Background: Homocysteinylated heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) orchestrates a posttranscriptional RNA operon during folate deficiency.
Results: Folate deficiency induced homocysteinylated hnRNP-E1 to bind HPV16 RNA, reduced both viral capsid proteins, promoted HPV16 DNA integration into genomic DNA, and rapidly transformed HPV16-organotypic rafts implanted in immunodeficient mice to cancer.
Conclusion: A likely molecular link between folate nutrition and HPV16 is established.
Significance: Folate/vitamin-B12 deficiency can promote HPV16 DNA integration and carcinogenesis.
Keywords: Cancer Prevention, Folate, Homocysteine, Papillomavirus, RNA-binding Protein, RNA-Protein Interaction, Viral Protein, Posttranscriptional RNA Operon, hnRNP-E1/PCBP1, Viral Integration
Abstract
Although HPV16 transforms infected epithelial tissues to cancer in the presence of several co-factors, there is insufficient molecular evidence that poor nutrition has any such role. Because physiological folate deficiency led to the intracellular homocysteinylation of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) and activated a nutrition-sensitive (homocysteine-responsive) posttranscriptional RNA operon that included interaction with HPV16 L2 mRNA, we investigated the functional consequences of folate deficiency on HPV16 in immortalized HPV16-harboring human (BC-1-Ep/SL) keratinocytes and HPV16-organotypic rafts. Although homocysteinylated hnRNP-E1 interacted with HPV16 L2 mRNA cis-element, it also specifically bound another HPV16 57-nucleotide poly(U)-rich cis-element in the early polyadenylation element (upstream of L2̂L1 genes) with greater affinity. Together, these interactions led to a profound reduction of both L1 and L2 mRNA and proteins without effects on HPV16 E6 and E7 in vitro, and in cultured keratinocyte monolayers and HPV16-low folate-organotypic rafts developed in physiological low folate medium. In addition, HPV16-low folate-organotypic rafts contained fewer HPV16 viral particles, a similar HPV16 DNA viral load, and a much greater extent of integration of HPV16 DNA into genomic DNA when compared with HPV16-high folate-organotypic rafts. Subcutaneous implantation of 18-day old HPV16-low folate-organotypic rafts into folate-replete immunodeficient mice transformed this benign keratinocyte-derived raft tissue into an aggressive HPV16-induced cancer within 12 weeks. Collectively, these studies establish a likely molecular linkage between poor folate nutrition and HPV16 and predict that nutritional folate and/or vitamin-B12 deficiency, which are both common worldwide, will alter the natural history of HPV16 infections and also warrant serious consideration as reversible co-factors in oncogenic transformation of HPV16-infected tissues to cancer.
Introduction
Human papillomaviruses (HPVs),3 which infect suprabasal keratinocytes and persist in differentiating epithelial cells of cutaneous, mucosal, and genital tissues, are causally implicated in several cancers as well as venereal and other skin warts (1–4). The Centers for Disease Control and Prevention (www.cdc.gov/hpv/cancer.html) estimates that HPV causes almost all cervical cancer and links HPV to about 50% of vulvar cancers, 65% of vaginal cancers, 35% of penile cancers, 95% of anal cancer, and up to 60% of oropharyngeal cancers. The prevalence of latent HPV infection is ∼40% worldwide, and whereas 5–10% of infected women will develop squamous intraepithelial lesions, less than 1% will develop cervical cancer (5). Among the various oncogenic HPV types, HPV16 is the commonest agent that is responsible for an estimated one-half of all HPV-associated cancers worldwide. There are established co-factors that accelerate HPV-induced transformation of tissues to cancer. Among these co-factors (early age at first intercourse, multiple partners, smoking, oral contraceptives, and reduced immunity with HIV/AIDS), the role of poor folate nutrition has been suspected for 3 decades, but no molecular linkage with HPV has been identified. Nevertheless, because of limited non-surgical options for eradication of established HPV infection in cervical and other epithelial tissues, identification of any reversible cofactor (such as a nutritional deficiency) that can modulate the expression of HPV is eminently worthy of study.
Following epithelial cell division, HPV-infected daughter cells migrate superficially from the basal region and begin differentiation. Upon terminal differentiation in the granulosa and cornified layers of infected epithelia, vegetative viral DNA replication or amplification is induced followed by activation of viral late gene expression, encoding HPV viral capsid proteins L1 and L2 at a 20:1 ratio, respectively, with subsequent assembly and encapsidation of HPV DNA into infectious HPV virions (3, 6). Although a comprehensive understanding of the precise molecular basis for the expression of L2 and L1 in differentiating cells is still incomplete, it has been known for over a decade that cellular hnRNP-E1 can interact with the 3′-coding region of HPV16 L2 mRNA (7). Furthermore, by engineering HeLa cells to express L2, Collier et al. (7) showed that transfection of hnRNP-E1 reduces expression of L2. However, it not clear if this interaction involves direct reduction of L2 mRNA translation and/or reduced stability of the mRNA-protein complex or another mechanism and if this occurs in cultured keratinocytes that can actively express all HPV genes. Moreover, the pathophysiologic context wherein hnRNP-E1 interacts with HPV16 RNA has also remained obscure.
Earlier we had determined that up-regulation of folate receptors in cervical cancer cells during folate deficiency involves the binding of hnRNP-E1 to an 18-base cis-element in the 5′-untranslated region of folate receptor-α mRNA, which triggers an increase in folate receptor biosynthesis at the translational level (8, 9). Because this RNA-protein interaction was stimulated by homocysteine, which accumulates intracellularly during folate deficiency, this work established a link between perturbed folate metabolism and coordinated translational regulation of folate receptors (10). Using purified components, we recently determined that the molecular mechanism of this posttranscriptional up-regulation of folate receptors during folate deficiency involves a concentration-dependent homocysteinylation of various cysteine residues within the (mRNA-binding) K-homology domains of hnRNP-E1, which probably leads to the sequential disruption of critical cysteine-S-S-cysteine bonds by the formation of multiple homocysteine-S-S-cysteine mixed disulfide bonds in hnRNP-E1 (11). This leads to a gradual unmasking of an underlying RNA-binding pocket in homocysteinylated hnRNP-E1 that progressively increases its affinity for folate receptor-α mRNA cis-element preparatory to folate receptor up-regulation. These data incriminated hnRNP-E1 as a physiologically relevant and sensitive candidate sensor of folate deficiency within cells (11), and because diverse mRNAs (including rabbit 15-lipoxygenase, murine tyrosine hydroxylase and intermediate neurofilament-middle molecular mass, and HPV16 L2 mRNA) also interacted with this RNA-binding domain (7, 8), we proposed that homocysteinylated hnRNP-E1 was well positioned to orchestrate a nutrition-sensitive (homocysteine-responsive) posttranscriptional RNA operon in folate-deficient cells.
Because this research had uncovered unexpected connections between folate deficiency, homocysteine, hnRNP-E1, and HPV16 (7, 10, 11), we determined the functional physiological consequence of folate deficiency to HPV16 viral capsid protein expression using a model of HPV16-harboring human keratinocytes ((HPV16)BC-1-Ep/SL cells) that expressed the full complement of HPV16 RNA when propagated as monolayers. Further extension of these studies to HPV16-organotypic rafts and additional animal studies suggests that folate deficiency is a likely (reversible) co-factor in HPV16-induced malignancies.
EXPERIMENTAL PROCEDURES
Adaptation of (HPV16)BC-1-Ep/SL Keratinocytes to Growth in High Folate and Low Folate F Medium in Absence of Feeder Layers
BC-1-Ep/SL cells were obtained from the American Type Culture Collection (Manassas, VA). BC-1-Ep/SL cells that were stably transfected with HPV16 (12) (referred to as (HPV16)BC-1-Ep/SL cells) were a generous gift from Professor Paul Lambert (University of Wisconsin). These cells grow in the presence of NIH 3T3 cell feeder layers in F-medium that was composed of 1 part of DMEM and 3 parts of Ham's F-12 medium (HyQ DME/high glucose, and HyQ Ham's F-12, respectively) (HyClone, Logan, UT), and supplemented with the following components: 5% fetal bovine serum (FBS), adenine (24 μg/ml), cholera toxin (8.4 ng/ml), epidermal growth factor (10 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml). The cells were first adapted to stable growth in the absence of feeder layers in high folate F-medium (F-HF) that contained a final folate concentration of 4.5 μm pteroylglutamic acid plus 6.8 nm 5-methyltetrahydrofolate; these cells are referred to hereafter as (HPV16)BC-1-Ep/SL-HF cells. Another aliquot of these cells were adapted stepwise over several weeks to stable growth in low folate F-medium (F-LF) which was similar in all other respects to supplemented F-HF except that it contained a final physiological low folate concentration of 6.8 nm 5-methyltetrahydrofolate that was contributed from 5% FBS; these cells are referred to hereafter as (HPV16)BC-1-Ep/SL-LF cells. There was no significant difference in doubling time between (HPV16)BC-1-Ep/SL cells stably propagated in either F-HF (29.5 h) or F-LF (30 h).
Effect of Homocysteine on Interaction of HPV16 57-nucleotide poly(U)-rich cis-Element with Purified Recombinant GST-hnRNP-E1
The HPV16 57-nucleotide cis-element (13) in the early polyadenylation element of the HPV16 genome was first cloned into the pSPT19 vector with EcoRI on the 5′-end and HindIII on the 3′-end. The two oligonucleotides (5′-AAT TCT TTT TTC TTT TTT ATT TTC ATA TAT AAT TTT TTT TTT TGT TTG TTT GTT TGT TTT TTG-3′ and 5′-AGC TTA AAA AAC AAA CAA ACA AAC AAA AAA AAA AAT TAT ATA TGA AAA TAA AAA AGA AAA AAA-3′) were annealed and subcloned into the pSPT19 vector that was linearized with EcoRI and HindIII. The new plasmid pYS57 was verified by restriction enzyme digestion. To prepare 32P-labeled HPV16 57-nucleotide poly(U)-rich cis-element, pYS57 was linearized by HindIII, and then [α-32P]UTP was included in the transcription reaction using the SP6/T7 Transcription Kit (Roche Applied Science), followed by purifying RNA transcripts with NucTrap Probe Purification Columns and a Push Column Beta Shield Device (Stratagene). RNA-protein binding assays were carried out using 1 μg of dialyzed, purified recombinant GST-hnRNP-E1 (11) and 1 × 105 cpm of [32P]HPV16 57-nucleotide poly(U)-rich cis-element in binding buffer containing increasing concentrations of l-homocysteine (0–100 μm) in a final volume of 15 μl. After incubation at 4 °C for 30 min, 1 μl of heparin solution (100 mg/ml) was added, and incubation continued for 15 min. Electrophoresis of RNA-protein complexes was carried out using 6% native PAGE (60:1), and dried gels were autoradiographed overnight.
Comparison of Dissociation Constant (KD) of RNA-Protein Interactions
[35S]HPV16 L2 RNA or [35S]HPV16 57-nucleotide RNA cis-element RNA fragments were prepared using the SP6 transcription kit (Roche Applied Science) in the presence of 250 μCi of [α-35S]CTP or [α-35S]TTP and 5 μg of pSPT3′L2/EcoRI or 5 μg of pYS57/HindIII. After transcription, template DNAs were digested with DNase I (RNase-free), and RNA transcripts were purified with Quick-Spin G-25 columns. Binding of the [35S]HPV16 L2 RNA or [35S]HPV16 57-nucleotide RNA cis-element to purified recombinant GST-hnRNP-E1 at 25 μm l-homocysteine was then tested. Each level was measured in triplicate. Briefly, except for the addition of l-homocysteine, all KD experiments were carried out in the absence of added reducing agents. The binding assays between 0.1 μg of purified recombinant GST-hnRNP-E1 and [35S]HPV16 L2 RNA or [35S]HPV16 57-nucleotide RNA cis-element (0.02, 0.05, 0.13, 0.32, 0.8, 2, 5, and 12.5 nm) were carried out in a final volume of 500 μl of binding buffer with 25 μm l-homocysteine at 4 °C for 0.5 h. Bovine serum albumin (BSA) (0.1 μg) was used as a control in place of hnRNP-E1 for background determination. The mixture was then filtered through a Microcon YM-30 column by centrifugation at 12,000 × g followed by three consecutive washes, each with 500 μl of binding buffer. The retentate containing RNA-protein complexes (150 μl) was counted in a β-scintillation counter. Counts from samples containing hnRNP-E1 were recorded as total binding, whereas counts from samples containing BSA reflected nonspecific binding. The specific binding for each concentration of radioligand was determined by subtracting the nonspecific binding from the results of total binding. The KD value was calculated from a Scatchard plot (14) using GraphPad Prism 4 from GraphPad Software (San Diego, CA).
Effect of RNA Interference of hnRNP-E1 mRNA on HPV16 L1 and L2 mRNA
Before the day of siRNA transfection, (HPV16)BC-1-Ep/SL-LF cells were trypsinized, counted, and plated in 6-well plates at 1.4 × 105 cells/well in 2.5 ml of F-LF medium, so that cells were 35–45% confluent after overnight culture. Cells were transfected with either 10 nm predesigned Stealth RNA (siRNA-hnRNP-E1/PCBP1) (15) or 10 nm scrambled negative stealth RNAi control (Invitrogen) using Lipofectamine RNAiMAX transfection reagent, as described (11). Cells were trypsinized and harvested at 1, 2, and 3 days after transfection for qRT-PCR analysis of hnRNP-E1, hnRNP-E2, and HPV L1, L2, E6, and E7 mRNA levels. In addition, the rate of biosynthesis of newly synthesized hnRNP-E1 was also determined in transfected cells (11).
HPV16-Organotypic Raft Cultures
Before development of HPV16-organotypic raft cultures, (HPV16)BC-1-Ep/SL cells were adapted to either high folate F-medium (F-HF) or physiologically low folate F-medium (F-LF) containing 10% FBS, which contributed 13.6 nm 5-methyltetrahydrofolate to the F-LF medium, for over 25 weeks. NIH 3T3 cells were likewise adapted to modified F-HF or F-LF medium for over 10 weeks to ensure stable intracellular folate levels (10). Before being placed on the raft, (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells were cultured on mitomycin C-treated F-HF or F-LF NIH3T3 feeder layers, respectively.
