Abstract
The evaluation of a comprehensive α-helix mimetic library for binding the gp41 NHR hydrophobic pocket recognizing an intramolecular CHR α-helix provided a detailed depiction of structural features required for binding and led to the discovery of small molecule inhibitors (Ki 0.6–1.3 µM) that not only match or exceed the potency of those disclosed over the past decade, but that also exhibit effective activity in a cell–cell fusion assay (IC50 5–8 µM).
Small molecule viral cell entry inhibitors provide an attractive approach for the development of HIV-1 treatments with better resistance profiles.1–4 The HIV-1 envelope glycoprotein gp41 has emerged as a promising target in this class since it contains a deep hydrophobic pocket in the N-heptad repeat (NHR) region that is critical to formation of the fusogenic state and amenable to small molecule binding.5 The viral fusion process is driven from a membrane-tethered state to fusion of the viral and host cell membranes by rearrangement of the C-heptad repeat (CHR) and NHR regions from linear trimeric coiled-coils to a composite six-helix bundle. Peptide or small molecule binding in the NHR hydrophobic groove precludes binding of the CHR helices, inhibiting six-helix bundle formation and fusion.2,4 The NHR pocket has been validated as a binding hotspot by using first peptide inhibitors and more recently several classes of small molecules. Simple or constrained peptides, and small molecules that bind the NHR hydrophobic pocket inhibit gp41-mediated cell–cell fusion with IC50’s as low as 250 pM for long α-peptides and 1–5 µM for small molecules.2,4,6–9 Although no small molecule therapeutic has emerged to date, the synthetic 36 amino acid peptide T20 (Enfuvirtide) acting as such an HIV fusion inhibitor was approved in 2003 for use against multidrug resistant HIV.10 The interaction of the CHR with the NHR hydrophobic pocket is a challenging intramolecular protein–protein interaction mediated principally by Trp628, Trp631, Asp632 and Ile635 on the CHR α-helix with the Trp residues constituting the anchor residues (Figure 1A).2 The hydrophobic residues at the i, i+3, and i+7 positions of the α-helix project downward into the NHR pocket, while Asp632 makes an electrostatic interaction with Lys574 at the pocket periphery. Because of the therapeutic potential and α-helical nature of the interaction, the gp41 hydrophobic pocket is an attractive target for small molecule α-helix mimetics.11–15
Figure 1.
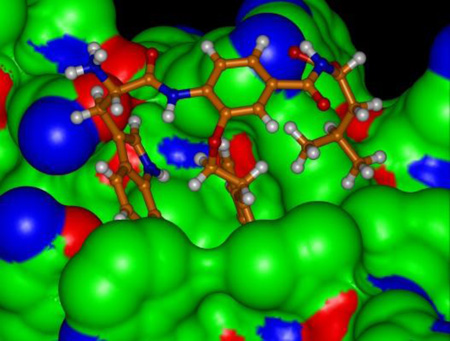
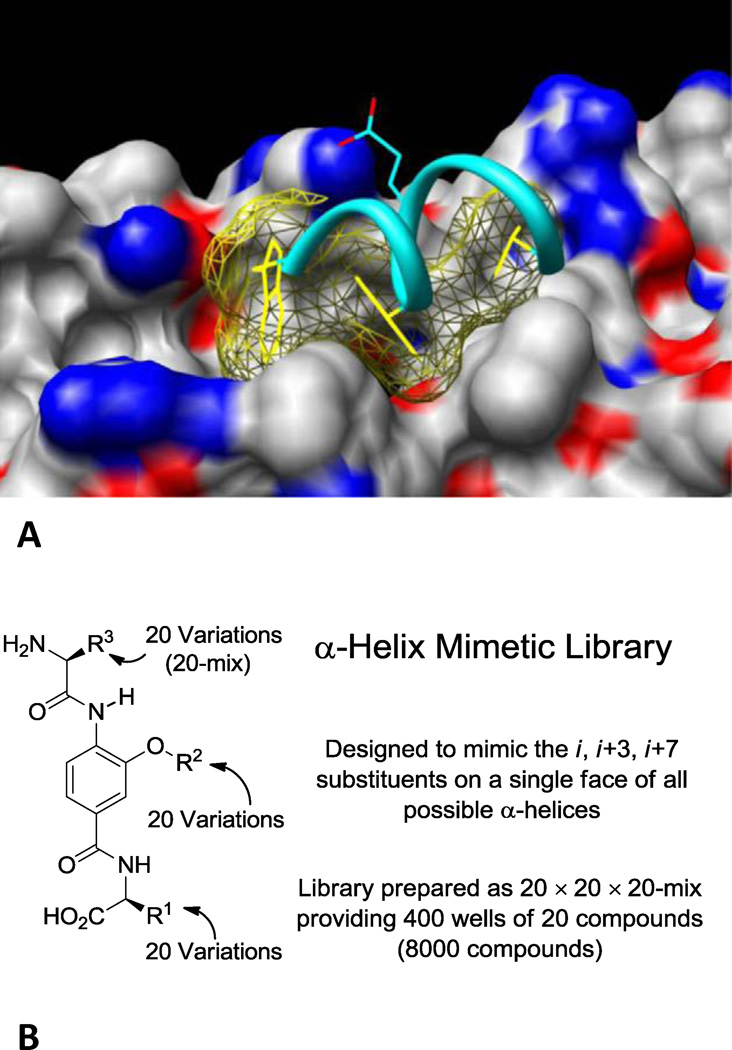
(A) The hydrophobic pocket of gp41 (Modeled structure starting with PDB 1IF3, R. Rizzo). The CHR hydrophobic (Trp628, Trp631, and Ile635, in yellow) and charged (Asp632, in cyan) residues are shown in the pocket. (B) The design and format of the α-helix mimetic library.
