Abstract
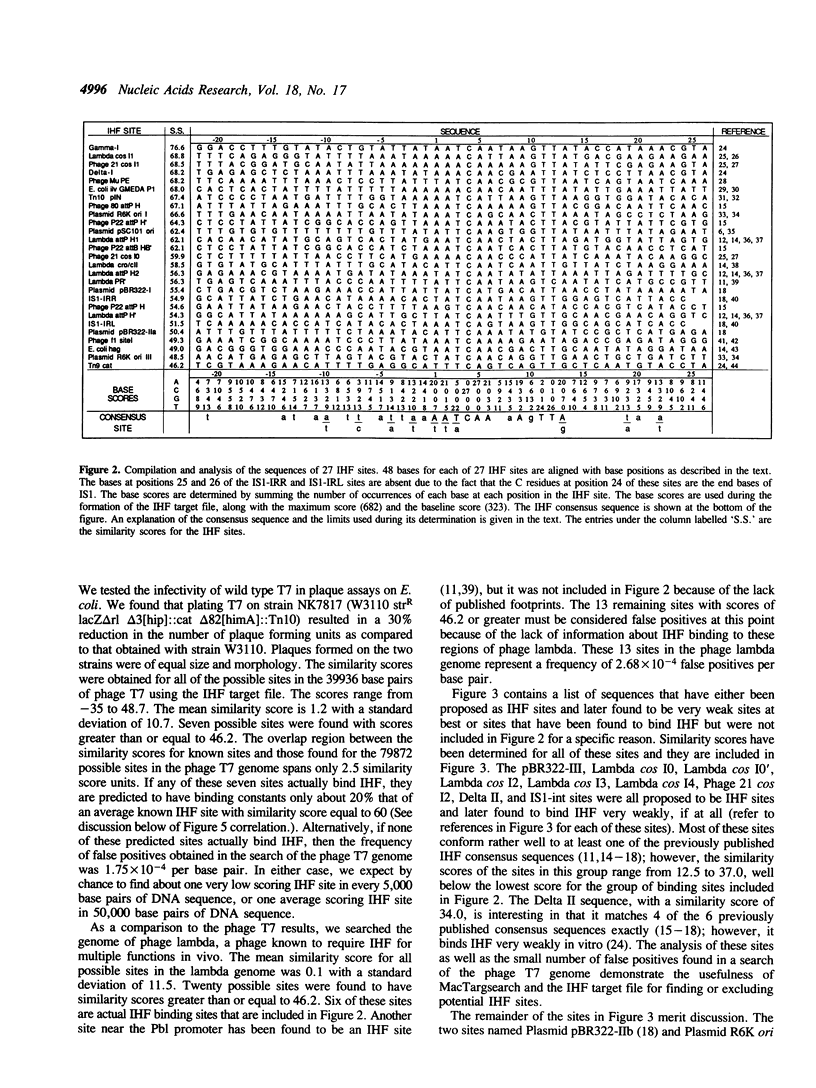
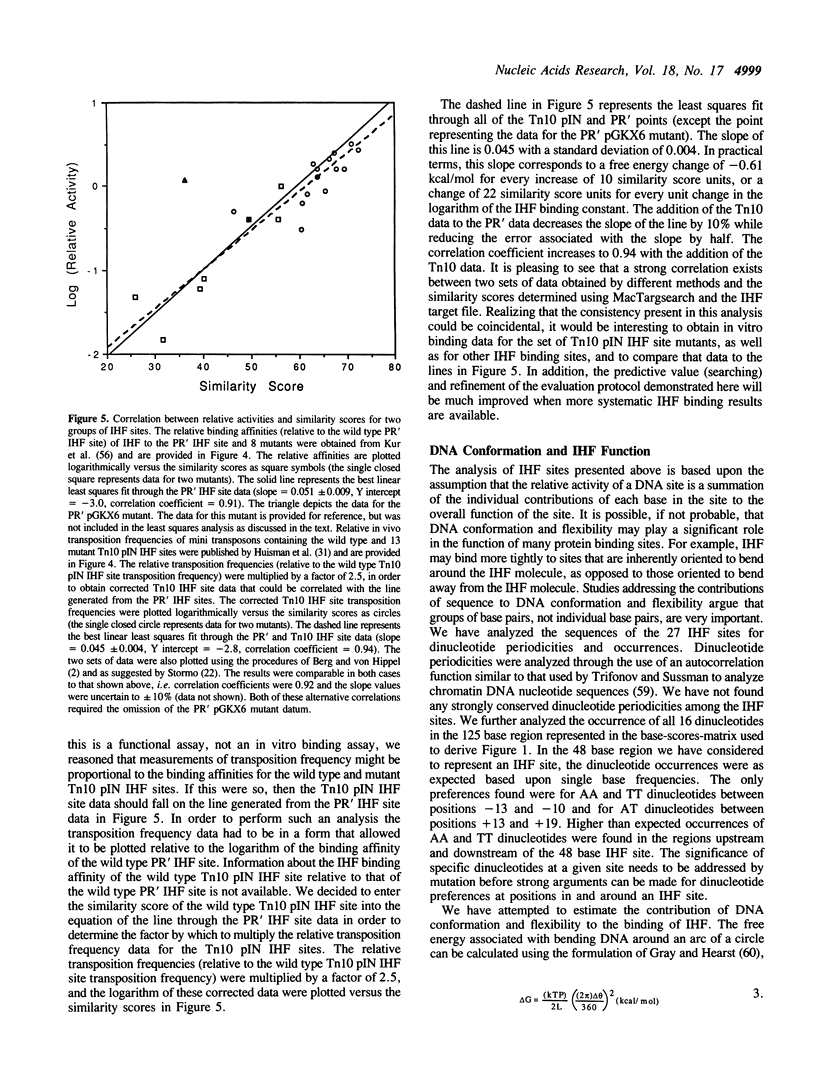
An analysis of the sequence information contained in a compilation of published binding sites for E. coli integration host factor (IHF) was performed. The sequences of twenty-seven IHF sites were aligned; the base occurrences at each position, the information content, and an extended consensus sequence were obtained for the IHF site. The base occurrences at each position of the IHF site were used with a program written for the Apple Macintosh computers in order to determine the similarity scores for published IHF sites. A linear correlation was found to exist between the logarithm of IHF binding and functional data (relative free energies) and similarity scores for two groups of IHF sites. The MacTargsearch program and its potential usefulness in searching for other sites and predicting their relative activities is discussed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Brody H., Hill C. W. Attachment site of the genetic element e14. J Bacteriol. 1988 May;170(5):2040–2044. doi: 10.1128/jb.170.5.2040-2044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B. Integration host factor and conjugative transfer of the antibiotic resistance plasmid R100. J Bacteriol. 1987 Sep;169(9):4391–4392. doi: 10.1128/jb.169.9.4391-4392.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Higgins C. F. Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J Bacteriol. 1987 Aug;169(8):3840–3843. doi: 10.1128/jb.169.8.3840-3843.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein B. I., Sweet D. S., Vaughn V., Friedman D. I. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M., Frackman S., Sippy J. Essential interaction between lambdoid phage 21 terminase and the Escherichia coli integrative host factor. J Mol Biol. 1985 May 25;183(2):239–246. doi: 10.1016/0022-2836(85)90216-5. [DOI] [PubMed] [Google Scholar]
- Filutowicz M., Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K gamma origin and is essential for replication. Nucleic Acids Res. 1988 May 11;16(9):3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Funnell B. E. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6657–6661. doi: 10.1073/pnas.85.18.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Gamas P., Galas D., Chandler M. DNA sequence at the end of IS1 required for transposition. Nature. 