly stimulated by PCNA and RFC, the clamp loader for PCNA (17) (Fig. 5A). To determine whether binding of ATM could influence PCNA-dependent DNA polymerase activity, we assayed DNA synthesis in vitro with recombinant proteins. In a primer extension assay containing pol δ and PCNA, DNA synthesis was stimulated by the addition of increasing amounts of an ATM-(2680–2700) peptide (compare lanes 1 to 3 in Fig. 5C). High concentrations of the same peptide inhibited pol δ in the same assay (lane 4, Fig. 5C). A quantitation of three independent experiments is shown in Fig. 5D.
FIGURE 5.
A, DNA synthesis assays with DNA pol δ, PCNA, and RFC. A primer extension assay was used to test the effect of PCNA and RFC on DNA pol δ activity. B, mutant ATM-(2680–2720)-2682E,2685E peptide has a lower affinity for PCNA than wild-type ATM-(2680–2720). Interaction assays with wild-type and mutant ATM PBP show the lower affinity of the mutant with PCNA (upper panel shows a GST pulldown assay of in vitro translated PCNA; the lower panel shows a Coomassie stain of the baits.). C, DNA synthesis assays with DNA pol δ and PCNA with or without RFC were done in the presence or absence of wild-type or mutant ATM peptides. The PCNA-binding peptide of ATM-(2680–2700) (ATMwt) stimulates DNA synthesis several-fold in the absence of RFC and inhibits it in the presence of RFC. D, quantitation of DNA synthesis shown in C. ATM-(2680–2700) (ATMwt) shows a much stronger ability to stimulate PCNA-dependent DNA pol δ activity than ATM-(2680–2720)-2682E,2685E (ATMmut). This figure shows ratios of fully extended primers over total primers as determined by autoradiography. Error bars represent standard deviations of three independent experiments.
Because the interaction of ATM PBP is not disrupted by 300 mm KCl (Fig. 3D), we speculate that the binding to PCNA is mediated by hydrophobic residues. We therefore hypothesized that mutating the ATM threonine 2682 and serine 2685 to the charged residue glutamic acid, which is also a phosphomimetic, would interfere with the binding of PBP to PCNA. We purified GST-ATM-(2680–2720) and GST-ATM-(2680–2720)-2682E,2685E (see Coomassie stain in Fig. 5B, lower panel) and tested the interaction of PCNA with these bead-immobilized proteins. The mutant ATM fragment bound PCNA 6–8 times weaker than wild-type (Fig. 5B, upper panel).
We then compared the ability of the wild-type and mutant ATM peptides, ATM-(2680–2700) and ATM-(2680–2700)-2682E,2685E, to stimulate pol δ in vitro. The wild-type ATM fragment stimulated DNA synthesis by pol δ to a greater extent than the mutant fragment compromised in PCNA binding (compare lanes 2 to 3 with lanes 5 to 6 in Fig. 5C). Similar results were observed when purified GST-ATM-(2680–2720) and GST-ATM-(2680–2720)-2682E,2685E were used in the DNA synthesis assay (data not shown). Unlike the wild-type ATM peptide, the PCNA-binding-impaired peptide did not inhibit pol δ at higher concentration (compare lanes 4 and 7 in Fig. 5C and see the quantitation in Fig. 5D).
Finally we tested the effect of wild-type and mutant ATM fragments on pol δ in the presence of both PCNA and RFC. Both wild-type and mutant ATM peptides inhibited DNA synthesis under these conditions, but the wild-type ATM peptide had a much stronger inhibitory effect than the mutant ATM peptide with the lower affinity for PCNA. In summary, these observations indicate that ATM does interact with PCNA and can influence PCNA activity in vitro. The interaction assays with PCNA deletion mutants suggest that ATM-(2680–2700) interacts with PCNA in the grove between two monomer domains (Fig. 4D). It could be that pol δ preferentially interacts with PCNA when one or more of three potential peptide binding sites on the trimer are unoccupied at low peptide peptide concentrations, whereas occupancy of the three binding sites at higher peptide concentration inhibits the interaction with pol δ. The maximum stimulatory effect on the peptide on DNA synthesis by pol δ and PCNA was 6- to 10-fold, whereas RFC stimulates the same reaction more than 100-fold as a consequence of active PCNA loading (Fig. 5D and data not shown). The inhibitory effect of the ATM peptide on RFC-stimulated DNA synthesis could be due to reduced loading of PCNA trimers bound by the peptide.