To construct HPV16-organotypic rafts, rat tail type 1 collagen (3.71 mg/ml) (BD Biosciences) was used to coat BD BioCoat inserts (6-well plates; 3-μm pore size). The remaining collagen was impregnated with either F-HF- or F-LF-adapted NIH3T3 (4.5 × 105 cells/ml) and plated on collagen-coated inserts, following which the collagen was allowed to contract in 10% FBS-containing F-HF or F-LF medium, respectively, for 5 days at 37 °C in a continuous CO2 (5%) incubator. Then 50 μl of 1.4 × 106 cells/ml (7 × 104 cells) in keratinocyte-plating medium (consisting of F-HF or F-LF medium containing 0.5% FBS, insulin (5 mg/ml), cholera toxin (8.4 ng/ml), adenine (24 mg/ml), and hydrocortisone (0.4 mg/ml)) were plated onto the collagen rafts. Four days after plating keratinocytes, the rafts were placed on two 1-inch2 cotton pads (VWR Scientific, Media, PA) in a BD BioCoat Deep Well 6-well plate (BD Biosciences Labware) to lift to the air-liquid interface. The rafts were fed from below the insert with cornification medium (consisting of F-HF or F-LF medium (containing 1.88 mm Ca2+) supplemented with 10% FBS, insulin (5 mg/ml), cholera toxin (8.4 ng/ml), adenine (24 mg/ml), and hydrocortisone (0.4 mg/ml)) every other day. Fourteen days after being lifted to the air-liquid interface, one part of the rafts was fixed in 4% formalin, embedded in 2% agar in 1% formalin followed by paraffin and cut into 4-μm-thick cross-sections; other parts of the rafts were used for RNA and DNA isolation or electron microscopic analysis.
Quantitative PCR (qPCR) for HPV16 Viral Load and Integration
To prepare HPV16-organotypic raft total DNA for analysis of the HPV16 viral load by qPCR, four 18-day-old HPV16-high folate-organotypic rafts as well as four HPV16-low folate-organotypic rafts were separately ground in 5 ml of Dulbecco's PBS with autoclaved sea sand (EMD Chemicals Inc.) using a mortar and pestle (Fisher). The resulting paste was clarified by centrifugation for 10 min at 4000 rpm in a refrigerated Beckman GPR centrifuge. After discarding the pellets, raft total DNA (including genomic and viral DNA) was extracted from the suspension using the NucleoSpin® blood kit (Macherey-Nagel GmbH & Co.). Briefly, the supernatant was incubated at 70 °C for 15 min in lysis buffer containing proteinase K, and after binding to the silica column and washing, the DNA was eluted in 100 μl of elution buffer. The DNA was stored at −20 °C until used for qPCR.
DNA copy numbers of HPV16 E6, E7, and E2 and GAPDH in 50 ng of raft total DNA were measured by qPCR in triplicate using the Platinum Quantitative PCR SuperMix-UDG kit (Invitrogen) and the ABI 7900 HT detection system (Applied Biosystems). Sequence-specific primers for GAPDH and HPV16 E6 and E7 were designed using the Invitrogen customer primer design online system; these sequences are shown in the supplemental data (under “Quantitative RT-PCR (qRT-PCR), Primers, and Validation”). The oligonucleotide primers used in the measurement of HPV16 DNA integration into genomic DNA were obtained from Integrated DNA Technologies (Coralville, IA). The assay for integration evaluated the ratio between HPV16 E2 DNA and HPV16 E6 DNA (16). An altered E2/E6 ratio is an established parameter that reflects integration into cellular DNA. The principle is that a unique region of the E2 open reading frame is most often deleted during HPV16 integration. This is targeted by one set of PCR primers and a probe, and another set targets the E6 open reading frame. In episomal form, both targets should be equivalent, whereas in integrated forms, the copy numbers of E2 would be less than those of E6. The primers for E2 were as follows: probe, 5′-6-FAM/CAC CCC GCC/ZEN/GCG ACC CAT A/3IABkFQ/-3′; forward primer, 5′-AAC GAA GTA TCC TCT CCT GAA ATT ATT AG-3′; and reverse primer, 5′-CCA AGG CGA CGG CTT TG-3′ (where 6-FAM represents 6-carboxyfluorescein; ZEN, an internal quencher 9 bases from the 5′ fluorophore, is an undisclosed proprietary agent; and IABkFQ is Dark Quencher Iowa Black® FQ). The primers for E6 were as follows: probe, 5′-6-FAM/CAG GAG CGA/ZEN/CCC AGA AAG TTA CCA CAG T/3IABkFQ/-3′; forward primer, 5′-GAG AAC TGC AAT GTT TCA GGA CC-3′; and reverse primer, 5′-TGT ATA GTT GTT TGC AGC TCT GTG C-3′. The probe/primer ratio in these experiments was 1:2.
Standard curves of HPV16 E6, E7, and E2 were generated for measurement of viral DNA copies, and standard curves of GAPDH were generated for normalizing the input DNA content. Briefly, standard curves were generated using 10-fold dilutions of HPV16 plasmid or amplified GAPDH DNA (from 101 to 107 copies). Amplifications were carried out using the ABI HT7900 sequence detection system; the cycle conditions for qPCR were 50 °C for 2 min and 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The actual copy numbers of HPV E6, E7, and E2 and GAPDH were determined by qPCR using 50 ng of raft total DNA and were then normalized by the copy numbers of GAPDH.
The viral load was calculated by dividing the normalized copy number of HPV16 E6 DNA by 50 ng of raft total DNA and expressed as copies of HPV16 E6 DNA per ng of raft total DNA, according to the formula by Carcopino et al. (17), with the exception that we used the “ng of raft total DNA” in place of “cells” in that formula. The amount of integrated HPV16 E6 DNA was calculated by subtracting the copy number of HPV16 E2 episomal DNA from the total copy number of HPV16 E6 DNA (episomal and integrated). The percentage of HPV16 DNA that was integrated into genomic DNA was then determined by dividing the integrated HPV16 E6 DNA copy number by total HPV16 E6 DNA copy number, and the result was multiplied by 100.
Model for Implantation of HPV16-organotypic Rafts in Mice
All animal studies were approved by the Institutional Animal Care and Use Committee of Indiana University-Purdue University at Indianapolis. Four female mice from each of three strains (supplemental Table S1), beige nude XID (Hsd:NIHS-Lystbg Foxn1nuBtkxid), athymic nude (Hsd:athymic nude-Foxn1nu), and SHrNTM SCID (NOD.Cg-prkdcscidHrhr/NCrHsd), 4–6 weeks old and weighing 16–18 g, were obtained from Harlan Sprague-Dawley Laboratories (Indianapolis, IN) and acclimatized for 1 week at the Laboratory Animal Resource Center at the Indiana University School of Medicine.
Previous methods used to implant fragments of HPV-infected human foreskins under the skin of mice (18, 19) were modified for implantation of either HPV16-high folate- or HPV16-low folate-organotypic rafts in all three species of immunodeficient mice. Briefly, 18-day-old HPV16-high folate-organotypic rafts and HPV16-low folate-organotypic rafts were washed with sterile Dulbecco's PBS, cut into 5 × 5 × 1-mm pieces, and transported in Petri dishes at 4 °C to the animal facility in either serum-free F-HF or F-LF medium, respectively. All mice remained deeply anesthetized before and throughout the surgical procedure. Under sterile conditions, a 1-cm midflank skin incision was made perpendicular to the spine below the costophrenic angle with scissors, and a subcutaneous pocket was created using a blunt forceps. A fragment from either an HPV16-high folate- or an HPV16-low folate-organotypic raft was then inserted subcutaneously and repositioned in a top-to-bottom orientation to facilitate angiogenesis of the raft from below, and the incision was closed with a surgical clip. After postoperative recovery from anesthesia, the mouse was transferred to a regular cage. The surgical clips were removed 1 week after surgery without anesthesia. The mice were then observed twice a week for tumor growth. Once tumors were established, measurements were taken on a regular basis (18, 19). Quantification of tumor growth was made using the formula described recently (20).
Following the transformation of an HPV16-low folate-organotypic raft into an aggressive tumor in a beige nude XID mouse, fragments (3 × 3 × 2 mm) from this primary tumor were transplanted into other immunodeficient mice. Following growth of these secondary tumors, additional immunodeficient mice were transplanted with fragments from these secondary tumors, and the subsequent growth of tertiary tumors was also monitored.
See supplemental data for details on preparation of unlabeled l-homocysteine and purified recombinant glutathione S-transferase (GST)-hnRNP-E1 fusion protein; determination of whether the RNA-protein interaction between the 3′-coding region of L2 mRNA and hnRNP-E1 is responsive to homocysteine; in vitro transcription-translation of HPV16 L2 mRNA; determination of whether HPV16 L2 single-stranded DNA binds to hnRNP-E1 in the presence of various concentrations of homocysteine; qRT-PCR, primers, and validation; measurement of intracellular homocysteine in (HPV16)BC-1-Ep/SL cells; transient transfection of wild-type hnRNP-E1 into (HPV16)BC-1-Ep/SL cells; determination of whether HPV16 57-nucleotide poly(T)-rich single-stranded DNA binds hnRNP-E1; interaction of endogenous homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide poly(U)-rich cis-element within cells and effects on downstream CAT reporter signal; specific binding between hnRNP-E1 and various mutations of HPV16 57-nucleotide RNA cis-element; effects of l-homocysteine on various CAT reporter constructs transfected into (HPV16)BC-1-Ep/SL-HF cells; analysis of the rates of biosynthesis and degradation of HPV16 L2 and L1 mRNA transcripts in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells by qRT-PCR; analysis of the rate of protein biosynthesis and degradation of HPV16 L2, L1, hnRNP-E1, and GAPDH in (HPV16)BC-1-Ep/SL-HF cells or (HPV16)BC-1-Ep/SL-LF cells; source of antibodies; and tissue histology, immunohistochemistry, fluorescence microscopy, Western blot analysis, and electron microscopy.
RESULTS
Homocysteine Responsiveness of Interaction of HPV16 L2 mRNA and Purified Recombinant GST-hnRNP-E1 in Vitro
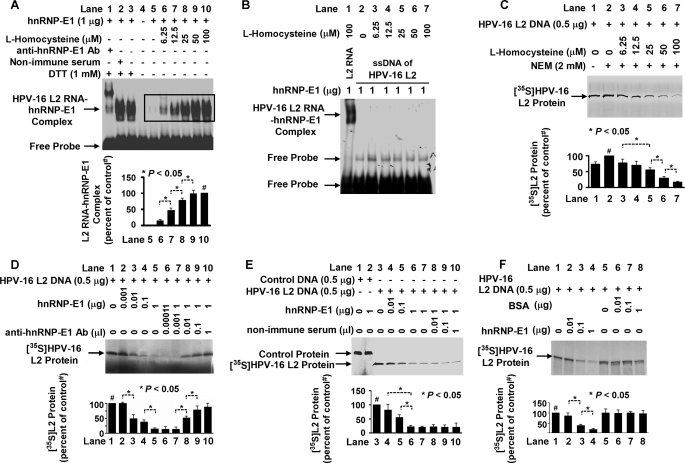
Because incubation of l-homocysteine with purified recombinant GST-hnRNP-E1 led to a covalent interaction with the protein (11), we tested the effect of homocysteinylated hnRNP-E1 in binding to HPV16 L2 mRNA and in altering its translation in vitro. Fig. 1A demonstrates a dose-dependent increase in generation of HPV16 L2 RNA-bound GST-hnRNP-E1 protein complexes with increasing physiologically relevant concentrations of l-homocysteine that achieved saturability and that exhibited a supershift only with specific anti-hnRNP-E1 antiserum (9) on gel shift assays. Because HPV16 L2 single-stranded sense DNA did not interact with l-homocysteine-derivatized GST-hnRNP-E1 (Fig. 1B), the locus for interaction between homocysteinylated hnRNP-E1 and HPV16 L2 was at the level of an RNA-protein interaction. As noted earlier for the binding interaction of hnRNP-E1 and folate receptor-α mRNA cis-element, there was a progressive increase in binding affinity between purified recombinant GST-hnRNP-E1 and HPV16 L2 RNA cis-element as the concentration of l-homocysteine increased in the reaction mixture; thus, there was a reduction in KD from basal values of 1.6–0.5 nm at 50 μm l-homocysteine.
FIGURE 1.
Characterization of RNA-protein interactions involving purified recombinant GST-hnRNP-E1 protein and HPV16 L2 RNA cis-element in the presence of increasing concentrations of l-homocysteine, leading to the translation of HPV16 L2 protein in vitro. A, gel shift analysis of the interaction of HPV16 L2 RNA cis-element (1 × 105 cpm) and GST-hnRNP-E1 protein in the absence or presence of various concentrations of l-homocysteine and the influence of nonimmune or anti-hnRNP-E1 antiserum on RNA-protein complex formation using 6% native PAGE and autoradiography. The pooled densitometric scanned data of RNA-protein complexes formed with increasing concentrations of l-homocysteine from three independent gel shift experiments are shown as a bar graph below one representative gel; these data are presented as the mean ± S.D. (error bars). The scanned area reflecting the L2 RNA-hnRNP-E1 protein signals formed with increasing concentrations of l-homocysteine is marked by a rectangle in the gel from lanes 5–10 and compared with the signal formed in the presence of 100 μm l-homocysteine in lane 10 (# signifies the 100% value). B, interaction between the single-stranded sense DNA (ssDNA) of HPV16 L2 and homocysteinylated hnRNP-E1. Lane 1 contains a positive control using HPV16 L2 RNA. C–F, in vitro translation of [35S]HPV16 L2 protein under various experimental conditions. In each of the following panels, the pooled densitometric scanned data of [35S]HPV16 L2 protein synthesized from three independent experiments are shown as a bar graph below one representative gel; these data are presented as the mean ± S.D. # in C, D, E, and F signifies the 100% value. C, effect of the addition of physiological concentrations of l-homocysteine on the biosynthesis of HPV16 L2 protein during in vitro translation. NEM, N-ethyl maleimide. D, effect of the addition of purified recombinant GST-hnRNP-E1 during in vitro biosynthesis of HPV16 L2 (lanes 2–5) and effect of increasing concentrations of anti-hnRNP-E1 antiserum (anti-hnRNP-E1 Ab) in quenching the inhibitory effect of GST-hnRNP-E1 on HPV16 L2. E, effect of increasing concentrations of nonimmune serum on the translation of HPV16 L2 in the presence of purified recombinant GST-hnNP-E1. F, comparison of the effect of the addition of purified recombinant GST-hnRNP-E1 versus BSA on HPV16 L2 protein synthesis.
