Background: Intestinal cell kinase (ICK), a ubiquitously expressed and highly conserved serine/threonine protein kinase, supports cell proliferation.
Results: Phosphorylation of Raptor Thr-908 by ICK is important for full activation of mTORC1.
Conclusion: ICK plays an important role in regulating the mTORC1 activity.
Significance: These findings provide a novel mechanistic insight into the signaling cascades by which ICK regulates proliferation.
Keywords: Enzyme Mechanisms, mTOR, Protein Kinases, Protein Phosphorylation, Signal Transduction, Intestinal Cell Kinase (ICK), Raptor, Raptor Phosphorylation, mTORC1
Abstract
Intestinal cell kinase (ICK), named after its cloning origin, the intestine, is actually a ubiquitously expressed and highly conserved serine/threonine protein kinase. Recently we reported that ICK supports cell proliferation and G1 cell cycle progression. ICK deficiency significantly disrupted the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) signaling events. However, the biological substrates that mediate the downstream signaling effects of ICK in proliferation and the molecular mechanisms by which ICK interacts with mTORC1 are not well defined. Our prior studies also provided biochemical evidence that ICK interacts with the mTOR/Raptor complex in cells and phosphorylates Raptor in vitro. In this report, we investigated whether and how ICK targets Raptor to regulate the activity of mTORC1. Using the ICK substrate consensus sequence [R-P-X-S/T-P/A/T/S], we identified a putative phosphorylation site, RPGT908T, for ICK in human Raptor. By mass spectrometry and a phospho-specific antibody, we showed that Raptor Thr-908 is a novel in vivo phosphorylation site. ICK is able to phosphorylate Raptor Thr-908 both in vitro and in vivo and when Raptor exists in protein complexes with or without mTOR. Although expression of the Raptor T908A mutant did not affect the mTORC1 integrity, it markedly impaired the mTORC1 activation by insulin or by overexpression of the small GTP-binding protein RheB under nutrient starvation. Our findings demonstrate an important role for ICK in modulating the activity of mTORC1 through phosphorylation of Raptor Thr-908 and thus implicate a potential signaling mechanism by which ICK regulates cell proliferation and division.
Introduction
The RCK (the tyrosine kinase gene v-ros cross-hybridizing kinase) family within the CMGC (CDK/MAPK/GSK3/CLK) group of the human kinome consists of male germ cell-associated kinase (MAK)2 (1), ICK/MRK (Intestinal cell kinase/MAK-related kinase) (2, 3) and MAPK/MAK/MRK-overlapping kinase (4). They all share significant homology with MAP kinases in the catalytic domain and contain a MAPK-like TXY motif in the activation T-loop (5, 6). However, they exhibit significant divergence in the composition of their C-terminal non-catalytic domains that may confer their functional specificities by mediating associations with distinct protein complexes and cellular machinery.
Similar to MAPKs, the catalytic activity of ICK can be regulated by dual phosphorylation of its TDY motif (2, 5). Within the TDY motif of ICK, autophosphorylation of Tyr-159 only confers basal kinase activity. Additional phosphorylation of Thr-157 is required for full activation (5). However, the regulatory mechanism for the activation of ICK appears to be very different from that of classic MAPKs. Unlike MAPKs, the ICK kinase activity cannot be acutely stimulated by serum or growth factors. Our prior studies suggest that protein phosphatase 5 (7) and CCRK/CDK20 (cell cycle-related kinase/cyclin-dependent kinase 20) (8) are putative upstream yin-yang regulators for the phosphorylation of ICK Thr-157 (9). We have shown that oxidative stress, which activates protein phosphatase 5, is able to inactivate ICK by dephosphorylating Thr-157 within the TDY motif (9). Although CCRK is closely related to yeast Cak1p (Cdk-activating kinase) and mammalian CDK7 (cyclin-dependent kinase 7), there is controversy as to whether CCRK has any intrinsic CDK activating kinase activity (8, 10). How the activity of CCRK is regulated, which may provide an important clue to the regulatory mechanism for the activation of ICK, remains unknown.
The biological functions of the ICK/MAK/MOK family have been elusive until recently. MAK is highly expressed in testis. However, the MAK-null mouse is viable and fertile, suggesting the existence of functional redundancy or compensation for the lack of MAK in testis (11). In the MAK-null retina, photoreceptors exhibit elongated cilia and progressive degeneration, suggesting that MAK is required for regulation of ciliary length and retinal photoreceptor survival (12). MAK is a coactivator for androgen receptor in prostate cancer cells and is required for androgen receptor-mediated signaling and cell proliferation (13, 14). A loss-of-function point mutation, R272Q, of ICK has been recently identified as the causative mutation in a neonatal lethal multiplex human syndrome endocrine-cerebro-osteodysplasia (ECO), implicating a key role for ICK in development of multiple organ systems (15). Using shRNA knockdown, we have shown that suppression of ICK expression in intestinal epithelial cells markedly impaired cell proliferation and G1 cell cycle progression (16). Furthermore, ICK deficiency led to a significant decrease in the mTORC1 activity, concomitant with reduced expression of specific mTORC1 downstream targets, cyclinD1 and c-Myc (16). These results suggest that ICK may target the mTORC1 signaling pathway to regulate cell proliferation and cell cycle progression.
The serine-threonine protein kinase mammalian target of rapamycin (mTOR) is the core catalytic component of two structurally and functionally distinct protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), that collectively integrates nutrient, hormonal, and energy signal inputs to control cell growth, proliferation, and survival (17–19). mTORC1, when activated by growth factors and nutrients, stimulates cell growth and proliferation by phosphorylating two key regulators of mRNA translation and ribosome biogenesis, S6K1 (ribosomal protein S6 kinase) and 4EBP1 (eukaryotic initiation factor 4E-binding protein 1) (20–23). In addition to the catalytic subunit mTOR, mTORC1 also contains four associated components, Raptor, mLST8/GβL, PRAS40, and Deptor (24–30). Raptor, a regulatory associated protein of mTOR, plays an important role as a scaffolding protein to recruit substrates S6K1 and 4EBP1 to mTOR (31). Upon growth factor stimulation, Raptor binding to substrates can be enhanced by the dissociation of the competitive inhibitor PRAS40 (the proline-rich Akt substrate of 40KDa) from mTORC1 (28, 32–34). Raptor can also positively regulate mTOR activity in response to nutrient sufficiency by directly interacting with Rag family GTPases to induce mTORC1 relocalization to an intracellular vesicular compartment containing RheB, a Ras-like GTP-binding protein that activates mTOR via an unknown mechanism (35–37). Another main upstream regulator of mTORC1 is the tuberous sclerosis complex (TSC), a heterodimer of TSC1 and TSC2. TSC functions as a GTPase activating protein to catalyze the conversion of Rheb-GTP to Rheb-GDP and thereby down-regulates mTOR activity (38, 39).