1985 Oct 3;317(6036):458–460. doi: 10.1038/317458a0. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Greenstein D., Zinder N. D., Horiuchi K. Integration host factor interacts with the DNA replication enhancer of filamentous phage f1. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6262–6266. doi: 10.1073/pnas.85.17.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffo G., Oppenheim A. B., Gottesman M. E. Repression of the lambda pcin promoter by integrative host factor. J Mol Biol. 1989 Sep 5;209(1):55–64. doi: 10.1016/0022-2836(89)90169-1. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Collier D. A., Kilpatrick M. W., Krause H. M. Supercoiling and integration host factor change the DNA conformation and alter the flow of convergent transcription in phage Mu. J Biol Chem. 1989 Feb 15;264(5):3035–3042. [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Chandler M., Lane D. Mapping and regulation of the pifC promoter of the F plasmid. Biochim Biophys Acta. 1988 May 6;950(1):75–80. doi: 10.1016/0167-4781(88)90075-9. [DOI] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Alterations in the p'R promoter of coliphage lambda modify both its activity and interaction with the integration host factor (IHF). Mol Gen Genet. 1990 May;221(3):411–420. doi: 10.1007/BF00259406. [DOI] [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Physical and biological consequences of interactions between integration host factor (IHF) and coliphage lambda late p'R promoter and its mutants. Gene. 1989 Sep 1;81(1):1–15. doi: 10.1016/0378-1119(89)90331-4. [DOI] [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Repression of transcription from the b2-att region of coliphage lambda by integration host factor. Virology. 1989 Feb;168(2):236–244. doi: 10.1016/0042-6822(89)90263-8. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Wek R. C., Lopes J. M., Pereira R., Taillon B. E., Hatfield G. W. The complete nucleotide sequence of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987 Mar 11;15(5):2137–2155. doi: 10.1093/nar/15.5.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- McIntire S. A., Dempsey W. B. oriT sequence of the antibiotic resistance plasmid R100. J Bacteriol. 1987 Aug;169(8):3829–3832. doi: 10.1128/jb.169.8.3829-3832.