Endogenous hnRNP-E1 is found in small quantities in the reticulocyte lysate of the translation reaction mixture (9, 11), so after quenching the excess (4.1 mm) β-mercaptoethanol extant in the commercial translation kit by 2 mm N-ethyl maleimide, the addition of increasing physiologically relevant concentrations of l-homocysteine led to a dose-dependent quenching of HPV16 L2 protein synthesis (Fig. 1C, lanes 3–7); these inhibitory effects were not due to premature degradation of HPV16 L2 mRNA during in vitro translation (supplemental Fig. S2). In addition, the in vitro translation of HPV16 L2 was mediated by hnRNP-E1 because there was a dose-dependent reduction in [35S]HPV16 L2 protein (Fig. 1D, lanes 2–5) upon the addition of increasing concentration of purified recombinant GST-hnRNP-E1, which was reversed in a dose-dependent manner by the addition of increasing concentrations of anti-hnRNP-E1 antiserum to the reaction mixture (Fig. 1D, lanes 6–10). By contrast, increasing concentrations of non-immune serum (Fig. 1E, lanes 8–10) had no effect in reversing the inhibitory effect of purified recombinant GST-hnRNP-E1 on HPV16 L2 synthesis. Moreover, an unrelated protein like bovine serum albumin (Fig. 1F, lanes 6–8) had no such effect in reducing the translation of [35S]HPV16 L2 protein in vivo. Collectively, these in vitro translation studies suggested that the enhanced binding between the cis-element in the 3′ coding region of HPV16 L2 mRNA and hnRNP-E1 that was induced by l-homocysteine (Fig. 1C) led to the inhibition of HPV16 L2 viral capsid protein synthesis.
Determination of Thiol Content of (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF Cells and Effects on RNA-Protein Interaction
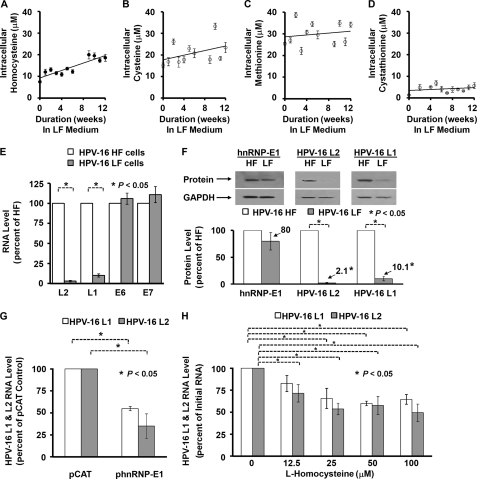
(HPV16)BC-1-Ep/SL cells propagated long term in F-HF and F-LF media had comparable cell doubling times of 29.5 and 30 h, respectively. Simultaneous measurements of various intracellular thiols in (HPV16)BC-1-Ep/SL-HF cells that were shifted to F-LF medium over 12 weeks confirmed a progressive increase in only homocysteine to nearly 3 times more than base line, from 7 to ∼20 μm by 12 weeks (Fig. 2, A–D). This suggested that the low folate environment, which led to the accumulation of more homocysteine, could influence the interaction of HPV16 L2 RNA cis-element and endogenous hnRNP-E1. Because (HPV16)BC-1-Ep/SL-HF expressed the full complement of HPV16 RNA (supplemental Fig. S1, discussed below), we compared the expression of HPV16 RNA between (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells.
FIGURE 2.
Determination of the extent of accumulation of intracellular thiols in (HPV16)BC-1-Ep/SL-HF cells as a function of time following transfer to low folate medium (A–D) and comparison of the expression of HPV16 RNA and protein in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells under various experimental conditions (E–H). Unless otherwise specified, the results are presented as the mean ± S.D. (error bars) from three independent sets of experiments (n = 3) with each data point performed in triplicate. A–D, concentration of intracellular homocysteine, cysteine, methionine, and cystathionine in (HPV16)BC-1-Ep/SL-HF cells transferred to low folate (F-LF) medium over 12 weeks. Due to prohibitive costs, this longitudinal experiment was carried out once; the results from each data point were derived from the mean of three samples. The curve-fitting analyses were determined by linear regression. E–H, comparison of expression of HPV16 RNA and proteins (L1, L2, E6, and E7) in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells (HPV16 HF and HPV16 LF cells, respectively) using qRT-PCR (E), Western blot analysis (F), following transfection of hnRNP-E1 (using a plasmid, phnRNP-E1) in (HPV16)BC-1-Ep/SL-HF cells or a pCAT (control) plasmid (G), and the effect of increasing concentrations of extracellular l-homocysteine on HPV16 L1 and L2 RNA levels in (HPV16)BC-1-Ep/SL-HF cells (H).
Measurement of Various HPV16 RNA in (HPV16)BC-1-Ep/SL-HF Cells
In order to validate the qRT-PCR method that was used to measure small quantities of HPV16 RNA in (HPV16)BC-1-Ep/SL-HF cells, we determined individual standard curves between target RNA (HPV16 L2, L1, E6, and E7 as well as human folate receptor-α, hnRNP-E1, and reference RNA (GAPDH), which demonstrated comparable high efficiency of amplification of the target RNA and the reference RNA. Supplemental Fig. S1 demonstrates qRT-PCR profiles with the standard curve of HPV16 L1 and GAPDH (supplemental Fig. S1, A and B) as well as HPV16 L2 and GAPDH (supplemental Fig. S1, C and D). Both uninfected BC-1-Ep/SL (supplemental Fig. S1E) and (HPV16)BC-1-Ep/SL-HF cells (supplemental Fig. S1F) expressed folate receptor-α and hnRNP-E1; however, only (HPV16)BC-1-Ep/SL-HF cells expressed both HPV16 L1 and L2 as well as HPV16 E6 and E7 RNA. Thus, qRT-PCR easily discriminated between cells that did and did not contain the HPV16 genome. Next, the RNA expression of HPV16 L2, L1, E6, and E7 in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells was compared.
Fig. 2E demonstrates a much lower amount of HPV16 L2 RNA in (HPV16)BC-1-Ep/SL-LF than (HPV16)BC-1-Ep/SL-HF cells, with no difference in the amplification plots of GAPDH; surprisingly, however, comparable data were also obtained with HPV16 L1 RNA (Fig. 2E). Thus, when compared with control (HPV16)BC-1-Ep/SL-HF cells, the RNA expression of both HPV16 L2 and L1 in (HPV16)BC-1-Ep/SL-LF cells was significantly decreased (97 and 90%, respectively) (Fig. 2E). By contrast, analysis of HPV16 E6 and E7 RNA did not show significant differences in expression in (HPV16)BC-1-Ep/SL-LF and (HPV16)BC-1-Ep/SL-HF cells. Fig. 2F also confirmed a greater inhibition in the expression of L2 and L1 proteins (98 and 90%, respectively) in (HPV16)BC-1-Ep/SL-LF cells when compared with (HPV16)BC-1-Ep/SL-HF cells. The observed reduction of HPV16 L2 and L1 was not due to up-regulation of hnRNP-E1 in (HPV16)BC-1-Ep/SL-LF cells because hnRNP-E1 protein expression was only 80% of that found in high folate cells (Fig. 2F). Nevertheless, hnRNP-E1 was incriminated in reducing HPV16 L1 and L2 mRNA expression because (HPV16)BC-1-Ep/SL-HF cells that were transiently transfected with a plasmid containing hnRNP-E1 exhibited ∼65% inhibition of expression of HPV16 L2 RNA and ∼45% inhibition of expression of HPV16 L1 RNA when compared with values obtained with pCAT (control) plasmid (Fig. 2G). However, because there was no interaction between hnRNP-E1 and single-stranded HPV16 L2 DNA (Fig. 1B), this suggested that the reduction in HPV16 L2 RNA (and HPV16 L1 RNA), following either stable propagation of cells in low folate medium (Fig. 2E) or following transfection of hnRNP-E1 (Fig. 2G), was through effects on a putative locus that was distinct from HPV16 L2 RNA. Moreover, the greater reduction of HPV16 L1 and L2 in low folate cells (Fig. 2, E and F) suggested that this locus was responsive to homocysteine-derivatized hnRNP-E1.
Because extracellular l-homocysteine can enter cultured human cells by an active cysteine transporter system (21), we determined if abruptly increasing the concentration of l-homocysteine in medium led to acute changes (over 2 h) in HPV16 L1 and L2 in (HPV16)BC-1-Ep/SL-HF cells. Fig. 2H confirmed a dose-dependent reduction in intracellular HPV16 L1 and L2 RNA when (HPV16)BC-1-Ep/SL-HF cells were exposed to increasing concentrations of l-homocysteine. Because effects were also noted at 12.5 and 25 μm l-homocysteine, this suggested that the interaction of endogenous hnRNP-E1 and HPV16 RNA occurred within cells at physiologically appropriate concentrations of homocysteine that would be found even in mild folate deficiency.
Interaction of hnRNP-E1 with HPV16 57-nucleotide Poly(U)-rich cis-Element
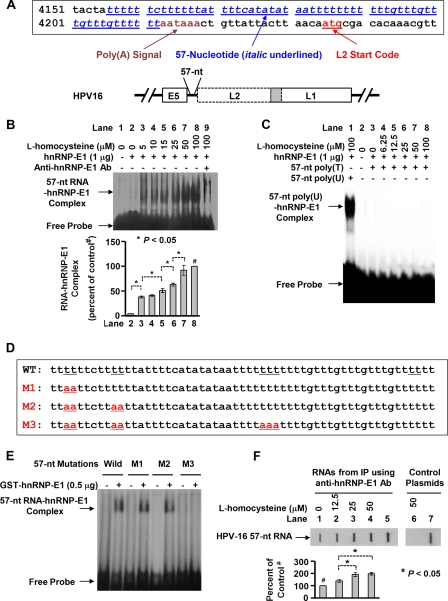
A potential candidate locus to mediate the profound dual reduction of HPV16 L2 and L1 expression in (HPV16)BC-1-Ep/SL-LF cells was a 57-nucleotide RNA domain in the early polyadenylation element of the HPV16 genome (Fig. 3A). This domain, which is located upstream of L2̂L1 genes, can control the expression of L2 and L1 genes (13) and is also known to bind other members of the hnRNP family (13). Fig. 3B shows that the addition of increasing concentrations of l-homocysteine to a mixture of purified recombinant GST-hnRNP-E1 and this putative HPV16 57-nucleotide RNA cis-element led to progressively greater RNA-protein complex signals in a dose-dependent, saturable manner. There was a supershift of the RNA protein signal with anti-hnRNP-E1 antibody (Fig. 3B, lane 9) with no supershift with non-immune serum (not shown), which confirmed the presence of hnRNP-E1 within the RNA-protein complexes. By contrast, there was no interaction between homocysteinylated hnRNP-E1 and HPV16 57-nucleotide poly(T)-rich single-stranded sense DNA (Fig. 3C), confirming that the HPV16 57-nucleotide RNA domain only interacted with homocysteinylated hnRNP-E1.
FIGURE 3.
Analysis of the specificity of interaction of the HPV16 57-nucleotide poly(U)-rich RNA cis-element with purified recombinant GST-hnRNP-E1 using gel shift assays, mutation studies in vitro, and slot blot hybridization analysis to detect enrichment of similar intracellular RNA-protein complexes following exposure of (HPV16)BC-1-Ep/SL-HF cells to increasing physiologically relevant concentrations of l-homocysteine. A, DNA sequence of the HPV16 57-nucleotide cis-element in the early polyadenylation element upstream of L2̂L1 genes (in italic type and underlined in blue) in the HPV16 genome in the upper panel; the position of the poly(A) signal is shown in brown type, and the L2 start code is indicated in red type. The lower panel shows the location of the 57-nucleotide sequence (57-nt) upstream of the HPV16 L2/L1 genes; the overlapping sequence between L2 and L1 is shown in the shaded portion. B, gel shift assays of the interaction of the HPV16 57-nucleotide RNA cis-element (57-nt RNA) with hnRNP-E1 in the presence of increasing concentrations of l-homocysteine (0–100 μm; lanes 2–9). The pooled densitometric scanned data of RNA-protein complexes formed with increasing concentrations of l-homocysteine from three independent gel shift experiments are shown as a bar graph below one representative gel; these data are presented as the mean ± S.D. All densitometric signals were compared with the control (lane 8), reflecting maximal signal obtained with 100 μm l-homocysteine (marked by # in the bar graph). There was a supershift of the RNA-protein complex signal with anti-hnRNP-E1 antibody in lane 9. C, gel shift assay of the interaction between HPV16 57-nucleotide poly(T)-rich single-stranded sense DNA (57nt poly(T)) and purified recombinant GST-hnRNP-E1 in the presence of increasing concentrations of l-homocysteine (lanes 3–8) compared with positive control employing HPV16 57-nucleotide poly(U) cis-element (lane 1). This gel is representative of three separate experiments that yielded comparable results. D, wild-type HPV16 57-nucleotide RNA cis-element and various mutations (M1–M3) within this sequence that are marked in red and underlined. E, gel shift analysis of the interaction of hnRNP-E1 with either wild type [α-32P]HPV16 57-nucleotide RNA cis-element (pWT) or various mutations of [α-32P]HPV16 57-nucleotide RNA cis-element (M1–M3) in the presence of 50 μm l-homocysteine. This gel is representative of three separate experiments that yielded comparable results. F, slot blot hybridization analysis to detect intracellular RNA-protein complexes of HPV16 57-nucleotide RNA cis-element bound to hnRNP-E1 within 2 h after the exposure of (HPV16)BC-1-Ep/SL-HF cells to increasing physiologically relevant concentrations of l-homocysteine (see “Experimental Procedures” for methodological details). Intracellular RNA-protein complexes were cross-linked and immunoprecipitated with anti-hnRNP-E1 antiserum, and, following RNase treatment and proteolysis of the immunoprecipitate, equal concentrations of the released small RNA fragments (RNAs from IP) were tested for the presence of HPV16 57-nucleotide RNA cis-element using a 35S-labeled antisense probe to the 57-nucleotide RNA cis-element (lanes 1–5) or a control 35S-labeled sense probe (lanes 6 and 7). Denatured pYS57 plasmid DNA used as additional positive controls (lanes 5 and 7) were also tested for hybridization with this antisense or sense probe. The hybridization signals in response to l-homocysteine are compared with base-line values (denoted as 100% detected with no addition of l-homocysteine). The result of densitometric scans of the signals is shown below the slot blot and is presented as the mean ± S.D. (error bars) from three independent sets of experiments (n = 3). The 100% control value is indicated by # in lane 1. IP, immunoprecipitation.