Given that Raptor plays such an essential role in modulating the mTOR activity, a more detailed characterization of its biochemical properties is necessary to provide further insights into the regulatory mechanisms underlying mTORC1 activation. Recently, multiple phosphorylation sites of Raptor have been identified, several of which are critical for the regulation of mTORC1 activity in response to insulin, nutrients, or energy stress. Phosphorylation of Raptor Ser-722 and Ser-792 by AMP-activated protein kinase (AMPK) is required for the inhibition of mTORC1 and cell cycle arrest induced by energy stress (40). RSK-mediated phosphorylation of Ser-719/721/722 enhances mTORC1 activity stimulated by the Ras/MAPK pathway (41). Phosphorylation of Ser-863 by either mTOR or ERK1/2 promotes mTORC1 activation in response to various stimuli, including growth factors, nutrients, and cellular energy (42–44). Taken together, these findings indicate that the complex phosphorylation status of Raptor is tightly associated with the activity of mTORC1.
We have demonstrated previously that suppression of ICK expression in cultured epithelial cells induced growth retardation and cell cycle delay at G1, concomitant with a marked down-regulation of mTORC1 activity (16). Because ICK can interact with mTOR/Raptor in cells and because Raptor is an in vitro substrate for ICK (16), we sought to investigate whether ICK can target Raptor in vivo to regulate the mTORC1 activity and signaling events in accordance with various environmental cues. Using the ICK phosphorylation consensus, we identified a putative phosphorylation site, Thr-908, for ICK in human Raptor. By site-directed mutagenesis, we proved that ICK can phosphorylate Raptor Thr-908 in vitro and that this phosphorylation event still occurs when Raptor is in complex with mTOR. Furthermore, using mass spectrometry and a phospho-specific antibody, we showed that ICK is able to significantly enhance Raptor Thr-908 phosphorylation in vivo. More importantly, we were able to demonstrate that phosphorylation of Raptor Thr-908 is important for promoting mTORC1 activation. Our findings not only uncover a novel phosphorylation site of Raptor that contributes to the regulation of mTORC1 activity but also provide an important regulatory mechanism by which ICK controls cell proliferation and division.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human insulin (Novolin R) was from Novo Nordisk. Rapamycin and Calyculin A were from Calbiochem EMD Biosciences. DNA plasmids encoding FLAG-Raptor, FLAG-Raptor (S863A), FLAG-RheB, FLAG-mTOR, FLAG-mTOR (rapamycin-resistant mutant S2035W), Myc-mTOR, AU1-mTOR, and HA-S6K1 were described in Ref. 44. Torin1 was a gift to T. E. Harris from Dr. David Sabatini (Whitehead Institute). Human Raptor mutants T908A and T908E were generated using the QuikChange kit II from Stratagene and confirmed by DNA sequencing.
Antibodies
Antibodies to mTOR, Raptor, S6K1, phospho-S6K1 Thr-389, Akt, phospho-Akt Ser-473, and phospho-Akt Thr-308 were all from Cell Signaling Technology. An affinity-purified ICK rabbit polyclonal antibody was previously described in Ref. 16. Anti-FLAG M2 monoclonal antibody and Anti-FLAG M2 affinity gel were from Sigma-Aldrich. Anti-Myc monoclonal antibody (9E10) and anti-AU1 tag polyclonal antibody were from Pierce.
The polyclonal Raptor Phospho-Thr-908 antibody was generated in rabbits against keyhole limpet hemocyanin-coupled phosphopeptides (CSRDLPSGRPGpTTGP) using the custom antibody service from Genscript. Anti-phosphopeptide-specific antibodies were affinity-purified through a positive selection over phosphopeptide antigens followed by negative selections over non-phosphopeptide antigens and FLAG-Raptor T908A protein antigens.
Cell Culture, Transfection, and Treatment
HEK293T and HEK293E cells were maintained at 37 °C and 5% CO2 in DMEM supplemented with 4.5 g/liter glucose and 10% FBS. At 30–40% confluence, cells were transfected using a calcium phosphate protocol as described in Refs. 5, 9. For insulin stimulation, cells were serum-starved overnight in DMEM containing 0.1% FBS, treated with 20 nm rapamycin for 20 min, and then with 100 nm insulin for 10, 30, 60 and 90 min respectively. For amino acid starvation, cells were incubated with Dulbecco's PBS (d-PBS) containing D-glucose (1 g/liter) and sodium pyruvate (1 mm) for 1 h.
Cell Extract Preparation and Immunoprecipitation
Cell extracts were prepared essentially as described in Ref. 9. Briefly, cells were harvested in ice-cold PBS and lysed in lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 2 mm EGTA, and supplemented with complete protease inhibitors (Roche), 1 mm Na3VO4, 1 μm microcystin LR, and 5 mm β-glycerophosphate). To preserve mTOR-Raptor interaction, cells were lysed in CHAPS lysis buffer (40 mm HEPES (pH 7.4), 150 mm NaCl, 0.3% CHAPS, 1 mm EDTA, 1 mm EGTA, and supplemented with complete protease inhibitors (Roche), 1 mm Na3VO4, 1 μm microcystin LR, and 5 mm β-glycerophosphate). After the cell lysate was cleared by centrifugation, the supernatant was incubated with antibodies bound to GammaBind Plus Sepharose beads (GE Healthcare) for 2 h at 4 °C. The beads were extensively washed with lysis buffer followed by PBS buffer. The beads were boiled in SDS sample buffer (0.5 m Tris (pH 6.8), 10% SDS, 10% glycerol, 0.1% bromphenol blue) for 5 min to elute binding proteins.
In Vitro Kinase Assay
FLAG-Raptor or FLAG-Raptor/Myc-mTOR complex were immunoprecipitated from whole cell lysates as described above. The beads were extensively washed in lysis buffer followed by kinase buffer (50 mm HEPES (pH 7.5), 10 mm MgCl2, 5 mm DTT supplemented with complete protease inhibitors (Roche), 1 mm Na3VO4, 1 μm microcystin LR, and 5 mm β-glycerophosphate). The bead samples were then incubated with 5 μCi [γ-32P]ATP (7000 Ci/mmol), 100 μm ATP, and 1 μg of affinity-purified His-ICK(1–296) in 50 μl of kinase buffer at 30 °C for 20 min with gentle agitation. The reaction was terminated by addition of 50 μl of 2× SDS sample buffer. The reaction sample was heated at 95 °C for 5 min and separated on a 10% SDS gel. The gel was dried and exposed for autoradiograph.
Affinity Gel Purification of FLAG-tagged Raptor
FLAG-Raptor (5 μg of plasmid DNA/10-cm plate) was transfected into HEK293E cells. Forty-eight hours after transfection, cells were treated with 20 nm Calyculin A for 30 min before cell harvesting and lysis. FLAG-Raptor protein was purified from the whole cell extract by the anti-FLAG M2 affinity gel (Sigma Aldrich) following the manufacturer's instructions.
LC-MS/MS Analysis of FLAG-tagged Raptor
FLAG-Raptor samples were reduced and carbamidomethylated at room temperature using dithiothreitol and iodoacetamide (Sigma Aldrich), respectively, and then digested proteolytically (1:20 enzyme to substrate) with trypsin (Promega), as reported previously (45). An aliquot of the resulting peptide mixture was pressure-loaded onto a fused silica capillary precolumn (360 μm outer diameter × 75 μm inner diameter) packed with 6 cm of irregular C18 reverse phase packing material (ODS-A, 120 Å pore size, 15 μm diameter). Following a desalting rinse using 0.1 m acetic acid, the precolumn was connected to a fused silica capillary analytical column (360 μm o.d. x 50 μm i.d) containing 6–8 cm of C18 reverse phase packing material (ODS-A, 120 Å pore size, 5 μm diameter) and equipped with an electrospray emitter tip. The tryptic peptides were gradient-eluted into the mass spectrometer at a flow rate of 60 nL/min, as reported previously (46).