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechulam Y., Blanquet S., Fayat G. Dual level control of the Escherichia coli pheST-himA operon expression. tRNA(Phe)-dependent attenuation and transcriptional operator-repressor control by himA and the SOS network. J Mol Biol. 1987 Oct 5;197(3):453–470. doi: 10.1016/0022-2836(87)90558-4. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Fayat G., Blanquet S. Sequence of the Escherichia coli pheST operon and identification of the himA gene. J Bacteriol. 1985 Aug;163(2):787–791. doi: 10.1128/jb.163.2.787-791.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. E., McClure W. R. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acids Res. 1986 Jan 10;14(1):109–126. doi: 10.1093/nar/14.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R. F., Ortuno M. J., Lawther R. P. Binding of integration host factor (IHF) to the ilvGp1 promoter of the ilvGMEDA operon of Escherichia coli K12. Nucleic Acids Res. 1988 Jul 11;16(13):5973–5989. doi: 10.1093/nar/16.13.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Snyder U. K., Thompson J. F., Landy A. Phasing of protein-induced DNA bends in a recombination complex. Nature. 1989 Sep 21;341(6239):255–257. doi: 10.1038/341255a0. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Simon M. DNA sequence adjacent to flagellar genes and evolution of flagellar-phase variation. J Bacteriol. 1983 Jul;155(1):74–81. doi: 10.1128/jb.155.1.74-81.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. J., Mosig G. Integration host factor (IHF) represses a Chlamydomonas chloroplast promoter in E. coli. Nucleic Acids Res. 1988 Apr 25;16(8):3313–3326. doi: 10.1093/nar/16.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui P., Helu V., Freundlich M. Altered osmoregulation of ompF in integration host factor mutants of Escherichia coli. J Bacteriol. 1988 Oct;170(10):4950–4953. doi: 10.1128/jb.170.10.4950-4953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke C., Bastia D. Primary structure of the essential replicon of the plasmid pSC101. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiater L. A., Grindley N. D. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988 Jun;7(6):1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin W. N., Feiss M. The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21. Nucleic Acids Res. 1988 Mar 25;16(5):2015–2030. doi: 10.1093/nar/16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]