To further evaluate the specificity of the HPV16 57-nucleotide RNA cis-element for binding purified recombinant GST-hnRNP-E1 in the presence of l-homocysteine, we induced specific mutations within the HPV16 57-nucleotide RNA cis-element (Fig. 3D). As shown in Fig. 3E, although there was interaction between purified recombinant GST-hnRNP-E1 and some mutations (pM1 and pM2) similar to wild-type (pWT) HPV16 57-nucleotide RNA cis-element in the presence of 50 μm l-homocysteine, there was no interaction when an additional mutation (pM3) was introduced, confirming the specificity of this RNA-protein interaction.
To determine if there was physiological interaction of hnRNP-E1 with this HPV16 57-nucleotide cis-element within cells in response to homocysteine, we captured these RNA-protein complexes in (HPV16)BC-1-Ep/SL-HF cells as a function of the addition of physiologically relevant concentrations of l-homocysteine, as described recently (11). This was achieved by first cross-linking the endogenous RNA-protein complexes with UV light, followed by specific immunoprecipitation using anti-hnRNP-E1 antiserum. Then after cleaving unprotected RNA that was not in close apposition to the immunoprecipitated hnRNP-E1 protein, the HPV16 57-nucleotide RNA cis-element (as well as other RNA cis-elements) that remained cross-linked to hnRNP-E1 were released by proteolysis of hnRNP-E1. Following this step, equivalent amounts of the remaining RNA were tested for the capacity to hybridize under stringent conditions with 35S-labeled antisense probe to the HPV16 57-nucleotide RNA cis-element using slot blot hybridization analysis. As shown in Fig. 3F, lanes 2–4, there was an l-homocysteine-induced dose-dependent increase in signal from hybridization with HPV16 57-nucleotide RNA cis-element antisense probe when compared with control samples to which l-homocysteine was not added (Fig. 3F, lane 1). Moreover, the specificity of this hybridization was shown by the lack of hybridization signal (Fig. 3F, lane 6) using the HPV16 57-nucleotide RNA cis-element sense probe. The positive controls using denatured pYS57 (Fig. 3F, lanes 5 and 7) confirmed signals of hybridization with both 35S-labeled antisense and 35S-labeled sense 57-nucleotide RNA cis-element probes. Collectively, these results reflected the capture of HPV16 57-nucleotide RNA cis-element-bound hnRNP-E1 protein complexes within (HPV16)BC-1-Ep/SL-HF cells as a function of increasing intracellular l-homocysteine concentrations.
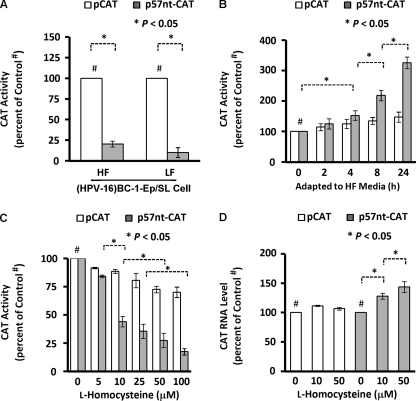
Next, we sought to define the functional significance of the interaction between endogenous homocysteinylated hnRNP-E1 and the HPV16 57-nucleotide RNA cis-element in cells. Transfecting a plasmid containing this HPV16 57-nucleotide RNA cis-element placed proximal to a CAT reporter, p57nt+CAT, into (HPV16)BC-1-Ep/SL-HF cells and (HPV16)BC-1-Ep/SL-LF cells resulted in p57nt+CAT signals of 20 and 10%, respectively, when compared with pCAT controls (Fig. 4A). This suggested that the greater interaction of endogenous homocysteinylated hnRNP-E1 with p57nt+CAT reporter plasmids in (HPV16)BC-1-Ep/SL-LF cells, which contained nearly 3-fold more intracellular homocysteine than (HPV16)BC-1-Ep/SL-HF cells (Fig. 2A), resulted in a functional effect. (The basis for a significant reduction in p57nt+CAT signals even in (HPV16)BC-1-Ep/SL-HF cells when compared with pCAT control is addressed in experiments leading to Table 1; see below). Earlier, we showed that abruptly exposing HeLa-IU1-LF cells (that were stably propagated in low folate medium) to high folate medium (containing over 250-fold more folate) progressively lowered the intracellular homocysteine toward basal values by 24 h (10). When (HPV16)BC-1-Ep/SL-LF cells were similarly exposed to high folate medium (Fig. 4B), the p57nt+CAT reporter signals progressively increased as a function of time to over 3-fold more than basal values by 24 h, but pCAT controls increased only marginally. This suggested that upon folate repletion, the reduced intracellular homocysteine led to a disinhibition of CAT signals compared with base-line controls, reflecting a reduced interaction of endogenous hnRNP-E1 with p57nt+CAT reporters. Next, we determined if increasing the concentration of l-homocysteine in medium led to acute changes (by 2 h) in p57nt+CAT reporter signals in (HPV16)BC-1-Ep/SL-HF cells. Fig. 4C confirmed that p57nt+CAT reporter signals progressively decreased with increasing physiologically relevant concentrations of l-homocysteine, whereas pCAT controls were not as affected. This was not because of any reduction in the level of p57nt+CAT mRNA in these cells. In fact, as shown in Fig. 4D, the measured mRNA levels of pCAT control were not significantly changed, whereas the mRNA levels of p57nt+CAT were slightly increased (up to 1.5-fold) with the addition of 0, 10, and 50 μm l-homocysteine. These data supported the likelihood that the observed inhibitory effects of l-homocysteine on the HPV16 57-nucleotide RNA cis-element-CAT construct-related CAT activity (Fig. 4C) were at a translational level.
FIGURE 4.
Comparison of CAT activity following the transfection of the HPV16 57-nucleotide cis-element-driven CAT reporter constructs into (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells and after the acute modulation of the intracellular homocysteine concentrations by altered culture conditions. The results are presented as the mean ± S.D. (error bars) from three independent sets of experiments (n = 3) with each data point performed in triplicate. A, CAT reporter activity following transfection of (HPV16)BC-1-Ep/SL-HF cells (HF) and (HPV16)BC-1-Ep/SL-LF cells (LF) with either control plasmids (pCAT; open bars) or plasmids in which the HPV16 57-nucleotide RNA cis-element was placed proximal to CAT reporter (p57nt-pCAT; shaded bars). pSV-β-gal was used as an internal control, and the chemiluminescent assay for quantitative determination of β-galactosidase activity in transfected cells was employed to monitor transfection efficiency (10). All data were normalized by internal standard and protein content of each treatment. B, CAT activity from (HPV16)BC-1-Ep/SL-LF cells that were transfected with either pCAT or p57nt+CAT plasmids, following which cells were abruptly transferred to high folate medium, and CAT activity was determined as a function of time. C, CAT activity from (HPV16)BC-1-Ep/SL-HF cells transfected with either pCAT or p57nt+CAT plasmid DNA. After a 48-h incubation in high folate medium, cells were exposed to increasing concentrations of l-homocysteine for 2 h and then assessed for CAT activity. D, expression of CAT mRNA in (HPV16)BC-1-Ep/SL-HF cells that were transfected with either pCAT or p57nt+CAT plasmid DNA and then exposed to increasing concentrations of l-homocysteine.
TABLE 1.
Comparison of the binding affinity between purified recombinant GST-hnRNP-E1 and HPV16 L2 RNA cis-element and HPV16 57-nucleotide RNA cis-element at equimolar concentrations of l-homocysteine and l-cysteine, and evidence of a lower dissociation constant (KD) with l-homocysteine than l-cysteine for the RNA-protein interaction involving the HPV16 57-nucleotide RNA cis-element
The results are presented as the mean ± S.D. from three independent sets of experiments with each data point performed in triplicate.
| Thiol amino acids | [35S]RNA | KD | p value |
|---|---|---|---|
| nm | |||
| l-Homocysteine (25 μm) | [35S]HPV16 L2 | 0.517 ± 0.039 | |
| l-Homocysteine (25 μm) | [35S]HPV16 57 nt | 0.223 ± 0.021 | 0.031 |
| l-Cysteine (15 μm) | [35S]HPV16 57 nt | 1.098 ± 0.207 | |
| l-Homocysteine (15 μm) | [35S]HPV16 57 nt | 0.404 ± 0.123 | 0.001 |
Taken together, these results suggested that interaction of endogenous homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide RNA cis-element in (HPV16)BC-1-Ep/SL-LF cells inhibited downstream genes and could explain, in part, the observed reduction in levels of HPV16 L2 and L1 mRNA and protein (Fig. 2, E and F).
Comparison of Dissociation Constants of Interaction between Homocysteinylated hnRNP-E1 and HPV16 L2 mRNA cis-element versus HPV16 57-nucleotide RNA cis-Element
Because there were two distinct loci in HPV16 RNA that interacted with homocysteinylated hnRNP-E1, we compared the binding affinity of these two RNA-protein interactions under physiological conditions of l-homocysteine. Formal dissociation constant studies using purified recombinant GST-hnRNP-E1 at physiological folate-deficient conditions (25 μm l-homocysteine) revealed a consistently lower KD in the interaction of homocysteinylated hnRNP-E1 with [35S]HPV16 57-nucleotide RNA cis-element (KD = 0.22 nm) when compared with [35S]HPV16 L2 RNA (KD = 0.52 nm) (Table 1). These studies suggest that the binding affinity of homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide RNA cis-element domain was greater under physiological conditions of folate deficiency and therefore more likely to contribute to the net reduction of HPV16 L1 and L2 mRNA and proteins when compared with the interaction of homocysteinylated hnRNP-E1 with HPV16 L2 mRNA.
Because of comparably high basal concentrations of cysteine and homocysteine in (HPV16)BC-1-Ep/SL-LF cells (Fig. 2, A and B), we determined if there were subtle differences in binding affinity between the HPV16 57-nucleotide RNA cis-element and purified recombinant GST-hnRNP-E1 protein at equimolar (15 μm) concentrations of l-cysteine and l-homocysteine. Table 1 shows that binding affinity was significantly greater in the presence of l-homocysteine (KD = 0.40 nm) than l-cysteine (KD = 1.1 nm). These data predicted that l-homocysteine had a greater capacity to stimulate interaction of endogenous hnRNP-E1 with the HPV16 57-nucleotide RNA cis-element than l-cysteine in (HPV16)BC-1-Ep/SL-LF cells. Parenthetically, these results can also explain, in part, why transient transfection of p57nt+CAT into (HPV16)BC-1-Ep/SL-HF cells (which contained an estimated 16 μm cysteine and 7 μm homocysteine) reduced CAT-signals to 20% of controls (Fig. 4A).
Effect of RNA Interference of hnRNP-E1 mRNA on HPV16 L1 and L2 mRNA in (HPV16)BC-1-Ep/SL-LF Cells
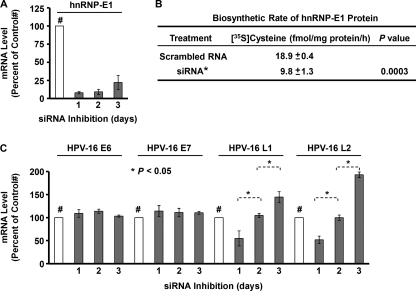
RNA interference (RNAi) revealed that siRNA elicited maximal effect in reducing hnRNP-E1 mRNA by over 90% within 1 day, which persisted over the ensuing 3 days (Fig. 5A). This siRNA had no effect on the closely related hnRNP-E2 mRNA (11). Evaluation of the biosynthetic rate of newly synthesized hnRNP-E1 protein 48 h after RNAi of hnRNP-E1 mRNA revealed a significant reduction when compared with controls using scrambled RNA (Fig. 5B). This predicted that RNAi of hnRNP-E1 mRNA could affect the level of HPV16 L2 and L1 in (HPV16)BC-1-Ep/SL-LF cells. As shown in Fig. 5C, there was no change in HPV16 E6 and E7 mRNA over the subsequent 3 days in (HPV16)BC-1-Ep/SL-LF cells following RNAi of hnRNP-E1 mRNA; this was consistent with a lack of effect of homocysteinylated hnRNP-E1 on E6 and E7 mRNA and protein (Fig. 2). By contrast, there was a significant time-dependent increase in levels of HPV16 L2 and L1 mRNA over the subsequent 3 days (Fig. 5C). This reflected an escape from the inhibitory effects of interaction of HPV16 RNA with homocysteinylated hnRNP-E1 protein and confirmed the specificity of effects of hnRNP-E1 on HPV16 L1 and L2.
FIGURE 5.
Effect of RNA interference against hnRNP-E1 mRNA on the mRNA level of hnRNP-E1 (A), the biosynthetic rate of newly synthesized hnRNP-E1 protein (B), and levels of HPV16 E6, E7, L1, and L2 RNAs in (HPV16)BC-1-Ep/SL-LF cells that were stably propagated in low folate medium (C). The results are presented as the mean ± S.D. (error bars) from three independent sets of experiments (n = 3) with each data point performed in triplicate. A, (HPV16)BC-1-Ep/SL-LF cells were transfected with either scrambled control RNA (open bar) or siRNA against hnRNP-E1 mRNA (shaded bars) and harvested at 1, 2, and 3 days after transfection for RNA purification and qRT-PCR to determine the mRNA level of hnRNP-E1 (A) and HPV16 E6, E7, L1, and L2 (C). The data using scrambled RNA are expressed as 100% and were unchanged over 3 days, and data on inhibited hnRNP-E1 mRNA or other HPV related RNA are expressed as a percentage of control values. #, the 100% control value. The maximum inhibition of hnRNP-E1 mRNA on the first day was consistently confirmed on four independent occasions.
Comparison of Rates of Biosynthesis and Degradation of HPV L1 and L2 mRNAs and HPV16 L1 and L2 Proteins in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF Cells
As shown in Table 2, the rates of biosynthesis of L2 and L1 RNA transcripts were significantly decreased in (HPV16)BC-1-Ep/SL-LF when compared with (HPV16)BC-1-Ep/SL-HF cells. Table 2 also shows that the rate of degradation of L2 and L1 RNA in (HPV16)BC-1-Ep/SL-LF cells was significantly increased when compared with (HPV16)BC-1-Ep/SL-HF cells. Together, these data were consistent with a conclusion that the binding of homocysteinylated hnRNP-E1 to the HPV16 57-nucleotide poly(U)-rich cis-element led to a reduction in the rate of biosynthesis of both L2 and L1 mRNA as well as an increase in degradation of these mRNA transcripts, and these events probably contributed to the net reduction in L2 and L1 RNA levels in (HPV16)BC-1-Ep/SL-LF cells (Fig. 2). (Because there was no interaction between homocysteinylated hnRNP-E1 and single stranded HPV16 DNA, the term “biosynthesis of RNA” used here refers to the rate of formation of mature HPV16 L2 and L1 RNA.)