Mass spectra were acquired on a front-end electron transfer dissociation-enabled LTQ-FT (Thermo Scientific). MS1 spectra were acquired in the high-resolution FT mass analyzer and MS2 spectra were acquired in the linear ion trap mass analyzer. Following each high resolution MS1 spectrum, eight data-dependent low resolution MS2 spectra were acquired in a collision activated disscociation/electron transfer dissociation toggle manner. All MS2 spectra were collected using the following instrument parameters: one microscan, 3 m/z mass isolation window, default charge state of +10, monoisotopic precursor selection “enabled,” and precursor ions in the +1 charge state were excluded from MS2 analysis. Full automated gain control targets were set to 1e6 for fourier transform mass spectrometry and 3e4 for ion trap mass spectrometry. ETD spectra utilized a 30-ms reaction time with a 2- to 4-ms electron transfer reagent injection time with azulene as the electron transfer reagent.
MS Data Analysis
All MS2 spectra were searched against a Raptor protein database, and the human, rat, and mouse non-redundant database using the open mass spectrometry search algorithm. Prior to searching the data, MS2 peak lists were generated using a Java-based program written in-house. All searches were completed using either the “no enzyme” or “trypsin” digestion parameter. Precursor and peptide mass tolerances were set to ± 0.05 and ± 0.35, respectively. All database searches included the following variable modifications: carbamidomethylation of Cys; oxidation of Met; mono-, di-, and tri-methylation of Lys; phosphorylation of Ser, Thr, and Tyr; and O-GlcNAcylation of Ser and Thr. The open mass spectrometry search algorithm removed the reduced charge species from the ETD peak lists prior to searching. Up to three missed cleavages were allowed for trypsin digest searches. Results from the complementary ETD/CAD searches were used as a guide for data analysis. All data were confirmed by manual interpretation of the MS2 spectra.
RESULTS
ICK Phosphorylates Raptor Thr-908 in Vitro
Previously we found that ICK promotes proliferation and G1 cell cycle progression of cultured epithelial cells (16). Furthermore, ICK can interact with mTOR/Raptor in vivo, and Raptor, but not mTOR, is an in vitro substrate for ICK (16). These observations raised the intriguing questions as to whether Raptor is an in vivo target for ICK and, if so, whether ICK phosphorylation of Raptor plays an important role in regulating the activity of mTORC1.
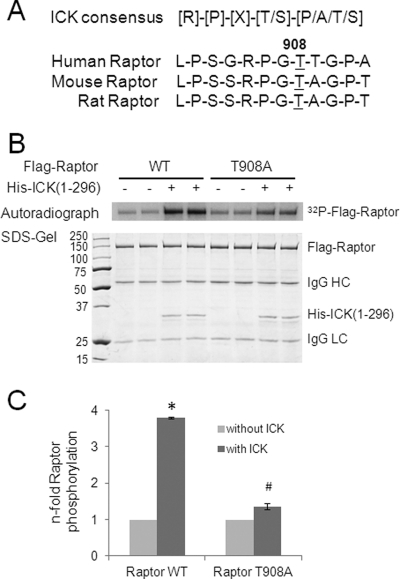
Using the ICK phosphorylation consensus motif (Fig. 1A) (9), we identified one putative ICK phosphorylation site, Thr-908, in human Raptor that is also conserved in mice and rats. To test whether Raptor Thr-908 can be phosphorylated by ICK, we performed an in vitro kinase assay using His-ICK(1–296), an active recombinant ICK protein including the entire catalytic domain of ICK (1–284) (9). Beads bound with nearly equal amounts of either FLAG-Raptor WT or FLAG-Raptor T908A mutant proteins, as shown in the SDS gel (Fig. 1B), were used as substrates. Although ICK was able to robustly phosphorylate the wild-type Raptor, the T908A mutation almost completely abolished the phosphorylation of Raptor by ICK (Fig. 1, B and C). This result indicates that Thr-908 is the predominant ICK phosphorylation site on Raptor in vitro.
FIGURE 1.
ICK phosphorylates Raptor Thr-908 in vitro. A, the substrate phosphorylation consensus for ICK is R-P-X-S/T-P/A/T/S, with the preference for arginine at P-3 and proline at P-2 being more stringent than for proline at p + 1. Raptor Thr-908 lies within the ICK consensus motif R-P-G-T-T/A that is conserved in human, mouse, and rat. B, FLAG-tagged human Raptor of either the WT or the T908A mutant (T908A) was expressed in HEK293T cells. FLAG-Raptor from the whole cell lysate was captured on beads through anti-FLAG immunoprecipitation. The beads sample was incubated with His-ICK(1–296) for an in vitro kinase assay. Also shown are the 32P-autoradiograph (B), the Coomassie-stained SDS gel (B), and the quantification data (C) showing the relative number of fold increase in 32P signals incorporated into FLAG-Raptor (WT or T908A) in the absence or presence of ICK. Data are mean ± S.D. n = 3. *, p < 0.001; #, p < 0.05; Student's t test.
ICK Is Capable of Phosphorylating Raptor Thr-908 When Raptor Exists in Complexes with or without mTOR
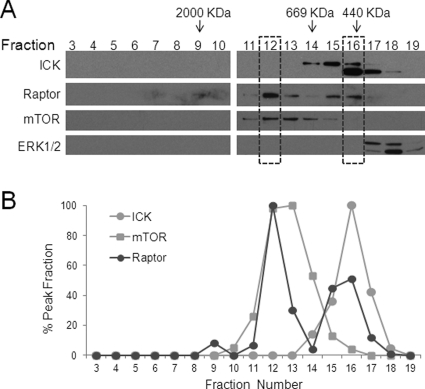
A previous study has demonstrated that Raptor was eluted off a gel filtration column in two major peaks, a high Mr peak that overlapped with the mTOR peak and a low Mr peak where very little, if any, mTOR was found (47). This observation suggests that Raptor may exist in distinct protein complexes in vivo. We reported previously that ICK could interact with Raptor with or without mTOR in complex (16). These results together prompted us to ask whether ICK cofractionates with Raptor in vivo and, more importantly, whether phosphorylation of Raptor Thr-908 by ICK occurs in protein complexes with or without mTOR. To address the first question, we examined the distribution pattern of the endogenous ICK protein in different fractions from a size-exclusion chromatography column that was used previously to characterize the fractionation profiles of mTOR and Raptor (47). We were able to recapitulate the original observation that Raptor displays two major elution peaks, with the high Mr peak coinciding with the peak of mTOR (Fig. 2, F12). Interestingly, we observed that the single peak of ICK largely overlaps with the low Mr peak of Raptor that contains very little (Fig. 2, F15) or an undetectable amount of mTOR (F16), suggesting that ICK may coexist with Raptor in protein complexes with or without mTOR.