As part of a program to prepare a comprehensive library designed to mimic the recognition motifs that mediate protein–protein interactions16,17 (α-helix, β-turn, β-strand), we described the design and synthesis of an α-helix mimetic library17,18 (Figure 1B). The library template was designed to mimic the side chain display of a single face of an α-helix, and was optimized from an original Hamilton design19 for a more flexible display of side chains and with incorporation of functionality that enhances solubility. Template substitution with all combinations of the 20 natural amino acid side chains or related derivatives yielded an 8,000-member library (20 × 20 × 20). The screening of this library is expected to provide a member capable of interrogating most α- helix mediated protein–protein interactions even if the nature of the interaction is unknown, define the key residues mediating the interaction, and provide an initial structure–activity relationship (SAR) study for lead optimization.
Utilizing a competitive metallopeptide-based fluorescence assay designed to directly detect inhibitors that bind the NHR pocket, we screened the 8,000 member α-helix mimetic library to validate the library design, assess its performance against a challenging intramolecular protein–protein interaction, and to gauge the quality of the resulting SAR data. Consistent with the design, the library yielded a new series of inhibitors not only capable of binding the gp41 hydrophobic pocket with affinities that match or exceed (Ki 0.7–1.3 µM) those of other small molecules disclosed to date, including an inhibitor that corresponds to the CHR α-helix residues, but that also exhibit effective activity in a cell–cell fusion assay (IC50 5–8 µM). We describe here these inhibitors as well as SAR insights provided by the library that serves to further validate the mimicry of the library and the power it possesses for interrogating α-helix mediated protein–protein interactions.
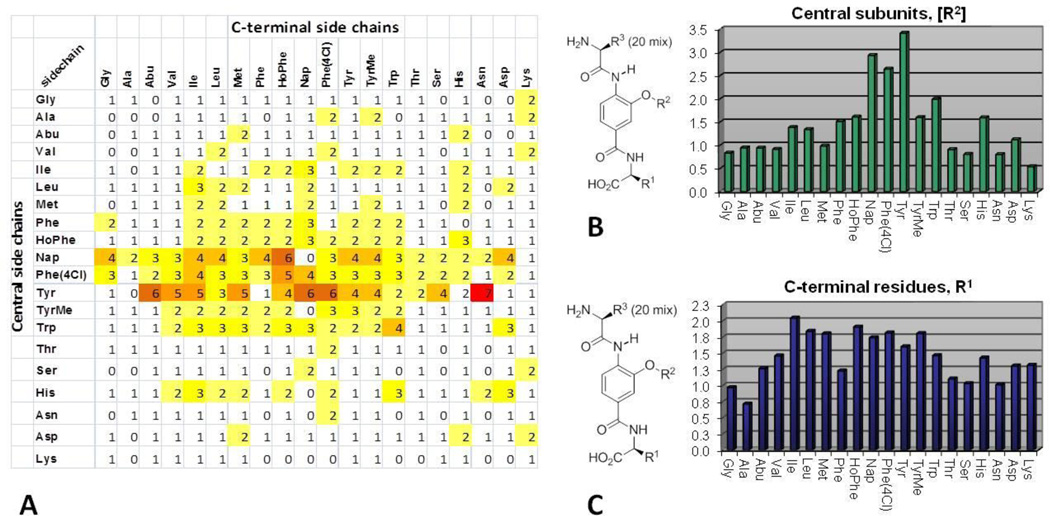
The competitive binding assay utilizes a receptor Fe(env2.0)3 containing three copies of residues 565– 579 of gp41 of the NHR flanked by five residues on each side, and a fluorescently labeled peptide derived from the C-peptide. By design, the assay only selects inhibitors that bind the hydrophobic pocket and inhibit CHR binding.20–22 The α-helix mimetic library, composed of 400 mixtures of 20 compounds, was screened in single point format and the fractional fluorescence values were used to compose a primary screen SAR (Figure 2A). As shown in the matrix analysis, the most active mixtures contained the [Tyr], [Phe(4Cl)], [Nap] and [Trp] central subunits. The strongest activity was observed when these central aromatic side chains were combined with aromatic or bulky aliphatic residues at the C-terminus.