TABLE 2.
Comparison of the biosynthetic and degradation rates of HPV16 L2 and L1 mRNAs in (HPV16)BC-1-Ep/SL cells that were stably propagated as monolayers in either HF or LF medium
The results are presented as the mean ± S.D. from three independent sets of experiments with each data point performed in triplicate.
| Cell type | Total RNA | p value |
|---|---|---|
| fmol/ng/h | ||
| Biosynthetic rate of HPV16 L2 mRNA | ||
| (HPV-16)BC-1-Ep/SL-HF | 8.49 ± 0.38 | |
| (HPV-16)BC-1-Ep/SL-LF | 4.95 ± 0.27 | 0.0002 |
| Biosynthetic Rate of HPV16 L1 mRNA | ||
| (HPV-16)BC-1-Ep/SL-HF | 13.33 ± 0.82 | |
| (HPV-16)BC-1-Ep/SL-LF | 7.07 ± 0.36 | 0.0003 |
| Degradation Rate of HPV16 L2 mRNA | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.34 ± 0.03 | |
| (HPV-16)BC-1-Ep/SL-LF | 0.74 ± 0.04 | 0.0002 |
| Degradation Rate of HPV16 L1 mRNA | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.42 ± 0.03 | |
| (HPV-16)BC-1-Ep/SL-LF | 0.71 ± 0.06 | 0.0023 |
The protein biosynthetic rates of GAPDH and L1 were comparable in both (HPV16)BC-1-Ep/SL-LF and (HPV16)BC-1-Ep/SL-HF cells (Table 3). However, the rate of protein biosynthesis of L2 in (HPV16)BC-1-Ep/SL-LF cells was about one-half the value obtained with (HPV16)BC-1-Ep/SL-HF cells. These data are compatible with Fig. 1, where the independent interaction of L2 mRNA and homocysteinylated hnRNP-E1 led to reduction in synthesis of L2 in vitro. In addition, the rate of biosynthesis of hnRNP-E1 was lower in (HPV16)BC-1-Ep/SL-LF cells (Table 3). Thus, although the biosynthetic rate of hnRNP-E1 in low folate media was lower (consistent with a lower net amount of hnRNP-E1, as noted in Fig. 2F), the accumulation of nearly 3-fold more intracellular homocysteine could result in the homocysteinylation of endogenous hnRNP-E1, leading to efficient reduction in HPV16 L1 and L2 via effects on the HPV16 57-nucleotide cis-element and the L2 mRNA cis-element.
TABLE 3.
Comparison of the protein biosynthetic rates of HPV16 L2, HPV16 L1, hnRNP-E1, and GAPDH proteins in HPV16 -harboring human keratinocytes that were stably propagated as monolayers in either high folate medium ((HPV16)BC-1-Ep/SL-HF) or low folate medium ((HPV16)BC-1-Ep/SL-LF)
The results are presented as the mean ± S.D. from three independent sets of experiments with each data point performed in triplicate.
| Cell type | [35S]Cysteine | p value |
|---|---|---|
| fmol/mg protein/h | ||
| Biosynthetic rate of HPV16 L2 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 108.45 ± 5.08 | |
| (HPV-16)BC-1-Ep/SL-LF | 54.99 ± 0.27 | 0.0001 |
| Biosynthetic rate of HPV16 L1 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 85.71 ± 1.46 | |
| (HPV-16)BC-1-Ep/SL-LF | 84.29 ± 1.89 | 0.3618 |
| Biosynthetic rate of hnRNP-E1 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 94.77 ± 3.88 | |
| (HPV-16)BC-1-Ep/SL-LF | 49.29 ± 4.45 | 0.0002 |
| Biosynthetic rate of GAPDH protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 77.85 ± 4.12 | |
| (HPV-16)BC-1-Ep/SL-LF | 80.22 ± 3.52 | 0.4921 |
The rates of protein degradation of hnRNP-E1 and GAPDH were comparably unchanged in (HPV16)BC-1-Ep/SL-HF and (HPV16)BC-1-Ep/SL-LF cells (Table 4). However, the rate of degradation of HPV16 L1 protein was 1.7-fold higher, whereas that of HPV16 L2 protein was 2.4-fold higher in (HPV16)BC-1-Ep/SL-LF cells when compared with (HPV16)BC-1-Ep/SL-HF cells. Thus, the increased degradation of HPV16 L2 and L1 protein that was independent of effects on HPV16 L2 and L1 RNA degradation could be yet another contributor to the net reduction in L2 and L1 in (HPV16)BC-1-Ep/SL-LF cells.
TABLE 4.
Comparison of the protein degradation rates of HPV16 L2, HPV16 L1, hnRNP-E1, and GAPDH proteins in HPV16 -harboring human keratinocytes that were stably propagated as monolayers in either high folate medium ((HPV16)BC-1-Ep/SL-HF) or low folate medium ((HPV16)BC-1-Ep/SL-LF)
The results are presented as the mean ± S.D. from three independent sets of experiments with each data point performed in triplicate.
| Cell type | [35S]Cysteine | p value |
|---|---|---|
| fmol/mg protein/h | ||
| Degradation rate of HPV16 L2 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.543 ± 0.082 | |
| (HPV-16)BC-1-Ep/SL-LF | 1.291 ± 0.129 | 0.001 |
| Degradation rate of HPV16 L1 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.945 ± 0.034 | |
| (HPV-16)BC-1-Ep/SL-LF | 1.555 ± 0.205 | 0.007 |
| Degradation rate of hnRNP-E1 protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.633 ± 0.032 | |
| (HPV-16)BC-1-Ep/SL-LF | 0.624 ± 0.027 | 0.723 |
| Degradation rate of GAPDH protein | ||
| (HPV-16)BC-1-Ep/SL-HF | 0.637 ± 0.025 | |
| (HPV-16)BC-1-Ep/SL-LF | 0.702 ± 0.042 | 0.081 |
Collectively, these biosynthesis and degradation studies at the mRNA and protein level (Tables 2–4) demonstrated that (HPV16)BC-1-Ep/SL-LF cells exhibited (i) reduced rates of biosynthesis of both HPV16 L1 and L2 RNA, (ii) increased degradation rates of both HPV16 L1 and L2 RNA, (iii) reduced rates of biosynthesis of HPV16 L2 protein, and (iv) increased rates of degradation of both HPV16 L1 and L2 proteins when compared with (HPV16)BC-1-Ep/SL-HF cells. Taken together, the combination of these changes can explain the basis for the profound reduction of HPV16 L1 and L2 in folate-deficient (HPV16)BC-1-Ep/SL-LF cells.
Comparison of HPV16 Viral Capsid Proteins, Viral Load, Viral Particles, and Viral Integration in HPV16-organotypic Rafts Developed in either Low Folate or High Folate Medium
In order to define the physiological consequences of the HPV16 RNA-hnRNP-E1 interaction, we switched studies from the use of (HPV16)BC-1-Ep/SL keratinocytes propagated as monolayers to (HPV16)BC-1-Ep/SL keratinocyte-derived HPV16-organotypic rafts (12). In this model, HPV16-harboring keratinocytes were stimulated to differentiate so that the HPV16 life cycle would be operative (3, 6); HPV16 L1 and L2 would be generated late in the HPV16 life cycle within (superficial) differentiated layers of these HPV16-organotypic rafts; and complete 55-nm HPV16 viral particles would be found in the topmost layers of these rafts. Accordingly, (HPV16)BC-1-Ep/SL-HF and -LF cells were developed into HPV16-high folate- and HPV16-low folate-organotypic rafts, respectively. The use of a slightly higher concentration of folate (13.6 nm 5-methyltetrahydrofolate) in HPV16-low folate-organotypic rafts (as opposed to the 6.8 nm 5-methyl-tetrathydrofolate that was used for (HPV16)BC-1-Ep/SL-LF monolayers) allowed for better quality rafts to be generated that did not compromise the capacity for differentiation (supplemental Fig. S3). Differentiation in HPV16-high folate- and HPV16-low folate-organotypic rafts was confirmed using histology (supplemental Fig. S3, A and B); immunofluorescence (supplemental Fig. S3, C and D) with filaggrin, a filament-associated protein that binds to keratin fibers in epithelial cells and is a bona fide differentiation marker (22); immunohistochemistry (supplemental Fig. S3, F, G, I, and J); and Western blots (supplemental Fig. S3, E and H) to measure keratin 10 and filaggrin. Although there were subtle architectural abnormalities arising from disordered proliferation in the basal layers of HPV16-low folate-organotypic rafts when compared with HPV16-high folate-organotypic rafts, this was comparable with that seen in folate-deficient murine fetal epithelial tissues (23), where differentiation of keratinocytes in the skin was also retained.
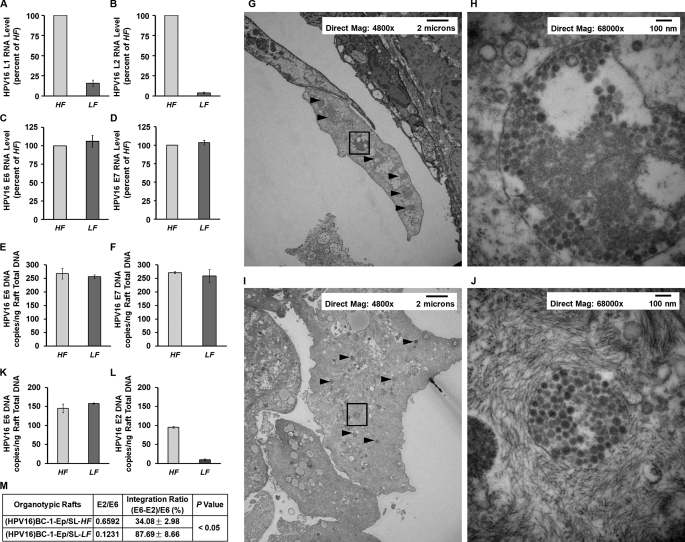
As noted with monolayers of HPV16-harboring keratinocytes in low folate medium (Fig. 2), there was a marked reduction of HPV16 L1 and L2 mRNA in HPV16-low folate-organotypic rafts (Fig. 6, A and B) without changes in HPV16 E6 or E7 RNA (Fig. 6, C and D) when compared with HPV16-high folate-organotypic rafts. In addition, there were no differences in HPV16 DNA viral load when comparing HPV16-high folate- and HPV16-low folate-organotypic rafts (Fig. 6, E and F), indicating no perturbation of the amplification step in the HPV16 life cycle of these rafts. The inability of HPV16-low folate-organotypic rafts to generate HPV16 L1 and L2 viral capsid proteins despite a large amount of amplified HPV DNA predicted a net reduction in the number of HPV16 55-nm viral particles when compared with HPV16-high folate-organotypic rafts. This was confirmed by electron microscopy of the topmost differentiated layers of HPV16-high folate-organotypic rafts (Fig. 6, G and H) and HPV16-low folate-organotypic rafts (Fig. 6, I and J). The arrowheads point to several large clumps of viral particles in the ×4,800 magnification panel of HPV16-high folate-organotypic rafts (Fig. 6G), and a higher magnification of ×68,000 of a boxed square from this panel is shown in Fig. 6H, where an abundance of 55 nm HPV16 viruses were seen. This sharply contrasts with comparable magnification views from HPV16-low folate-organotypic rafts, which exhibited a marked reduction of clumps of viruses (arrowheads in the ×4,800 magnification panel; Fig. 6I). A boxed square from Fig. 6I that was magnified in Fig. 6J revealed 55-nm viruses, confirming that differentiation of HPV16-low folate-organotypic rafts was not compromised; however, the net number of these viral particles was less than that in HPV16-high folate-organotypic rafts (Fig. 6, compare I and G).
FIGURE 6.
Comparison of HPV16 RNA (A–F), HPV16 viral load (E and F), HPV16 viral particles (G–J), and HPV16 DNA integration into genomic DNA (K–M) in HPV16-high folate-organotypic rafts (HF) and HPV16-low folate-organotypic rafts (LF). Unless otherwise specified, the results are presented as the mean ± S.D. from three independent sets of experiments (n = 3) with each data point performed in triplicate. A–D, expression of HPV16 L1 RNA (A), HPV16 L2 RNA (B), HPV16 E6 RNA (C), and HPV16 E7 RNA (D) from HPV16-high folate-organotypic rafts (light shaded bars) and HPV16-low folate-organotypic rafts (dark shaded bars). E and F, determination of HPV16 E6 DNA (E) and HPV16 E7 DNA (F) viral load from HPV16-high folate-organotypic rafts (light shaded bars) and HPV16-low folate-organotypic rafts (dark shaded bars). G–J, photomicrographs of electron microscopy frames from the uppermost layer of HPV16-high folate-organotypic rafts (G and H) and HPV16-low folate-organotypic rafts (I and J) at lower magnification of ×4,800 (G and I) and higher magnification of ×68,000 (H and J). Clumps of HPV16 viral particles are identified by arrowheads in G and I; one of these clumps in G and I identified by a boxed square is shown at a higher magnification of ×68,000 in H and J, respectively. The magnification and scale in each of these frames are shown in the top. Note that HPV16-low folate-organotypic rafts contained smaller clumps of HPV16 viruses and overall lesser numbers of 55-nm HPV16 viruses per clump in higher magnification frames. K–M, comparison of HPV16 E6 and E2 DNA ratios as a reflection of the extent of integration of HPV16 DNA into the genomic DNA of rafts in HPV16-high folate-oganotypic rafts and HPV16-low folate-organotypic rafts (K and L). The amount of integrated HPV16 E6 DNA was calculated by subtracting the copy number of HPV16 E2 episomal DNA from the total copy number of HPV16 E6 DNA (episomal and integrated). The percentage of HPV16 DNA that was integrated into genomic DNA was then determined by dividing the integrated HPV16 E6 DNA copy number by total HPV16 E6 DNA copy number, and the result was multiplied by 100 (M).