FIGURE 2.
Cell fractionation analysis of endogenous ICK, Raptor and mTOR. A, cell extracts from 3T3-L1 adipocytes were prepared and fractionated by using a Superose 6 column as described in Ref. 47. Samples of equal volume from each fraction were immunoblotted with antibodies against ICK, Raptor, mTOR, and ERK1/2, respectively. B, the relative abundance of ICK, mTOR and Raptor signals from different fractions were plotted in a line graph. Note that the high Mr peak of Raptor (fraction 12) largely coincides with the mTOR peak (fractions 12 and 13), whereas the low Mr peak of Raptor (fractions 15 and 16) partially overlaps with the ICK peak (fractions 15–17). Anti-ICK recognized two bands on Western blot analysis. The lower band may represent either an alternatively spliced variant (60 KDa) or a truncated product of the full-length ICK (70 KDa, upper band).
To address the question whether ICK is able to phosphorylate Raptor when it is in complex with mTOR, we either expressed FLAG-Raptor alone (positive control) or coexpressed FLAG-Raptor and Myc-mTOR in HEK293T cells (Fig. 3). After immunoprecipitation, the immune complexes captured on beads were used as substrates for ICK in an in vitro kinase assay. FLAG-Raptor alone was robustly phosphorylated by ICK (Fig. 3A, left panel). ICK was able to increase the phosphorylation of FLAG-Raptor in the Myc-mTOR immune complexes (Fig. 3, A, right panel, and B, left panel), although it appears that FLAG-Raptor was phosphorylated in the Myc-mTOR immunoprecipitates in the absence of ICK. Given that mTOR is known to phosphorylate Raptor Ser-863, we used an mTOR-specific inhibitor, Torin 1, in our in vitro kinase assay to suppress Raptor Ser-863 phosphorylation. As expected, Torin 1 was able to effectively abolish FLAG-Raptor phosphorylation in the Myc-mTOR immune complexes in the absence of ICK (Fig. 3B, right panel), suggesting that Raptor Ser-863 is the major phosphorylation site of FLAG-Raptor in the Myc-mTOR immunoprecipitates. In the presence of Torin 1, it is apparent that ICK was able to further enhance the phosphorylation of FLAG-Raptor when Raptor is in complex with mTOR. These results demonstrate that ICK is able to phosphorylate Raptor in vitro when Raptor is either alone or associated with mTOR.
FIGURE 3.
ICK is able to phosphorylate Raptor Thr-908 either with or without mTOR in the same complex. A, FLAG-Raptor was either transfected alone or cotransfected with Myc-mTOR in HEK293T cells. After immunoprecipitation (IP) using anti-FLAG or anti-Myc antibodies, recombinant proteins of Raptor or Raptor bound to mTOR on beads were used as substrates for an in vitro kinase assay. Shown are the 32P-autoradiograph (Autorad) and the Coomassie-stained SDS gel. An aliquot of the beads samples from the above in vitro kinase assay was Western blotted against anti-Myc and anti-FLAG antibodies. B, Myc-mTOR and FLAG-Raptor were coexpressed in HEK293T cells. The recombinant mTOR/Raptor complex was isolated from the whole cell lysate by the anti-Myc immunoprecipitation. The immune complex captured on beads was used as substrates for an in vitro kinase assay in the absence or presence of 100 nm Torin 1. Shown are the 32P-autoradiograph and the Coomassie-stained SDS gel (7.5%). An aliquot of the beads samples from the above in vitro kinase assay was separated on a 10% SDS gel and Western-blotted against anti-Myc and anti-FLAG antibodies.
Raptor Thr-908 Phosphorylation Does Not Prevent the Association between Raptor and mTOR
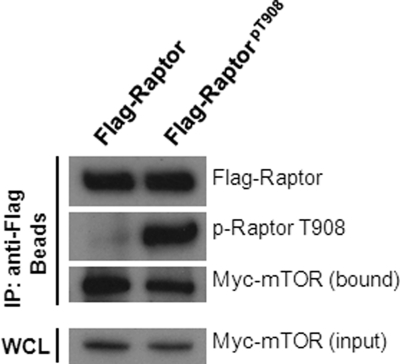
Because Raptor Thr-908 can be phosphorylated by ICK when Raptor is not in complex with mTOR, we asked the question whether the binding capacity of Raptor to mTOR is affected by Raptor Thr-908 phosphorylation. FLAG-Raptor proteins were isolated from the whole cell lysate by anti-FLAG immunoprecipitation and were phosphorylated in vitro by His-ICK(1–296). Phosphorylation of Thr-908 on FLAG-Raptor was confirmed by the phospho-Thr-908 antibody signal on Western blot analysis (Fig. 4). Our mass spectrometry analysis also indicated that nearly 100% of FLAG-Raptor is Thr-908-phosphorylated after in vitro reaction with His-ICK(1–296) (data not shown). The bead samples containing FLAG-Raptor were incubated with the whole cell lysate containing Myc-mTOR, and the amount of Myc-mTOR bound to the beads was assessed on Western blot analysis by the anti-Myc antibody. FLAG-Raptor that is fully phosphorylated at Thr-908 by ICK appears to be able to bind Myc-mTOR, albeit to a lesser extent than FLAG-Raptor that contains very little phospho-Thr-908 signals (Fig. 4). This result indicates that Raptor, once phosphorylated on Thr-908 by ICK, is still able to associate with mTOR.
FIGURE 4.
Raptor Thr-908 phosphorylation does not prevent Raptor from association with mTOR. FLAG-Raptor was expressed in HEK293T cells. After anti-FLAG immunoprecipitation (IP), FLAG-Raptor was captured on affinity gel beads. The bead samples were subjected to an in vitro kinase reaction in the absence (negative control) or presence of His-ICK(1–296). The phosphorylation status of Thr-908 on FLAG-Raptor was assessed by blotting the beads samples with the anti-phospho-Thr-908 antibody. The bead samples were extensively rinsed and then incubated with the whole cell lysate (WCL) containing Myc-mTOR for 2 h at 4 °C. Following rigorous rinses, the bead samples were analyzed for the presence of Myc-mTOR and FLAG-Raptor by blotting with anti-Myc and anti-FLAG antibodies respectively. Also shown is the total Myc-mTOR input from the whole cell lysate.