DISCUSSION
Following our initial observation that an ATM-specific inhibitor influences DNA synthesis in an ATM-dependent manner, we investigated whether ATM binds to PCNA, a docking platform for many factors involved in DNA replication and DNA repair. Surprisingly, we found that ATM and PCNA physically interact both in vivo and in vitro. We were able to define the ATM site sufficient for PCNA binding to a small region near the ATM kinase domain. This small peptide, ATM PBP, stimulates DNA pol δ in a PCNA-dependent manner in an in vitro assay.
ATM Binding to PCNA Does Not Involve a PIP Box
Unlike many other PCNA interacting proteins, the ATM fragment sufficient to bind PCNA does not have a motif that would fit the consensus of either a PIP box or a related KA box (described in Ref. 30). Consistent with the absence of a PIP box, this interaction is not mediated by the PCNA interconnector loop that plays an essential role in p21 (22), pol δ3 (p66), and FEN1 binding (23). ATM is a 350-kDa protein that forms a dimer in solution when in its inactive state (10). Future studies will determine whether ATM binds to PCNA as a monomer or as a dimer. Our interaction data with ATM fragments suggest that ATM dimerization is not needed for PCNA binding, and it is probable that monomerization is needed to expose the region close to the kinase domain for the interaction. Of note, theoretically trimeric PCNA contains three identical binding sites for a partner. Nevertheless, given ATM's size it is unlikely that three ATM molecules could simultaneously bind the trimeric ring. Instead it is possible that ATM and the DNA polymerase share PCNA as a docking platform during DNA synthesis.
Regulation of PCNA-ATM Interaction
Recent studies have shown that PCNA activity is highly regulated by post-translational modification (31). For instance, mono-ubiquitylation of PCNA at Lys-164 facilitates the recruitment of translesion synthesis polymerases that enable DNA damage bypass, whereas poly-ubiquitylation of the same residue dislodges translesion synthesis polymerases from PCNA enabling a template-switching mechanism in the error-free pathway of post-replication repair. At least in yeast, PCNA is also SUMOylated at Lys-164 and Lys-127 resulting in the inhibition of recombination via recruitment of Srs2 or inhibition of cohesion establishment via repulsion of Eco1 respectively (32, 33). Future studies will show whether the interaction of PCNA with ATM is also regulated by post-translational modification of PCNA or - as in the case of p21, RFC1 and Fen1 (31) by phosphorylation of its binding partner.
PCNA as a Recruiter of ATM
Previously identified interacting partners of PCNA include proteins involved in DNA replication, DNA repair, cell cycle control, sister chromatid cohesion, and chromatin assembly and maintenance (30). Our novel finding that ATM binds PCNA suggests that PCNA plays a role in the early stage of DNA damage signaling by ATM.
PCNA is a key factor of several cellular processes involving DNA synthesis, such as DNA replication, DNA repair, and homologous recombination. Our initial observation that ATM inhibition leads to decreased DNA synthesis is likely due to an inhibition of DNA replication during S phase, given the extent of change in thymidine incorporation observed. In view of the physical interaction of PCNA with ATM and the resulting stimulation of DNA pol δ, it is tempting to speculate that ATM recruitment to the replication fork plays a role in the surveillance of ongoing replication. Interestingly, several reports on the early characterization of A-T cells found that those cells had a decreased DNA synthesis rate and prolonged S phase compared with normal cells (3, 4). Yet, an alternative possibility would place the actual function of the interaction of PCNA with ATM after replication forks stalls or collapses, particularly at the stage of homologous recombination following double strand break formation. In this regard, PCNA was found to be required for recombination-associated DNA synthesis by pol δ (28), and PCNA and pol δ specifically were reported to be needed for break-induced replication (34), a homologous recombination pathway used when only one end of the double strand break shares homology with a template. The idea that ATM regulates PCNA function at the recombination step is particularly attractive, because we found ATM inhibitors to decrease both DNA synthesis and DNA damage-induced sister chromatid exchange (13). Furthermore, in both cases kinase inhibition does not phenocopy protein loss, leading us to speculate that ATM inhibition leads to ATM physically blocking the dynamic repair processes involved in homologous recombination (35).
Acknowledgment
We thank Dr. Titia de Lange for some of the GST-fused ATM expression vectors.
This work was supported, in whole or in part, by National Institutes of Health Grants CA148644 (to C. J. B.), GM57479 (to A. E. T. ), and P30CA047904. This work was also supported by the Frieda G. and Saul F. Shapira Breast Cancer Research Program.