Because of this significant discordance between HPV16 DNA viral load and the number of actual HPV16 viral particles observed on electron microscopy in HPV16-low folate-organotypic rafts, we investigated if there was a greater degree of integration of these apparently “capsid-less” HPV16 DNA into genomic DNA. The method to determine the extent of integration of HPV16 DNA into genomic DNA was based on comparative measurement of the absolute values of the HPV16 E2 and E6 open reading frames in DNA samples from HPV16-high folate- and HPV16-low folate-organotypic rafts. In episomal form, the ratio of E2 to E6 is 1. However, ratios of E2 to E6 of less than 1 indicate the presence of both integrated and episomal forms. This is because the E2 primers and probe locations were selected to recognize the E2 hinge region, which is the part of the E2 open reading frame that is most often deleted upon HPV16 viral integration into cellular DNA (16, 24). Fig. 6K depicts the data on HPV16 E6 DNA copies/ng of raft total DNA, whereas Fig. 6L shows HPV16 E2 DNA copies/ng of raft total DNA. When compared with HPV16-high folate-organotypic rafts, the HPV16-low folate-organotypic rafts exhibited a very low E2/E6 ratio. Thus, low folate rafts contained ∼88% HPV16 DNA integration into genomic DNA when compared with ∼34% in high folate rafts (Fig. 6M). (The basal value of 34% integration in the host DNA of HPV16-high folate-organotypic rafts may have arisen during earlier immortalization of keratinocytes by HPV16 (12).)
Evaluation of Potential for Malignant Transformation of HPV16-High Folate- or HPV16-Low Folate-organotypic Rafts Implanted Subcutaneously in Different Species of Immunodeficient Mice
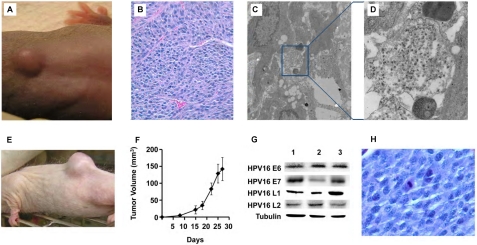
To define the consequences of high level integration of HPV16 into the genomic DNA of HPV16-low folate-organotypic rafts, we tested the hypothesis that this degree of integration was a potentially dangerous implant in a susceptible immunodeficient host. Among the three species of mice with varying degrees of immunodeficiency studied (supplemental Table S1), athymic nude mice have no T cells, but they do have B cells as well as natural killer cells; SHrNTM SCID mice have no B or T cells but do have some (albeit impaired) natural killer cell activity (the immunodeficiency is “leaky” because mice can develop some immunoglobulins as they age (19)); and, finally, beige nude XID mice appear to have no B, T, or natural killer cells and are the most immunodeficient. Accordingly, we evaluated the growth potential of a small fragment of either HPV16-high folate-organotypic rafts or HPV16-low folate-organotypic rafts that was subcutaneously implanted in either folate-replete athymic nude or SHrNTM SCID or beige nude XID mice (2 mice per raft variable). By the 12th week, a small subcutaneous tumor (Fig. 7A) developed in one of the two beige nude XID mice implanted with a fragment of an HPV16-low folate-organotypic raft (which contained 88% HPV16 DNA integration into cellular DNA). This tumor grew steadily over the next 2 weeks to 2 cm3 but then rapidly increased in size over the next 10 days to 10 cm3, which mandated euthanasia. On light microscopy, histological evaluation of cells revealed a high nuclear/cytoplasmic ratio with an open chromatin, mitotic figures, and significant angiogenesis (Fig. 7B). We could also identify viral particles within the tumor on electron microscopy (Fig. 7, C and D). By contrast, there was no growth in HPV16-high folate-organotypic rafts implanted subcutaneously in two other beige nude XID mice; nor was there growth of HPV16-organotypic rafts propagated in either low folate or high folate among the other two species of athymic nude or SHrNTM SCID mice. We then tested whether subcutaneous transplantation of a small fragment of the HPV16-low folate-organotypic raft-derived primary tumor could grow in other folate-replete immunodeficient mice. The results revealed that rapidly growing secondary tumors developed in all three beige nude mice and in one of one athymic nude mouse within 2 weeks. Furthermore, a fragment from a secondary tumor from a beige nude XID mouse also developed into a tertiary tumor within 2 weeks when implanted in SHrNTM SCID mice. One example is shown in Fig. 7E, and the growth profile of four such tumors in SHrNTM SCID mice is shown in Fig. 7F. Of additional significance, all tumors expressed HPV16 E6 and E7 as well as L1 and L2 proteins, as noted in the original tumor (Fig. 7G, lane 1) and in secondary tumors that developed in beige nude XID and athymic nude mice (Fig. 7G, lanes 2 and 3, respectively), thereby pointing to the HPV16 origin of the tumors. Histological evaluation of tertiary tumors revealed a monotonous population of cells with a high nuclear/cytoplasmic ratio and several mitotic figures (Fig. 7H). Thus, the growth characteristics of this tumor fulfilled all criteria for an aggressive HPV16-derived cancer in that it could be transplanted and retransplanted over two generations of mice and also retained expression of HPV16 oncogenes. Therefore, these studies showed that an 18-day-old HPV16-low folate-organotypic raft had the capacity to be transformed into a high grade malignant cancer within only 12 weeks, provided the host was severely immunodeficient. Taken together, these findings are consistent with the probability that folate deficiency functioned as a co-factor in HPV16-induced carcinogenesis.
FIGURE 7.
Transformation of an 18-day-old HPV16-low folate-organotypic raft into a malignant tumor within 12 weeks after subcutaneous implantation in a folate-replete beige nude XID mouse (A–D) and demonstration of highly aggressive growth characteristics and HPV16-derived protein expression in retransplanted tumors in immunodeficient mice (E–H). Due to prohibitive costs, this long term experiment involving the use of three species of mice with varying extents of immunodeficiency was conducted once; see “Experimental Procedures” and “Results” for details. A, photograph of a beige nude XID mouse at 12 weeks following the subcutaneous implantation of a fragment obtained from an HPV16-low folate-organotypic raft. After steadily increasing to 2 cm3 by the 14th week, the tumor rapidly enlarged to 10 cm3 in the ensuring 10 days, mandating euthanasia. There was no growth of HPV16-organotypic rafts propagated in either low folate or high folate among the other two species of athymic nude or SHrNTM SCID mice. B, histological section of the primary tumor from A stained with hematoxylin-eosin that highlights significant angiogenesis within a population of cells with a high nuclear/cytoplasmic ratio, suggesting an underlying malignancy. Magnification was ×44. C and D, clumps of HPV16 viral particles within the primary tumor (C); the area in the blue square is further magnified to demonstrate the presence of 55-nm HPV16 viral particles (D). E, photograph of an aggressive tertiary tumor that developed within 2 weeks in an SHrNTM SCID mouse following the subcutaneous implantation of a small fragment from a secondary tumor that developed in an athymic nude mouse, which was earlier implanted with a fragment of the primary tumor in the beige nude XID mouse in A. F, determination of growth pattern of four tertiary tumors that developed in SHrNTM SCID mice. The data are plotted as tumor volume as a function of time following subcutaneous implantation. Error bars, S.D. G, Western blots to determine the presence of HPV16-specific proteins within the primary tumor in the beige nude XID mouse (lane 1) as well as from secondary tumors from another beige nude XID mouse (lane 2) and an athymic nude mouse (lane 3). Note the presence of HPV16 E6, E7, L1, and L2 proteins in all tumors. There was no signal using an unrelated antiserum (data not shown). H, histological section of a tertiary tumor stained with hematoxylin-eosin that highlights a monotonous population of malignant cells that exhibited a high nuclear/cytoplasmic ratio, open chromatin, and several cells captured at various stages of replication. Magnification was ×100.
DISCUSSION
This report provides mechanistic insight into the functional consequences of linkage involving folate deficiency, homocysteine, hnRNP-E1, HPV16 RNA, HPV16 DNA integration into genomic DNA, and HPV16-induced cancer in mice. Although homocysteinylated hnRNP-E1 bound to HPV16 L2 mRNA and decreased its translation in vitro (Fig. 1), this interaction could not entirely explain the observation of a very significant co-reduction of HPV16 L1 mRNA and protein that was identified in HPV16-harboring BC-1-Ep/SL-LF keratinocytes, which were stably propagated as monolayers in physiologically low folate medium (Fig. 2). However, the interaction of homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide poly(U)-rich cis-element in the upstream early polyadenylation element of the HPV16 genome (13) was an attractive possibility to explain this observation for three reasons. (i) Deletion of this domain, which is upstream of overlapping L2̂L1 genes in the HPV16 genome, reduced utilization of the HPV16 early polyadenylation signal, resulting in read-through into the HPV16 late region and increased production of L1 and L2 mRNAs (13); therefore, we reasoned that the binding of hnRNP-E1 to this region could reduce L2 and L1. (ii) This domain binds other members of the hnRNP family, including hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract-binding protein (13). (iii) Earlier, we purified hnRNP-E1 using poly(U)-Sepharose affinity chromatography (9), suggesting that hnRNP-E1 would bind the HPV16 57-nucleotide poly(U)-rich cis-element. Subsequent studies on the binding of purified recombinant GST-hnRNP-E1 to HPV16 57-nucleotide poly(U)-rich cis-element in the presence of physiological concentrations of l-homocysteine, as well as mutation studies involving this cis-element, indicated the specificity and importance of this interaction (Fig. 3). Subsequent CAT reporter studies (Fig. 4) predicted that a reduction in translation of the downstream sequences of HPV16 L2̂L1 mRNA following the interaction of HPV16 57-nucleotide RNA cis-element with homocysteinylated hnRNP-E1 would negatively impact net protein levels in (HPV16)BC-1-Ep/SL-LF cells, which contain more homocysteine; this was experimentally demonstrated (Tables 2–4). Taken together, the dual interaction of homocysteinylated hnRNP-E1 with HPV16 L2 mRNA and the HPV16 57-nucleotide RNA cis-element was probably responsible for the reduced levels of HPV16 L2 and L1 mRNA and protein in (HPV16)BC-1-Ep/SL-LF cells when compared with (HPV16)BC-1-Ep/SL-HF cells (Fig. 2). Because there was a greater affinity of purified recombinant GST-hnRNP-E1 for the HPV16 57-nucleotide RNA cis-element than for HPV16 L2 mRNA, as well as a greater affinity of purified recombinant GST-hnRNP-E1 for the HPV16 57-nucleotide RNA cis-element in the presence of l-homocysteine when compared with equimolar concentrations of l-cysteine (Table 1), it appears that the interaction of homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide RNA cis-element in the upstream early polyadenylation element may be the more important physiological interaction within HPV16-harboring keratinocytes. The direct link involving l-homocysteine in inducing the binding of hnRNP-E1 to the HPV16 57-nucleotide RNA cis-element was demonstrated by capture of these intracellular RNA-protein complexes as a function of the concentration of l-homocysteine added to (HPV16)BC-1-Ep/SL-HF cells (Fig. 3F). These studies required us to separate these complexes from other complexes that could also arise following the interaction of homocysteinylated hnRNP-E1 with other mRNA cis-elements that share common poly(rC)- or poly(U)-rich sequence signatures, which also allows them to also bind to hnRNP-E1 under these conditions (7, 10, 25–34). Because hnRNP-E1 may have cross-linked some of these other mRNA (in addition to HPV16 57-nucleotide RNA cis-element) in cells containing higher than basal concentrations of l-homocysteine, we employed a similar strategy that was recently employed to detect folate receptor-α mRNA cis-element·hnRNP-E1 complexes (11). Accordingly, we tested equal amounts of small RNA fragments that were released from endogenous (immunoprecipitated) homocysteinylated hnRNP-E1 for the capacity to hybridize under stringent conditions with antisense probe to the HPV16 57-nucleotide RNA cis-element. Our results, which revealed progressively increased hybridization signals, clearly reflected the capture of HPV16 57-nucleotide RNA cis-element-bound hnRNP-E1 protein complexes within cells as a function of increasing intracellular l-homocysteine concentrations that mimicked various grades of physiological folate deficiency (Fig. 3F).
The interaction of homocysteinylated hnRNP-E1 with poly(rC)-rich HPV16 L2 mRNA and HPV16 57-nucleotide poly(U)-rich sequences, but not with corresponding poly-T sequences (Fig. 1B), suggests that the decreased HPV16 L1 and L2 mRNA levels observed in low folate cells are not explained by altered rates of DNA transcription. However, it is possible that posttranscriptional splicing of newly transcribed heterogeneous nuclear pre-mRNA may have been affected upon binding by homocysteinylated hnRNP-E1; this is because earlier studies demonstrated that the splicing in vitro of an mRNA precursor (pre-mRNA) was inhibited by a monoclonal antibody to hnRNP C proteins (35). This antibody inhibits an early step of the reaction: cleavage at the 3′-end of the upstream exon and the formation of the intron lariat. This finding is relevant because Schwartz's laboratory (13) demonstrated that the HPV16 57-nucleotide poly(U)-rich sequence is also bound by hnRNP C1 (just like homocysteinylated hnRNP-E1 under physiologic conditions). Thus, it is possible that interaction of homocysteinylated hnRNP-E1 with the HPV16 57-nucleotide poly(U)-rich pre-mRNA led to interference with pre-mRNA processing, resulting in the reduced L1 and L2 mRNA biosynthetic rates (Table 2). Our studies also demonstrated reduced stability (increased rate of degradation of mRNA) of HPV16 L2 and L1 mRNA (Table 2), which explained, in part, the reduced L1 and L2 mRNA measured in low folate cells (Fig. 2). Because the HPV DNA of both L2 and L1 overlap, posttranscriptional splicing is required to release L2 from L1 mRNA. Thus, it is possible that the observed increased L2 and L1 mRNA degradation rates (that were measured in minutes) may have occurred before the splicing event, because the data on the rate of reduction of both mRNAs showed that it was relatively rapid when compared with the other measured metabolic parameters in low folate cells (Table 2). It is possible that additional binding of homocysteinylated hnRNP-E1 to the 3′-coding region of HPV16 L2 RNA in the L2̂L1 transcript before the splicing event that separates these overlapping L2̂L1 mRNA (13) favors degradation of these unspliced L2̂L1 transcripts (as noted by RNA degradation measurements in (HPV16)BC-1-Ep/SL-LF cells in Table 2). Despite this, the level of HPV16 L2 mRNA was slightly lower than HPV16 L1 mRNA in low folate cells (Fig. 2E). This can be partly explained by independent direct binding of homocysteinylated hnRNP-E1 to HPV16 L2 mRNA (Fig. 1), which could have further contributed to its reduction (after splicing) in low folate cells. Moreover, the relatively greater reduction in HPV16 L2 than in L1 protein expression can be explained by the additional direct effect of homocysteinylated hnRNP-E1 in independently reducing the translation of HPV16 L2 mRNA (Fig. 1).