Phosphorylation of Raptor Thr-908 Can Be Catalyzed by ICK in Vivo
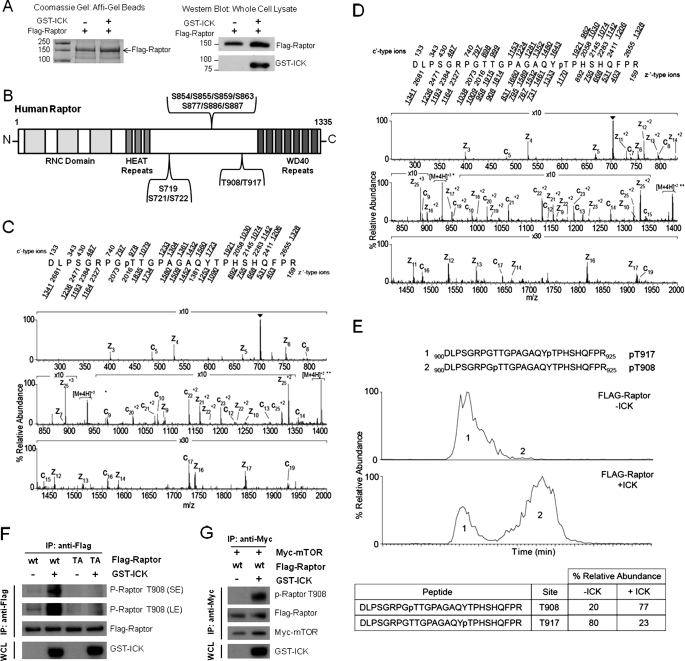
To determine whether Raptor Thr-908 can be phosphorylated in vivo by ICK, we conducted mass spectrometry analyses of FLAG-Raptor expressed in HEK293E cells with or without recombinant ICK (Fig. 5A). To enrich the sample with phospho-signals that are prone to quick turnover, we treated cells with Calyculin A, an inhibitor of the protein phosphatase 1/2A family of protein phosphatases, before harvesting and isolation of FLAG-Raptor for mass spectrometry analyses. Using this method, we identified a total of 12 phosphorylation sites on Raptor that form three clusters located in the region between the N-terminal HEAT repeats and the C-terminal WD repeats Ser-719/Ser-721/Ser-722 (cluster 1), Ser-854/Ser-855/Ser-859/Ser-863/Ser-877/Ser-886/Ser-887 (cluster 2), and Thr-908/Thr-917 (cluster 3) (Fig. 5B and supplemental table). Although the presence of phospho-sites within clusters 1 and 2 has been reported previously (40, 41, 43, 44, 48), this study reveals the presence of two novel phospho-sites, Thr-908 (Fig. 5C) and Thr-917 (D), within cluster 3. Coexpression with ICK significantly increased the percentage of mono-phosphorylated peptide containing Thr-908 as compared with that of mono-phosphorylated peptide containing Thr-917 (Fig. 5E), suggesting that ICK selectively targets Thr-908 in vivo.
FIGURE 5.
Phosphorylation of Raptor Thr-908 by ICK occurs in vivo. A, FLAG-Raptor was expressed alone or with GST-ICK in HEK293E cells. FLAG-Raptor was captured from the whole cell lysate and purified by anti-FLAG affinity gel. Affinity gel beads were extensively rinsed by lysis buffer followed by Tris buffer (50 mm Tris, 150 mm NaCl (pH 7.6)). Purified FLAG-Raptor on beads was shown on a Coomassie-stained SDS gel (left panel). A portion of the whole cell lysate was blotted with anti-FLAG and anti-GST antibodies to confirm the expression of FLAG-Raptor and GST-ICK, respectively. Bead samples were analyzed by mass spectrometry to determine the relative percentage of the unphosphorylated and phosphorylated signals. B, schematic illustration of the distribution of in vivo Raptor phosphorylation sites identified by mass spectrometry in this study (see details in supplemental table). C and D, The ETD MS/MS spectrum acquired in the ion trap of a front end electron transfer dissociation-enabled LTQ-FT on the [M+4H]4+ ions (m/z 704.0) of the tryptic peptide DLPSGRPGTTGPAGAQYTPHSHQFPR phosphorylated on Thr-908 and Thr-917, respectively. Predicted product ions of the c′- and z′·-types are listed above and below the peptide sequence, respectively. Singly charged ions are listed as monoisotopic masses, and doubly charged ions are listed as average masses. Fragment ions observed in the spectrum are underlined within the peptide sequence and are sufficient to localize the site of phosphorylation to Thr-908. Ions in the precursor isolation window are labeled with a triangle (▾). Brackets enclose ions that correspond to charge-reduced species and fragments derived from them by loss of small, neutral molecules. E, extracted ion chromatograms of monophosphorylated FLAG-Raptor peptide Asp-900-Arg-925 and the table displaying % relative abundances of species. The top chromatogram is FLAG-Raptor, and the bottom chromatogram is FLAG-Raptor coexpressed with ICK. The earlier eluting species (species 1) is phosphorylated on Thr-917, and the later-eluting species (species 2) is phosphorylated on Thr-908. F, FLAG-Raptor WT or T908A mutant (TA) was either expressed alone or coexpressed with GST-ICK in HEK293T cells. The anti-FLAG immunoprecipitates (IP) were blotted with anti-FLAG and anti-phospho-Thr-908 antibodies to assess the total FLAG-Raptor and the phospho-Thr-908 signals, respectively. G, FLAG-Raptor WT was coexpressed with Myc-mTOR in the absence or presence of GST-ICK in HEK293T cells. After anti-Myc immunoprecipitation, FLAG-Raptor associated with Myc-mTOR was assessed for the total FLAG-Raptor and the phospho-Thr-908 signals, respectively, as described in F.
To further confirm this result, we also assessed Raptor Thr-908 phosphorylation on FLAG-Raptor with and without coexpression with GST-ICK using an affinity-purified rabbit polyclonal phospho-Thr-908 antibody (Fig. 5F). In the absence of GST-ICK, very few phospho-Thr-908 signals were detected on the WT FLAG-Raptor. Strikingly, coexpression of GST-ICK was able to markedly enhance the phospho-Thr-908 signals on FLAG-Raptor WT. This significant increase in the phospho-Thr-908 reactivity was completely abolished by the Raptor T908A (TA) mutant, confirming the specificity of the phospho-Thr-908 signal. Similar results were obtained when Raptor associated with mTOR was analyzed for Thr-908 phosphorylation in the absence and presence of ICK coexpression in vivo (Fig. 5G), consistent with the results from our in vitro kinase assay experiment (Fig. 3), suggesting that ICK is able to phosphorylate Raptor Thr-908 when Raptor is associated with mTOR. Together, these results indicate that phosphorylation of Raptor Thr-908 occurs in vivo and can be markedly enhanced by ICK overexpression.
Phosphorylation of Raptor Thr-908 Is Able to Promote mTORC1 Activation without Altering Raptor Binding to mTOR or Its Substrate
Given the region between the N-terminal HEAT repeats and the C-terminal WD repeats of Raptor contains several mTOR activity-modifying phosphorylation sites, we sought to investigate whether Raptor Thr-908 plays an important role in regulating the activation of mTORC1. To determine the activity of mTORC1 in vivo upon insulin stimulation, we expressed recombinant mTOR, Raptor, and S6K1 proteins in HEK293E cells and assessed the phosphorylation status of two well known mTORC1 phosphorylation sites that are sensitive to insulin, S6K1 Thr-389 and 4EBP1 Ser-64. To minimize the interference of endogenous mTORC1, the mTOR rapamycin-resistant mutant S2035W, instead of the wild-type mTOR, was used so that the endogenous activity of mTORC1 can be suppressed by rapamycin treatment (shown in supplemental Fig. 1) before insulin stimulation. Because the Raptor S863A mutant has been reported to attenuate the activation of mTORC1 by insulin (42–44), it was thus included in our assay as a positive control. Similar to Raptor S863A, Raptor T908A also failed to increase the phosphorylation level of S6K1 Thr-389 to the same extent as that shown by the wild-type Raptor (Fig. 6A), which is particularly evident at later time points of insulin treatment (Fig. 6B, qualification data). Similarly, the Raptor mutant T908A was not able to elevate the level of phospho-4EBP1 Ser-64 signal to the same extent as that of the wild-type Raptor (Fig. 6B). Unfortunately, substitution of Glu for Thr at the 908 position was not able to mimic phosphorylation in that the Raptor T908E mutant yielded similar results as the Raptor T908A mutant when the mTORC1 activity was assessed (data not shown). These results, taken together, implicate a critical role for Raptor Thr-908 phosphorylation in the full activation of mTORC1 in response to insulin.