- A-T
- ataxia telangiectasia
- ATM
- A-T mutated
- PCNA
- proliferating cell nuclear antigen
- UTR
- untranslated region
- pol
- polymerase
- PIKK
- phosphatidylinositol 3-kinase-related kinase
- PIP
- PCNA-interaction protein
- PBP
- PCNA-binding peptide
- RFC
- replication factor C.
REFERENCES
- 1. Lavin M. F. (2008) Ataxia-telangiectasia. From a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 2. Painter R. B., Young B. R. (1980) Radiosensitivity in ataxia-telangiectasia. A new explanation. Proc. Natl. Acad. Sci. U.S.A. 77, 7315–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murnane J. P., Painter R. B. (1982) Complementation of the defects of DNA synthesis in irradiated and unirradiated ataxia-telangiectasia cells. Proc. Natl. Acad. Sci. U.S.A. 79, 1960–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen M. M., Simpson S. J. (1980) Growth kinetics of ataxia telangiectasia lymphoblastoid cells. Evidence for a prolonged S period. Cytogenet. Cell Genet. 28, 24–33 [DOI] [PubMed] [Google Scholar]
- 5. Cornforth M. N., Bedford J. S. (1985) On the nature of a defect in cells from individuals with ataxia-telangiectasia. Science 227, 1589–1591 [DOI] [PubMed] [Google Scholar]
- 6. Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M. J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., Löbrich M. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16, 715–724 [DOI] [PubMed] [Google Scholar]
- 7. Berkovich E., Monnat R. J., Jr., Kastan M. B. (2007) Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9, 683–690 [DOI] [PubMed] [Google Scholar]
- 8. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 9. Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D. A., Smith S., Uziel T., Sfez S., Ashkenazi M., Pecker I., Frydman M., Harnik R., Patanjali S. R., Simmons A., Clines G. A., Sartiel A., Gatti R. A., Chessa L., Sanal O., Lavin M. F., Jaspers N. G., Taylor A. M., Arlett C. F., Miki T., Weissman S. M., Lovett M., Collins F. S., Shiloh Y. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268, 1749–1753 [DOI] [PubMed] [Google Scholar]
- 10. Bakkenist C. J., Kastan M. B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 11. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 12. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- 13. White J. S., Choi S., Bakkenist C. J. (2010) Transient ATM kinase inhibition disrupts DNA damage-induced sister chromatid exchange. Sci. Signal 3, ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 15. Maga G., Hubscher U. (2003) Proliferating cell nuclear antigen (PCNA). A dancer with many partners. J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 16. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 17. Fazlieva R., Spittle C. S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. (2009) Proofreading exonuclease activity of human DNA polymerase δ and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 37, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takai H., Wang R. C., Takai K. K., Yang H., de Lange T. (2007) Tel2 regulates the stability of PI3K-related protein kinases. Cell 131, 1248–1259 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. H., Paull T. T. (2004) Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304, 93–96 [DOI] [PubMed] [Google Scholar]
- 20. Lempiäinen H., Halazonetis T. D. (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 22. Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 23. Bruning J. B., Shamoo Y. (2004) Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-δ p66 subunit and flap endonuclease-1. Structure 12, 2209–2219 [DOI] [PubMed] [Google Scholar]
- 24. Kontopidis G., Wu S. Y., Zheleva D. I., Taylor P., McInnes C., Lane D. P., Fischer P. M., Walkinshaw M. D. (2005) Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U.S.A. 102, 1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bowman G. D., O'Donnell M., Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 26. Hishiki A., Hashimoto H., Hanafusa T., Kamei K., Ohashi E., Shimizu T., Ohmori H., Sato M. (2009) Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284, 10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kunkel T. A., Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X., Stith C. M., Burgers P. M., Heyer W. D. (2009) PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell 36, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lange S. S., Takata K., Wood R. D. (2011) DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoimenov I., Helleday T. (2009) PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37, 605–613 [DOI] [PubMed] [Google Scholar]
- 31. López de Saro F. J. (2009) Regulation of interactions with sliding clamps during DNA replication and repair. Curr. Genomics 10, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fox J. T., Lee K. Y., Myung K. (2011) Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett. 585, 2780–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 34. Lydeard J. R., Lipkin-Moore Z., Sheu Y. J., Stillman B., Burgers P. M., Haber J. E. (2010) Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 24, 1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi S., Gamper A. M., White J. S., Bakkenist C. J. (2010) Inhibition of ATM kinase activity does not phenocopy ATM protein disruption: implications for the clinical utility of ATM kinase inhibitors. Cell Cycle 9, 4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]