Because hnRNP-E1 has multiple effects within cells (11, 36), prolonged RNA interference posed the risk of additional off-target effects. Moreover, because RNA interference led to 90% inhibition of hnRNP-E1 mRNA (without significant effects on hnRNP-E2 (11)), and the measured degradation rate of hnRNP-E1 had a t½ of over 48 h, we confirmed the effect of RNA interference against hnRNP-E1 mRNA on hnRNP-E1 protein by measuring a reduction in the rate of biosynthesis of newly synthesized hnRNP-E1 protein (Fig. 5). Then we evaluated the effects of this RNA interference on altered HPV16 L2 and L1 mRNA in (HPV16)BC-1-Ep/SL-LF cells, which afforded an assessment of the direct connection between hnRNP-E1 protein and HPV16 L1 and L2 mRNA. We found that following reduction of hnRNP-E1 mRNA and protein, there was a progressive increase (reflecting a release following disinhibition) of HPV16 L1 and L2 mRNA levels as a function of days following RNA interference (Fig. 5). The relatively greater increase of HPV16 L2 mRNA when compared with L1 mRNA following RNA interference of hnRNP-E1 mRNA was the converse of combined inhibitory effects of homocysteinylated hnRNP-E1 on both the HPV16 L2 RNA and 57-nucleotide poly(U)-rich cis-elements.
The availability of a purified cell line of HPV16-harboring BC-1-Ep/SL cells (that could be propagated long term as monolayers (12) in defined high folate versus low folate medium without changes in the doubling time of cells or spontaneous differentiation, and which had the ability to measure HPV16 early and late RNA in these cells) provided an unprecedented opportunity to mechanistically study the effect of hnRNP-E1 on HPV16 L1 and L2 RNA and proteins. However, to better define the physiological role of the HPV16 RNA interaction with homocysteinylated hnRNP-E1, we stimulated (HPV16)BC-1-Ep/SL cells to develop into HPV16-organotypic rafts in which the entire HPV16 cycle is operative (12). We determined that folate deficiency had a similar influence on HPV16 viral capsid proteins in HPV16-low folate-organotypic rafts as noted with keratinocyte monolayers (Fig. 6). Moreover, the presence of a similar HPV16 DNA viral load in 18-day-old HPV16-organotypic rafts developed in either high or low folate medium predicted fewer 55-nm HPV16 viral particles; this was confirmed by electron microscopy. However, the existence of high level integration of HPV16 DNA into the genome of HPV16-low folate-organotypic rafts was unexpected and requires an explanation. There were two potential variables that could have influenced these findings. First, in HPV16-low folate-organotypic rafts, there was a relative overabundance of amplified HPV16 DNA that could not be encapsidated into authentic HPV16 viral particles to complete the HPV16 life cycle (because of reduced HPV16 L1 and L2 viral capsid proteins). It is therefore possible that these apparently “capsid-less” HPV16 DNA, with no “escape route” out of the cell, could have somehow integrated into genomic DNA. Second, it is well known that folate deficiency inhibits thymidylate synthase, which leads to lowered deoxythymidine monophosphate synthesis and accumulation of deoxyuridine monophosphate (37). This higher deoxyuridine monophosphate/deoxythymidine monophosphate ratio leads to increased misincorporation of deoxyuridine triphosphate into DNA by DNA polymerase (38–41). Although increased uracil misincorporation into DNA in folate deficiency (37, 38, 41–43) leads to removal by uracil-DNA glycosylase (44) and refilling of the missing base by DNA polymerase β (45), with repetition over several cycles, multiple single strand nicks are introduced into DNA predisposing to chromosome breaks (46) that can contribute to an increased risk of cancer associated with folate deficiency (42, 47, 48). In addition, folate deficiency can also lead to double strand breaks in DNA, which are difficult to repair when the two nicks are close to one another (within 12 bp of each other) on opposite strands (49). Collectively, such double strand DNA breaks in folate-deficient cells predispose to the development of acentric chromosomes, DNA fragments, and micronuclei (46, 50) and could have rendered folate-deficient tissues more permissive to the integration of HPV16 DNA. Distinguishing between these two distinct variables and defining the extent of their relative (independent) contribution to HPV16 DNA integration into HPV16-low folate-organotypic raft DNA is currently under study.
In order to define the long term consequences of high level HPV16 DNA integration into folate-deficient HPV16-organotypic rafts warranted study in an animal model; however, there were no established models related to implantation of such rafts. Infection of human foreskin by oncogenic HPV that is implanted into immunodeficient mice has nevertheless been used in the past to confirm the role of HPV in growth promotion. However, none of these earlier models actually led to high grade malignancy in the host. For example, implantation of HPV16-infected human foreskin implanted in immunodeficient mice has only led to the formation of papillomas, not to high grade cancer (18). Although these models used SCID or athymic nude mice, because beige nude XID mice are severely immunodeficient, we determined if the latter were more likely to support the development of a malignancy provided that another co-factor (like folate deficiency) was also present. Thus, we tested whether the added variable of folate deficiency could tip the balance in favor of developing a cancer out of HPV16-low folate-organotypic rafts implanted subcutaneously in these mice. The transformation of benign 18-day old HPV16-low folate-organotypic raft-tissue into an aggressive malignancy within only 12 weeks in beige nude XID mice supports the likelihood that folate deficiency is a co-factor in HPV16-induced carcinogenesis. These findings raise additional questions on three interrelated issues: these include the importance of HPV16 DNA integration into the genome and carcinogenesis; the locus of integration into the host DNA; and the relationship of genomic DNA instability with folate deficiency (42). Although it is controversial whether HPV DNA integration is a critical event in HPV-induced carcinogenesis clinically (51), integration does disrupt the viral regulatory gene E2; this, in turn, interferes with negative feedback control of oncogene expression and leads to increased stability of E6 and E7 mRNAs (52) and a selective growth advantage of these cells (53). Several studies have documented that the locus of integration of HPV16 DNA within the human genome is frequently found within common fragile sites (54–56). Although the majority of rare fragile sites in genomic DNA are induced by folate deficiency (57–60), among the common fragile sites, the one at 3p14.2 (FRA3B) (the most sensitive site on normal human chromosomes, which also contains a spontaneous HPV16 integration site (54)) as well as several other common fragile sites can also develop gaps and breaks when DNA replication is perturbed by “folate stress” (involving depletion of cellular deoxynucleotide pools by either folate deficiency or methotrexate) (54, 60–62). Because there is direct evidence for the coincidence of viral integration sites and fragile sites (54), it is possible that HPV16-low folate-organotypic rafts contained integration of HPV16 DNA into these folate-sensitive or “folate-stressed” fragile sites in genomic DNA. With regard to the timing of HPV16 integration, Peitsaro et al. (16) have identified that integration of HPV16 DNA into the host genome can occur early in the course of HPV16-induced cervical dysplasia; this parallels our finding of high level integration HPV16 DNA in the cellular DNA of 18-day-old HPV16-low folate-organotypic rafts. It is still unclear whether women infected with HPV16 who have clinical folate deficiency (or vitamin-B12 deficiency, which also induces a functional intracellular folate deficiency (63)) have a greater extent of HPV16 DNA integration into the genomic DNA. This will be the focus of future clinical translational studies.
As suggested recently (11), the existence of accessible cysteine residues within KH or RNA recognition domains among other hnRNP family members and RNA-binding proteins, respectively, would probably be the primary determinant as to whether there is homocysteinylation of additional hnRNPs within folate-deficient cells; this would result in the co-activation of several additional nutrition-sensitive posttranscriptional RNA operons during clinical folate and/or vitamin-B12 deficiency. One strong candidate for homocysteinylation includes hnRNP-E2 (also called αCP2 or poly(C)-binding protein 2 (PCBP2)), which has 93% identity in the mRNA-binding KH domains with hnRNP-E1 and 100% identity in the position of cysteine residues within the three KH domains (that are amenable for interaction with homocysteine) (11). Acting together, these (and other homocysteinylated hnRNP-activated) posttranscriptional RNA operons would comprise a novel, higher order, nutrition-sensitive (homocysteine-responsive), posttranscriptional RNA regulon (64) that could have diverse effects on cellular and as well as on viral genes. In addition, apart from HPV16, homocysteinylated hnRNP-E1 and hnRNP-E2 may also bind to homologous RNA sequences in other benign and oncogenic HPV types, thereby potentially influencing their expression of late viral capsid proteins and infectivity of these viruses (65, 66). Furthermore, there could be additional effects on other viruses. For example, the binding of hnRNP-E1 to the 5′-terminal cloverleaf structure of poliovirus RNA, which facilitates interaction with the viral protein 3CD, a precursor of protease 3C and RNA polymerase 3D, promotes viral RNA replication (25, 67). Blyn et al. (68) have also demonstrated that hnRNP-E2 is an essential factor that is required for efficient translation of poliovirus RNA. Such interactions of homocysteinylated hnRNP-E1 and hnRNP-E2 with the poliomyelitis cis-element would lead to increased binding and augment viral proliferation during clinical poliomyelitis in developing countries, where hyperhomocysteinemia (due to nutritional folate and/or vitamin-B12 deficiency) is widely prevalent (63, 69–71), and sporadic poliomyelitis is still an important public health problem. Significantly, Woolaway et al. (34) have also shown that both hnRNP-E1 and hnRNP-E2 have a modulating role on the human immunodeficiency virus-1 (HIV-1); however, they elicit different responses because only overexpression of hnRNP-E1 inhibited the expression of several HIV-1 genes. Collectively, therefore, our study predicts that HPV16, poliomyelitis, and HIV-1 RNAs are members of this posttranscriptional nutrition-sensitive RNA operon that is modulated by homocysteinylated hnRNP-E1 (and probably hnRNP-E2). Because there are hundreds of millions of individuals in the developing world with nutritional deficiencies of folate and/or vitamin-B12 (10, 63, 70, 72, 73), the net effect of these deficiencies on the natural history of HPV16, poliomyelitis, and HIV-1 infections is an important new area for urgent study.
In summary, this work provides molecular evidence of a link between folate deficiency and HPV a full 30 years after an association was first suggested (74, 75) (and further reinforced in the ensuing years by several epidemiologic studies that have also suggested an adverse role for hyperhomocysteinemia (76–81)). Our studies predict that clinical nutritional folate and/or vitamin-B12 deficiency (i) will activate the nutrition-sensitive posttranscriptional RNA operon orchestrated by hnRNP-E1; (ii) will have a negative impact on generation of authentic HPV16 virions and alter the natural history of HPV16-induced infection; and (iii), will probably adversely influence the oncogenic transformation of epithelial dysplasia to cancer by facilitating HPV16 integration into unstable genomic DNA. Further evaluation among populations where both HPV16-induced cervical dysplasia and cancer as well as the deficiency of folate/vitamin-B12 is common (63, 69–71) will help to clarify whether these micronutrient deficiencies are bona fide co-factors in the clinical transformation of HPV16-infected tissues to cancer.
Supplementary Material
Acknowledgments
We are indebted to Professor Paul F. Lambert (University of Wisconsin, Madison, WI) for generously providing HPV16-harboring BC-1-Ep/SL cells, and we thank Professors Ann Roman and Darron R. Brown (Indiana University) for encouragement and sage advice during early phases of this study. We also thank Professors Ann Roman and Harikrishna Nakshatri (Indiana University) for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA120843 and R01HD39295 (to A. C. A.) and AG09834 (to S. P. S.). This work was also supported by a Veterans Affairs Merit Review Award (to A. C. A. and H. N. J.). One of the authors (S. P. S.) and the University of Colorado hold patents on the use of assays for total homocysteine and other metabolites to diagnose vitamin-B12 (cobalamin) and folate deficiencies, and a company has been formed at the University of Colorado to perform such assays.

This article contains supplemental data, Table S1, and Figs. S1–S3.
- HPV
- human papillomavirus
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- (HPV16)BC-1-Ep/SL cells
- HPV16-harboring BC-1-Ep/SL cells
- NEM
- N-ethyl maleimide
- F-HF
- high folate F-medium
- F-LF
- physiologic low folate F-medium
- HF
- high folate
- LF
- low folate
- qRT-PCR
- quantitative RT-PCR
- qPCR
- quantitative PCR
- CAT
- chloramphenicol acetyltransferase
- nt
- nucleotide(s).