FIGURE 6.
ICK-mediated phosphorylation of Raptor Thr-908 is important for activation of mTORC1 by insulin or by Rheb overexpression. A, FLAG-Raptor of WT or mutants (S863A, T908A) was cotransfected with FLAG-mTOR (rapamycin-resistant mutant S2035W) and HA-S6K1 into HEK293E cells. Thirty-six hours after transfection, cells were serum-starved overnight, treated with 20 nm rapamycin for 20 min, and then stimulated with 100 nm insulin for various time points as indicated. From cell extracts, the expression levels of total proteins of S6K1, mTOR, and Raptor as well as the phospho-S6K1 Thr-389 signals were assessed by immunoblotting. B, FLAG-Raptor WT or the T908A mutant was coexpressed with recombinant mTOR and S6K1 in HEK293E cells. Cells were treated essentially the same as described in A. The activities of mTORC1 toward substrates S6K1 and 4EBP1 were assessed by immunoblotting with phospho-specific antibodies recognizing S6K1 phospho-Thr-389 and 4EBP1 phospho-Ser-64, respectively. SE, short exposure; LE, long exposure. The values of phospho-specific signals normalized by the total protein signals are shown. Similar results were obtained from three independent experiments. C, FLAG-Raptor WT or the T908A mutant, the rapamycin-resistant FLAG-mTOR, and HA-S6K1 were coexpressed in HEK293E cells in the absence or presence of FLAG-Rheb. Forty-eight hours after transfection, cells were treated with 20 nm rapamycin for 30 min followed by amino acid starvation for 60 min. The mTORC1 activity toward its substrate S6K1 was assessed by immunoblotting with the phospho-S6K1 Thr-389 antibody. SE, short exposure; LE, long exposure. The values of the phospho-specific signals normalized by the total S6K1 protein signals were shown.
Because overexpression of the GTP-binding protein RheB is capable of rescuing S6K1 from inactivation by amino acid withdrawal (49), we tested whether Raptor T908A mutation affects RheB-dependent activation of mTORC1 in response to amino acid starvation. Rapamycin-resistant mTOR, Raptor wild-type or the T908A mutant, and S6K1 were coexpressed in the absence or presence of RheB. After rapamycin pretreatment to suppress endogenous mTORC1 activity, cells were amino acid-starved. As reported previously (49), overexpression of RheB is able to rescue S6K1 from inactivation by amino acid depletion in cell culture medium (Fig. 6C). The Raptor T908A mutant, however, exhibited reduced capability of RheB-dependent rescue of S6K1 phosphorylation as compared with the wild-type Raptor (Fig. 6C), suggesting that Raptor Thr-908 phosphorylation is important for RheB-dependent activation of mTORC1 in response to amino acid starvation.
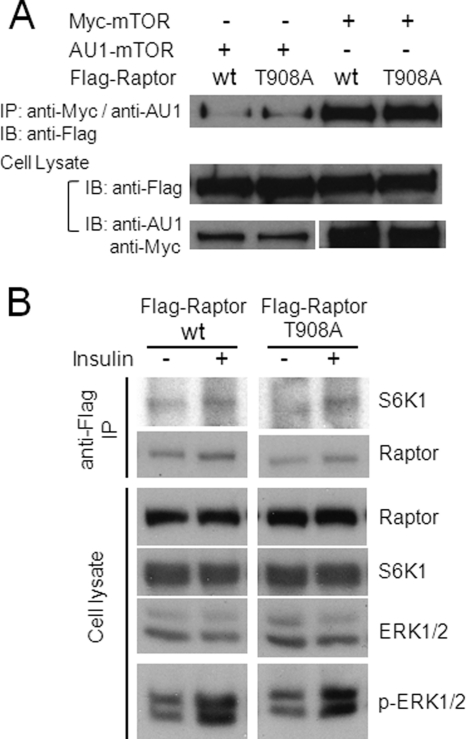
To investigate the possible causes for the biological effects of the Raptor T908A mutation, we assessed whether the T908A mutation alters the association of Raptor with mTOR or its substrates. We coexpressed recombinant mTOR with either Raptor WT or the Raptor T908A mutant in cells. After mTOR was pulled down by immunoprecipitation, the levels of Raptor WT and the Raptor T908A mutant associated with mTOR were analyzed by Western blot analysis. No significant change in the amount of Raptor bound to mTOR was detected between the wild type and the T908A mutant (Fig. 7A). In addition, we also tested whether the Raptor T908A mutant possesses a binding capacity to S6K1 that is very different from that of the wild-type Raptor. The Raptor T908A mutant bound as efficiently as the wild-type Raptor to the mTOR substrate S6K1 (Fig. 7B). Taken together, our results indicate that the Raptor T908A mutant does not appear to affect the binding capacity of Raptor to mTOR and S6K1.
FIGURE 7.
Raptor T908A mutation does not significantly alter binding of Raptor to mTOR and S6K1. A, HEK293E cells were cotransfected with Myc- or AU1-tagged mTOR and FLAG-tagged Raptor (WT or T908A), serum-starved, and stimulated with 100 nm insulin for 30 min. Myc-mTOR or AU1-mTOR was immunoprecipitated (IP) from the whole cell extract using anti-Myc or anti-AU1 antibodies, respectively. The level of FLAG-Raptor associated with either Myc-mTOR or AU1-mTOR immunoprecipitates was determined by immunoblotting (IB) with anti-FLAG. Total amounts of FLAG-Raptor, Myc-mTOR, and AU1-mTOR from the whole cell extract were indicated on Western blot analysis by using anti-FLAG, anti-Myc, and anti-AU1 antibodies, respectively. B, HEK293E cells were cotransfected with AU1-mTOR, FLAG-Raptor (WT or T908A), and HA-S6K1; serum-starved; and stimulated with 100 nm insulin for 30 min. FLAG-Raptor was immunoprecipitated from the whole cell extract by anti-FLAG, and the level of HA-S6K1 associated with the FLAG-Raptor immune complex was assessed on Western blot analysis using anti-HA. Total signal inputs of S6K1 and Raptor from the whole cell extract were also indicated on Western blot analysis. Insulin effect was indicated by the increase of phospho-ERK1/2 signals in the whole cell lysate.
DISCUSSION
In summary, we identified Raptor Thr-908 as an in vitro and in vivo phosphorylation site for ICK and demonstrated the importance of Raptor Thr-908 phosphorylation in promoting the activity of mTORC1. Our study thus provided a novel mechanistic insight into the regulatory mechanism by which ICK modulates the mTORC1 activity.
Emerging evidence from several previous studies have defined two clusters of phosphorylation sites on Raptor (40, 41, 43, 44, 48), several of which (Ser-863, Ser-719, Ser-721, and Ser-722) are critical for regulating the mTORC1 activity under different biological contexts. We hereby reported a cluster of two new in vivo phosphorylation sites (Thr-908 and Thr-917) on Raptor that is located in the same region as the other two groups. Among the two new sites, Thr-908 can be specifically targeted by ICK in vivo and is important for full activation of mTORC1. The biological significance and the protein kinase associated with phosphorylation of Thr-917 are still unknown.
In contrast to the strong signal of phospho-Ser-863, the signal of phospho-Thr-908 was weak on mass spectrometry, suggesting either a low stoichiometry of Thr-908 phosphorylation under steady-state conditions or that Thr-908 phosphorylation occurs only on a small fraction of Raptor because of specific subcellular compartmentalization. Furthermore, unlike Ser-863, phosphorylation of Thr-908 was not sensitive to insulin (data not shown), suggesting that the regulatory mechanism for the phosphorylation of Thr-908 is very different from that of Ser-863. This observation, however, is consistent with our finding that Thr-908 is a target for ICK because unpublished data from us (16) and others (2) have suggested that ICK is not activated by growth factors, including insulin.
Our findings here indicate that although Thr-908 phosphorylation is not sensitive to insulin treatment, it does exert a significant effect on the insulin-stimulated mTORC1 activity. An example demonstrating a similar regulatory paradigm comes from a recent study by Ekim et al. (50). Two phosphorylation sites, Ser-2159 and Thr-2164 within the mTOR kinase domain were identified, neither of which appears to be an mTOR autophosphorylation site. Both sites are of low stoichiometry and not sensitive to insulin treatment, yet they are important in promoting mTORC1 activity and signaling (50). Given that very little is known about how the activity of ICK is up-regulated, the regulatory mechanisms for Raptor Thr-908 phosphorylation catalyzed by ICK in vivo await further investigation.
Data from us and others have identified and placed the majority of the phosphorylation sites of Raptor in the linker region between the N-terminal Heat domain and the C-terminal WD domain. The study of the yeast TOR-Raptor complex by electron microscopy has revealed that TOR and Raptor are arranged in a head-to-toe, anti-parallel form. The structural model on the basis of such organization predicts that 1) the N-terminal RND domain of Raptor, which binds to the TOS motif of TORC1 substrates, is in close proximity to the C-terminal kinase domain of TOR; 2) the Heat domain, the linker region, and the WD40 domain of Raptor are predicted to be in direct contact with the Heat domain of TOR. Similar to Ser-863, mutation of Thr-908 did not appear to change the ability of raptor binding to either mTOR or its substrates. These results argue that the Raptor S863A and T908A mutations did not significantly disrupt the binding interface between Raptor and mTOR or mTOR substrates. Instead, these mutations may achieve their regulatory effects on the mTOR activity by inducing subtle conformational changes that affect the catalytic core of mTOR.
Our studies confirmed a previous report (47) where endogenous Raptor exists in protein complexes with and without endogenous mTOR. Because we were not able to detect endogenous phospho-Thr-908 signals either by mass spectrometry or by the phospho-Thr-908 specific antibody, the endogenous Raptor Thr-908 phosphorylation status in different protein complexes is not yet clear. However, our data have provided strong evidence suggesting that ICK is capable of targeting Thr-908 when Raptor is either with or without mTOR as a partner. In addition, once phosphorylated at Thr-908, Raptor can still associate with mTOR. Therefore, we speculate that the endogenous Thr-908 signal may exist in Raptor complexes with and without mTOR. In comparison, the catalysis of Raptor Ser-863 phosphorylation may also occur when Raptor is either with or without mTOR in complex, depending upon whether Raptor is targeted by mTOR (43, 44) or ERK1/2 (42) under a different cell stimulus.
Carriere et al. (42) reported that ERK1/2 can interact and phosphorylate Raptor in an mTOR-independent manner. Both ICK and ERK1/2 seem to be able to target Raptor in an mTOR-independent state but somehow affect the mTOR activity. One possible mechanism is that Raptor may be modified by phosphorylation through a variety of protein kinases such as ICK and ERK1/2 in the low Mr protein complexes and then is dynamically relocated to the high Mr protein complexes to associate with mTOR and affect the activity of mTORC1. This potential scenario could offer an opportunity for Raptor to be modified and regulated through insulin-PI3K-mTOR-independent pathways, such as Ras/MAPK for Raptor Ser-863 and ICK for Raptor Thr-908.
Although expression of the T908A mutant exhibited a significant compromise in the increase of mTOR substrate phosphorylation following insulin stimulation, the Thr-908 mutation by itself was not able to completely abolish the activation of mTORC1, suggesting that mutating additional phosphorylation sites are required for a more complete inactivation of mTORC1. This is in agreement with a previous report that multisite phosphorylation of Raptor was able to achieve a much stronger effect in promoting the activation of mTORC1 than any single site, such as Ser-863 (43). It remains a big challenge for future investigations to determine systematically how the complex phosphorylation events on Raptor are coordinated temporally and spatially to regulate the activity and the biological functions of mTORC1 in response to different environmental cues.
By identifying a novel phosphorylation site, Thr-908, on Raptor that plays an important role in regulating the activity of mTORC1, our study has provided further support to the current view that Raptor is a major target of multiplex phosphorylation events and phosphorylation of Raptor is an important mechanism to achieve the dynamic regulation of the mTORC1 activity and functions in response to diverse environmental cues. More importantly, by uncovering the biological functions of Raptor Thr-908 and its relationship with ICK, we have provided an unprecedented insight into the molecular and signaling mechanisms underlying the biological functions and regulations of ICK.
Supplementary Material
Acknowledgments
We thank Drs. David Sabatini and Diane Fingar for sharing reagents. We also thank Drs. Thomas W. Sturgill, Michael J. Weber, Steven M. Cohn, and Chi-Wing Chow for advice and encouragement.
This work was supported, in whole or in part, by National Institutes of Health Grants DK082614 (to Z. F.) and GM37537 (to D. F. H.).

This article contains supplemental Fig. 1 and table.
- MAK
- male germ cell-associated kinase
- ICK
- intestinal cell kinase
- Raptor
- regulatory associated protein of mTOR
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- S6K1
- ribosomal protein S6 protein kinase 1
- 4EBP1
- eukaryotic initiation factor 4E (eIF4E)- binding protein 1
- Rheb
- Ras homolog enriched in brain
- RSK
- p90 ribosomal S6 kinase
- CCRK
- cell cycle-related kinase
- HEAT
- Huntingtin, elongation factor 3 (EF3), protein phosphates 2A (PP2A), and the yeast P13-kinase TOR1.