REFERENCES
- 1. Flores E. R., Allen-Hoffmann B. L., Lee D., Lambert P. F. (2000) The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74, 6622–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bosch F. X., Lorincz A., Muñoz N., Meijer C. J., Shah K. V. (2002) The causal relation between human papillomavirus and cervical cancer. J. Clin. Path. 55, 244–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laimins L. A. (1993) The biology of human papillomaviruses. From warts to cancer. Infect. Agents Dis. 2, 74–86 [PubMed] [Google Scholar]
- 4. Longworth M. S., Laimins L. A. (2004) Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68, 362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston C. (2000) Quantitative tests for human papillomavirus. Lancet 355, 2179–2180 [DOI] [PubMed] [Google Scholar]
- 6. Schwartz S. (2000) Regulation of human papillomavirus late gene expression. Upsala J. Med. Sci. 105, 171–192 [PubMed] [Google Scholar]
- 7. Collier B., Goobar-Larsson L., Sokolowski M., Schwartz S. (1998) Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273, 22648–22656 [DOI] [PubMed] [Google Scholar]
- 8. Sun X. L., Antony A. C. (1996) Evidence that a specific interaction between an 18-base cis-element in the 5′-untranslated region of human folate receptor-α mRNA and a 46-kDa cytosolic trans-factor is critical for translation. J. Biol. Chem. 271, 25539–25547 [PubMed] [Google Scholar]
- 9. Xiao X., Tang Y. S., Mackins J. Y., Sun X. L., Jayaram H. N., Hansen D. K., Antony A. C. (2001) Isolation and characterization of a folate receptor mRNA-binding trans-factor from human placenta. Evidence favoring identity with heterogeneous nuclear ribonucleoprotein E1. J. Biol. Chem. 276, 41510–41517 [DOI] [PubMed] [Google Scholar]
- 10. Antony A., Tang Y. S., Khan R. A., Biju M. P., Xiao X., Li Q. J., Sun X. L., Jayaram H. N., Stabler S. P. (2004) Translational up-regulation of folate receptors is mediated by homocysteine via RNA-heterogeneous nuclear ribonucleoprotein E1 interactions. J. Clin. Invest. 113, 285–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Y. S., Khan R. A., Zhang Y., Xiao S., Wang M., Hansen D. K., Jayaram H. N., Antony A. C. (2011) Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J. Biol. Chem. 286, 39100–39115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flores E. R., Allen-Hoffmann B. L., Lee D., Sattler C. A., Lambert P. F. (1999) Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262, 344–354 [DOI] [PubMed] [Google Scholar]
- 13. Zhao X., Oberg D., Rush M., Fay J., Lambkin H., Schwartz S. (2005) A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 79, 4270–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Motulsky H., Christopoulos A. (2004) Fitting Models to Biological Data using Linear and Nonlinear Regression., 1st Ed., pp. 296–338, Oxford University Press, New York [Google Scholar]
- 15. Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peitsaro P., Johansson B., Syrjänen S. (2002) Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 40, 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carcopino X., Henry M., Benmoura D., Fallabregues A. S., Richet H., Boubli L., Tamalet C. (2006) Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J. Med. Virol. 78, 1131–1140 [DOI] [PubMed] [Google Scholar]
- 18. Brandsma J. L., Brownstein D. G., Xiao W., Longley B. J. (1995) Papilloma formation in human foreskin xenografts after inoculation of human papillomavirus type 16 DNA. J. Virol. 69, 2716–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonnez W. (2005) The HPV xenograft severe combined immunodeficiency mouse model. Methods Mol. Med. 119, 203–216 [DOI] [PubMed] [Google Scholar]
- 20. Feldman J. P., Goldwasser R., Mark S., Schwartz J., Orion I. (2009) A mathematical model for tumor volume evaluation using two-dimensions. J. Appl. Quant. Methods 4, 455–462 [Google Scholar]
- 21. Büdy B., O'Neill R., DiBello P. M., Sengupta S., Jacobsen D. W. (2006) Homocysteine transport by human aortic endothelial cells. Identification and properties of import systems. Arch. Biochem. Biophys. 446, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuspa S. H., Kilkenny A. E., Steinert P. M., Roop D. R. (1989) Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J. Cell Biol. 109, 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiao S., Hansen D. K., Horsley E. T., Tang Y. S., Khan R. A., Stabler S. P., Jayaram H. N., Antony A. C. (2005) Maternal folate deficiency results in selective up-regulation of folate receptors and heterogeneous nuclear ribonucleoprotein-E1 associated with multiple subtle aberrations in fetal tissues. Birth Defects Res. A Clin. Mol. Teratol. 73, 6–28 [DOI] [PubMed] [Google Scholar]
- 24. Kalantari M., Karlsen F., Kristensen G., Holm R., Hagmar B., Johansson B. (1998) Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int. J. Gynecol. Pathol. 17, 146–153 [DOI] [PubMed] [Google Scholar]
- 25. Ostareck-Lederer A., Ostareck D. H., Hentze M. W. (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci 23, 409–411 [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Kiledjian M., Weiss I. M., Liebhaber S. A. (1995) Detection and characterization of a 3′-untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol. Cell Biol. 15, 1769–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang Y., Xu X. S., Russell J. E. (2006) A nucleolin-binding 3′-untranslated region element stabilizes β-globin mRNA in vivo. Mol. Cell Biol. 26, 2419–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Czyzyk-Krzeska M. F., Bendixen A. C. (1999) Identification of the poly(C)-binding protein in the complex associated with the 3′-untranslated region of erythropoietin messenger RNA. Blood 93, 2111–2120 [PubMed] [Google Scholar]
- 29. Giles K. M., Daly J. M., Beveridge D. J., Thomson A. M., Voon D. C., Furneaux H. M., Jazayeri J. A., Leedman P. J. (2003) The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 278, 2937–2946 [DOI] [PubMed] [Google Scholar]
- 30. Paulding W. R., Czyzyk-Krzeska M. F. (1999) Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J. Biol. Chem. 274, 2532–2538 [DOI] [PubMed] [Google Scholar]
- 31. Stefanovic B., Hellerbrand C., Holcik M., Briendl M., Liebhaber S. A., Brenner D. A. (1997) Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol. Cell Biol. 17, 5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeap B. B., Voon D. C., Vivian J. P., McCulloch R. K., Thomson A. M., Giles K. M., Czyzyk-Krzeska M. F., Furneaux H., Wilce M. C., Wilce J. A., Leedman P. J. (2002) Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 277, 27183–27192 [DOI] [PubMed] [Google Scholar]
- 33. Thyagarajan A., Szaro B. G. (2004) Phylogenetically conserved binding of specific K homology domain proteins to the 3′-untranslated region of the vertebrate middle neurofilament mRNA. J. Biol. Chem. 279, 49680–49688 [DOI] [PubMed] [Google Scholar]
- 34. Woolaway K., Asai K., Emili A., Cochrane A. (2007) hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression. Retrovirology 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. (1986) Heterogeneous nuclear ribonucleoproteins. Role in RNA splicing. Science 231, 1534–1539 [DOI] [PubMed] [Google Scholar]
- 36. Chaudhury A., Chander P., Howe P. H. (2010) Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes. Focus on hnRNP E1's multifunctional regulatory roles. RNA 16, 1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Das K. C., Herbert V. (1989) In vitro DNA synthesis by megaloblastic bone marrow. Effect of folates and cobalamins on thymidine incorporation and de novo thymidylate synthesis. Am. J. Hematol. 31, 11–20 [DOI] [PubMed] [Google Scholar]
- 38. Wickramasinghe S. N., Fida S. (1994) Bone marrow cells from vitamin B12- and folate-deficient patients misincorporate uracil into DNA. Blood 83, 1656–1661 [PubMed] [Google Scholar]
- 39. Goulian M., Bleile B., Tseng B. Y. (1980) Methotrexate-induced misincorporation of uracil into DNA. Proc. Natl. Acad. Sci. U.S.A. 77, 1956–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goulian M., Bleile B., Tseng B. Y. (1980) The effect of methotrexate on levels of dUTP in animal cells. J. Biol. Chem. 255, 10630–10637 [PubMed] [Google Scholar]
- 41. Reidy J. A. (1988) Role of deoxyuridine incorporation and DNA repair in the expression of human chromosomal fragile sites. Mutat. Res. 200, 215–220 [DOI] [PubMed] [Google Scholar]
- 42. Duthie S. J., Hawdon A. (1998) DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. FASEB J. 12, 1491–1497 [PubMed] [Google Scholar]
- 43. Mashiyama S. T., Courtemanche C., Elson-Schwab I., Crott J., Lee B. L., Ong C. N., Fenech M., Ames B. N. (2004) Uracil in DNA, determined by an improved assay, is increased when deoxynucleosides are added to folate-deficient cultured human lymphocytes. Anal. Biochem. 330, 58–69 [DOI] [PubMed] [Google Scholar]
- 44. Barnes D. E., Lindahl T. (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 45. Lindahl T. (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res. 462, 129–135 [DOI] [PubMed] [Google Scholar]
- 46. Blount B. C., Mack M. M., Wehr C. M., MacGregor J. T., Hiatt R. A., Wang G., Wickramasinghe S. N., Everson R. B., Ames B. N. (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage. Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 94, 3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ciappio E., Mason J. B. (2010) Folate and Carcinogenesis. Basic Mechanisms. in Folate in Health and Disease, 2nd Ed. (Bailey L. B., ed) pp. 235–262, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 48. Ames B. N., Wakimoto P. (2002) Are vitamin and mineral deficiencies a major cancer risk? Nat. Rev. Cancer 2, 694–704 [DOI] [PubMed] [Google Scholar]
- 49. Dianov G. L., Timchenko T. V., Sinitsina O. I., Kuzminov A. V., Medvedev O. A., Salganik R. I. (1991) Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 225, 448–452 [DOI] [PubMed] [Google Scholar]
- 50. Kim Y. I., Pogribny I. P., Basnakian A. G., Miller J. W., Selhub J., James S. J., Mason J. B. (1997) Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am. J. Clin. Nutr. 65, 46–52 [DOI] [PubMed] [Google Scholar]
- 51. Woodman C. B., Collins S. I., Young L. S. (2007) The natural history of cervical HPV infection. Unresolved issues. Nat. Rev. Cancer 7, 11–22 [DOI] [PubMed] [Google Scholar]
- 52. Jeon S., Lambert P. F. (1995) Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs. Implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 92, 1654–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeon S., Allen-Hoffmann B. L., Lambert P. F. (1995) Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69, 2989–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilke C. M., Hall B. K., Hoge A., Paradee W., Smith D. I., Glover T. W. (1996) FRA3B extends over a broad region and contains a spontaneous HPV16 integration site. Direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 5, 187–195 [DOI] [PubMed] [Google Scholar]
- 55. Thorland E. C., Myers S. L., Persing D. H., Sarkar G., McGovern R. M., Gostout B. S., Smith D. I. (2000) Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 60, 5916–5921 [PubMed] [Google Scholar]
- 56. Thorland E. C., Myers S. L., Gostout B. S., Smith D. I. (2003) Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 22, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 57. Sutherland G. R., Richards R. I. (1995) The molecular basis of fragile sites in human chromosomes. Curr. Opin. Genet. Dev. 5, 323–327 [DOI] [PubMed] [Google Scholar]
- 58. Savelyeva L., Sagulenko E., Schmitt J. G., Schwab M. (2006) Low-frequency common fragile sites. Link to neuropsychiatric disorders? Cancer Lett. 232, 58–69 [DOI] [PubMed] [Google Scholar]
- 59. Schwartz M., Zlotorynski E., Kerem B. (2006) The molecular basis of common and rare fragile sites. Cancer Lett. 232, 13–26 [DOI] [PubMed] [Google Scholar]
- 60. Barbi G., Steinbach P., Vogel W. (1984) Nonrandom distribution of methotrexate-induced aberrations on human chromosomes. Detection of further folic acid-sensitive fragile sites. Hum. Genet. 68, 290–294 [DOI] [PubMed] [Google Scholar]
- 61. Glover T. W., Berger C., Coyle J., Echo B. (1984) DNA polymerase α inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67, 136–142 [DOI] [PubMed] [Google Scholar]
- 62. Yunis J. J., Soreng A. L. (1984) Constitutive fragile sites and cancer. Science 226, 1199–1204 [DOI] [PubMed] [Google Scholar]
- 63. Antony A. C. (2009) Megaloblastic Anemias. in Hematology: Basic Principles and Practice, 5th Ed (Hoffman R., Benz E. J., Jr., Shattil S. J., Furie B., Silberstein L. E., McGlave P., Heslop H., eds) pp. 491–524, Churchill Livingstone-Elsevier, Philadelphia [Google Scholar]
- 64. Keene J. D. (2007) RNA regulons. Coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 65. White W. I., Wilson S. D., Bonnez W., Rose R. C., Koenig S., Suzich J. A. (1998) In vitro infection and type-restricted antibody-mediated neutralization of authentic human papillomavirus type 16. J. Virol. 72, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Campos S. K., Ozbun M. A. (2009) Two highly conserved cysteine residues in HPV16 L2 form an intramolecular disulfide bond and are critical for infectivity in human keratinocytes. PLoS One 4, e4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gamarnik A. V., Andino R. (1997) Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3, 882–892 [PMC free article] [PubMed] [Google Scholar]
- 68. Blyn L. B., Towner J. S., Semler B. L., Ehrenfeld E. (1997) Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71, 6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chakravarty I., Sinha R. (2002) Prevalence of micronutrient deficiency based on results obtained from the national pilot program on control of micronutrient malnutrition. Nutr. Rev. 60, S53–S58 [DOI] [PubMed] [Google Scholar]
- 70. Antony A. C. (2003) Vegetarianism and vitamin B-12 (cobalamin) deficiency. Am. J. Clin. Nutr. 78, 3–6 [DOI] [PubMed] [Google Scholar]
- 71. Chitambar C. R., Antony A. C. (2006) Nutritional Aspects of Hematologic Diseases. in Modern Nutrition in Health and Disease, 10th Ed (Shils M. E., Shike M., Ross A. C., Caballero B., Cousins R. J., eds) pp. 1436–1461, Lippicott Williams & Wilkins, Baltimore [Google Scholar]
- 72. Antony A. C. (2001) Prevalence of cobalamin (vitamin B-12) and folate deficiency in India. Audi alteram partem. Am. J. Clin. Nutr. 74, 157–159 [DOI] [PubMed] [Google Scholar]
- 73. Stabler S. P., Allen R. H. (2004) Vitamin B12 deficiency as a worldwide problem. Annu. Rev. Nutr. 24, 299–326 [DOI] [PubMed] [Google Scholar]
- 74. Butterworth C. E., Jr., Hatch K. D., Gore H., Mueller H., Krumdieck C. L. (1982) Improvement in cervical dysplasia associated with folic acid therapy in users of oral contraceptives. Am. J. Clin. Nutr. 35, 73–82 [DOI] [PubMed] [Google Scholar]
- 75. Butterworth C. E., Jr., Hatch K. D., Macaluso M., Cole P., Sauberlich H. E., Soong S. J., Borst M., Baker V. V. (1992) Folate deficiency and cervical dysplasia. JAMA 267, 528–533 [PubMed] [Google Scholar]
- 76. Weinstein S. J., Ziegler R. G., Selhub J., Fears T. R., Strickler H. D., Brinton L. A., Hamman R. F., Levine R. S., Mallin K., Stolley P. D. (2001) Elevated serum homocysteine levels and increased risk of invasive cervical cancer in US women. Cancer Causes Control 12, 317–324 [DOI] [PubMed] [Google Scholar]
- 77. Thomson S. W., Heimburger D. C., Cornwell P. E., Turner M. E., Sauberlich H. E., Fox L. M., Butterworth C. E. (2000) Effect of total plasma homocysteine on cervical dysplasia risk. Nutr. Cancer 37, 128–133 [DOI] [PubMed] [Google Scholar]
- 78. Butterworth C. E., Jr. (1993) Folate status, women's health, pregnancy outcome, and cancer. J. Am. Coll. Nutr. 12, 438–441 [DOI] [PubMed] [Google Scholar]
- 79. Kwanbunjan K., Saengkar P., Cheeramakara C., Thanomsak W., Benjachai W., Laisupasin P., Buchachart K., Songmuaeng K., Boontaveeyuwat N. (2005) Low folate status as a risk factor for cervical dysplasia in Thai women. Nutr. Res. 25, 641–654 [Google Scholar]
- 80. Piyathilake C. J., Badiga S., Paul P., Vijayaraghavan K., Vedantham H., Sudula M., Sowjanya P., Ramakrishna G., Shah K. V., Partridge E. E., Gravitt P. E. (2010) Indian women with higher serum concentrations of folate and vitamin B12 are significantly less likely to be infected with carcinogenic or high-risk (HR) types of human papillomaviruses (HPVs). Int. J. Womens Health 2, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Piyathilake C. J., Henao O. L., Macaluso M., Cornwell P. E., Meleth S., Heimburger D. C., Partridge E. E. (2004) Folate is associated with the natural history of high-risk human papillomaviruses. Cancer Res. 64, 8788–8793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.