REFERENCES
- 1. Matsushime H., Jinno A., Takagi N., Shibuya M. (1990) A novel mammalian protein kinase gene (mak) is highly expressed in testicular germ cells at and after meiosis. Mol. Cell. Biol. 10, 2261–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Togawa K., Yan Y. X., Inomoto T., Slaugenhaupt S., Rustgi A. K. (2000) Intestinal cell kinase (ICK) localizes to the crypt region and requires a dual phosphorylation site found in map kinases. J. Cell. Physiol. 183, 129–139 [DOI] [PubMed] [Google Scholar]
- 3. Abe S., Yagi T., Ishiyama S., Hiroe M., Marumo F., Ikawa Y. (1995) Molecular cloning of a novel serine/threonine kinase, MRK, possibly involved in cardiac development. Oncogene 11, 2187–2195 [PubMed] [Google Scholar]
- 4. Miyata Y., Akashi M., Nishida E. (1999) Molecular cloning and characterization of a novel member of the MAP kinase superfamily. Genes Cells 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 5. Fu Z., Schroeder M. J., Shabanowitz J., Kaldis P., Togawa K., Rustgi A. K., Hunt D. F., Sturgill T. W. (2005) Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol. Cell. Biol. 25, 6047–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyata Y., Nishida E. (1999) Distantly related cousins of MAP kinase. Biochemical properties and possible physiological functions. Biochem. Biophys. Res. Commun. 266, 291–295 [DOI] [PubMed] [Google Scholar]
- 7. Hinds T. D., Jr., Sánchez E. R. (2008) Protein phosphatase 5. Int. J. Biochem. Cell Biol. 40, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y., Wu C., Galaktionov K. (2004) p42, a novel cyclin-dependent kinase-activating kinase in mammalian cells. J. Biol. Chem. 279, 4507–4514 [DOI] [PubMed] [Google Scholar]
- 9. Fu Z., Larson K. A., Chitta R. K., Parker S. A., Turk B. E., Lawrence M. W., Kaldis P., Galaktionov K., Cohn S. M., Shabanowitz J., Hunt D. F., Sturgill T. W. (2006) Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol. Cell. Biol. 26, 8639–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wohlbold L., Larochelle S., Liao J. C., Livshits G., Singer J., Shokat K. M., Fisher R. P. (2006) The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle 5, 546–554 [DOI] [PubMed] [Google Scholar]
- 11. Shinkai Y., Satoh H., Takeda N., Fukuda M., Chiba E., Kato T., Kuramochi T., Araki Y. (2002) A testicular germ cell-associated serine-threonine kinase, MAK, is dispensable for sperm formation. Mol. Cell. Biol. 22, 3276–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Omori Y., Chaya T., Katoh K., Kajimura N., Sato S., Muraoka K., Ueno S., Koyasu T., Kondo M., Furukawa T. (2010) Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl. Acad. Sci. U.S.A. 107, 22671–22676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma A. H., Xia L., Desai S. J., Boucher D. L., Guan Y., Shih H. M., Shi X. B., deVere White R. W., Chen H. W., Tepper C. G., Kung H. J. (2006) Male germ cell-associated kinase, a male-specific kinase regulated by androgen, is a coactivator of androgen receptor in prostate cancer cells. Cancer Res. 66, 8439–8447 [DOI] [PubMed] [Google Scholar]
- 14. Xia L., Robinson D., Ma A. H., Chen H. C., Wu F., Qiu Y., Kung H. J. (2002) Identification of human male germ cell-associated kinase, a kinase transcriptionally activated by androgen in prostate cancer cells. J. Biol. Chem. 277, 35422–35433 [DOI] [PubMed] [Google Scholar]
- 15. Lahiry P., Wang J., Robinson J. F., Turowec J. P., Litchfield D. W., Lanktree M. B., Gloor G. B., Puffenberger E. G., Strauss K. A., Martens M. B., Ramsay D. A., Rupar C. A., Siu V., Hegele R. A. (2009) A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am. J. Hum. Genet. 84, 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Z., Kim J., Vidrich A., Sturgill T. W., Cohn S. M. (2009) Intestinal cell kinase, a MAP kinase-related kinase, regulates proliferation and G1 cell cycle progression of intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall M. N. (2008) mTOR. What does it do? Transplant. Proc. 40, S5–8 [DOI] [PubMed] [Google Scholar]
- 18. Bhaskar P. T., Hay N. (2007) The two TORCs and Akt. Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 19. Laplante M., Sabatini D. M. (2009) mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma X. M., Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 21. Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. (2004) mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24, 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hara K., Yonezawa K., Kozlowski M. T., Sugimoto T., Andrabi K., Weng Q. P., Kasuga M., Nishimoto I., Avruch J. (1997) Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 272, 26457–26463 [DOI] [PubMed] [Google Scholar]
- 23. Proud C. G. (2004) mTOR-mediated regulation of translation factors by amino acids. Biochem. Biophys. Res. Commun. 313, 429–436 [DOI] [PubMed] [Google Scholar]
- 24. Kim D. H., Sabatini D. M. (2004) Raptor and mTOR. Subunits of a nutrient-sensitive complex. Curr. Top. Microbiol. Immunol. 279, 259–270 [DOI] [PubMed] [Google Scholar]
- 25. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 26. Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 27. Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 28. Wang L., Harris T. E., Lawrence J. C., Jr. (2008) Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J. Biol. Chem. 283, 15619–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., Sabatini D. M. (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Two TOR complexes, only one of which is rapamycin-sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 31. Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. (2003) The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 [DOI] [PubMed] [Google Scholar]
- 32. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 33. Nascimento E. B., Ouwens D. M. (2009) PRAS40. Target or modulator of mTORC1 signalling and insulin action? Arch. Physiol. Biochem. 115, 163–175 [DOI] [PubMed] [Google Scholar]
- 34. Fonseca B. D., Smith E. M., Lee V. H., MacKintosh C., Proud C. G. (2007) PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J. Biol. Chem. 282, 24514–24524 [DOI] [PubMed] [Google Scholar]
- 35. Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanrahan J., Blenis J. (2006) Rheb activation of mTOR and S6K1 signaling. Methods Enzymol. 407, 542–555 [DOI] [PubMed] [Google Scholar]
- 38. Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 40. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 42. Carriere A., Romeo Y., Acosta-Jaquez H. A., Moreau J., Bonneil E., Thibault P., Fingar D. C., Roux P. P. (2011) ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 286, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Foster K. G., Acosta-Jaquez H. A., Romeo Y., Ekim B., Soliman G. A., Carriere A., Roux P. P., Ballif B. A., Fingar D. C. (2010) Regulation of mTOR complex 1 (mTORC1) by raptor Ser-863 and multisite phosphorylation. J. Biol. Chem. 285, 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang L., Lawrence J. C., Jr., Sturgill T. W., Harris T. E. (2009) Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J. Biol. Chem. 284, 14693–14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 46. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang L., Rhodes C. J., Lawrence J. C., Jr. (2006) Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J. Biol. Chem. 281, 24293–24303 [DOI] [PubMed] [Google Scholar]
- 48. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 49. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 50. Ekim B., Magnuson B., Acosta-Jaquez H. A., Keller J. A., Feener E. P., Fingar D. C. (2011) mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol. Cell. Biol. 31, 2787–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.