Figure 2.
(A) 20 × 20 matrix of fractional fluorescence values from the binding assay on the 400 α-helix mimetic library mixtures. (B) The sum average scores of individual central subunits. (C) The sum average scores of C-terminal residues.
These preferences were also depicted by scoring the individual subunits using the sum average of the 20 mixtures (Figure 2B and 2C). This analysis highlights the dominant role played by the central [Tyr], [Phe(4Cl)], [Nap] and [Trp] subunits and also that the Ile side chain ranked highest among the C-terminal residues, in line with expectations for mimicry of the CHR Trp-Trp-Ile helical face. Good activity was also observed with other bulky aliphatic (Leu, Met) or a variety of aromatic (HoPhe, Nap, Phe(4Cl) or TyrMe) residues in the C-terminal position, although their impact is more muted than the dominant central subunit. The most active mixtures were also evaluated in a cell–cell fusion (CCF) assay (Figure S1), validating the binding assay SAR and establishing that the mixtures that performed well in the binding assay were also effective inhibitors of gp41-mediated cell–cell fusion (IC50 3–12 µM), Figure S1.
The CHR α-helix residues that interact with the NHR pocket have been ranked by alanine scanning for a C34 peptide (residues 628–661 of CHR) in a fusion assay (IC50 0.55 nM) and viral entry assay (IC50 2.1 nM).5 The change of Trp631 to Ala had the most deleterious effect (30-fold) followed by Trp628 (5–8 fold) and then Ile635 (2-fold). As with Trp631, the SAR demonstrated that the central subunits in the α-helix mimetics played the dominant role in the interaction giving rise to active mixtures with a variety of C-terminal residues. While [Nap], [Phe(4Cl)] and [Trp] can be seen as interchangeable in their ability to provide an extended hydrophobic interaction with the deep pocket, the activity of [Tyr] suggested this residue may be capable of supporting a unique binding mode. The fact that Ile performed best in the C-terminal position also suggested that mimicry of the CHR α-helix was being achieved, that the relative contributions of the three side chains are in accordance with the known contributions of Trp628, Trp631, and Ile635, and that other hydrophobic residues can mimic the Ile635 interaction. Thus, immediately following the screening and even without the examination of individual compounds, key insights into the binding interaction with the protein pocket were provided.
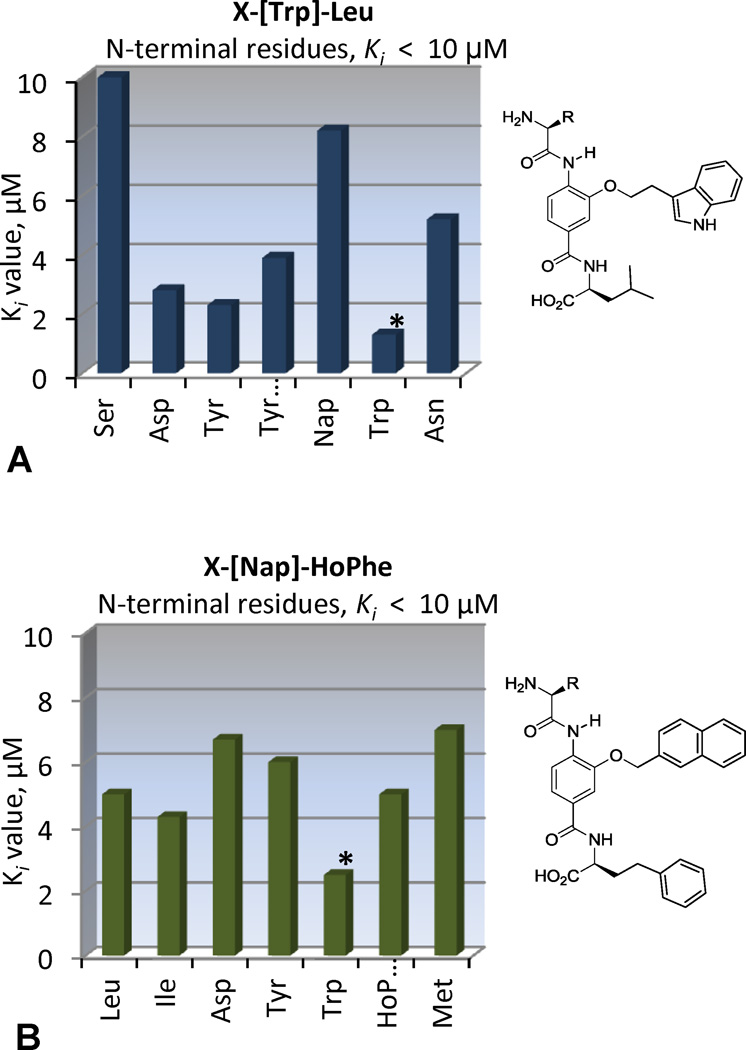
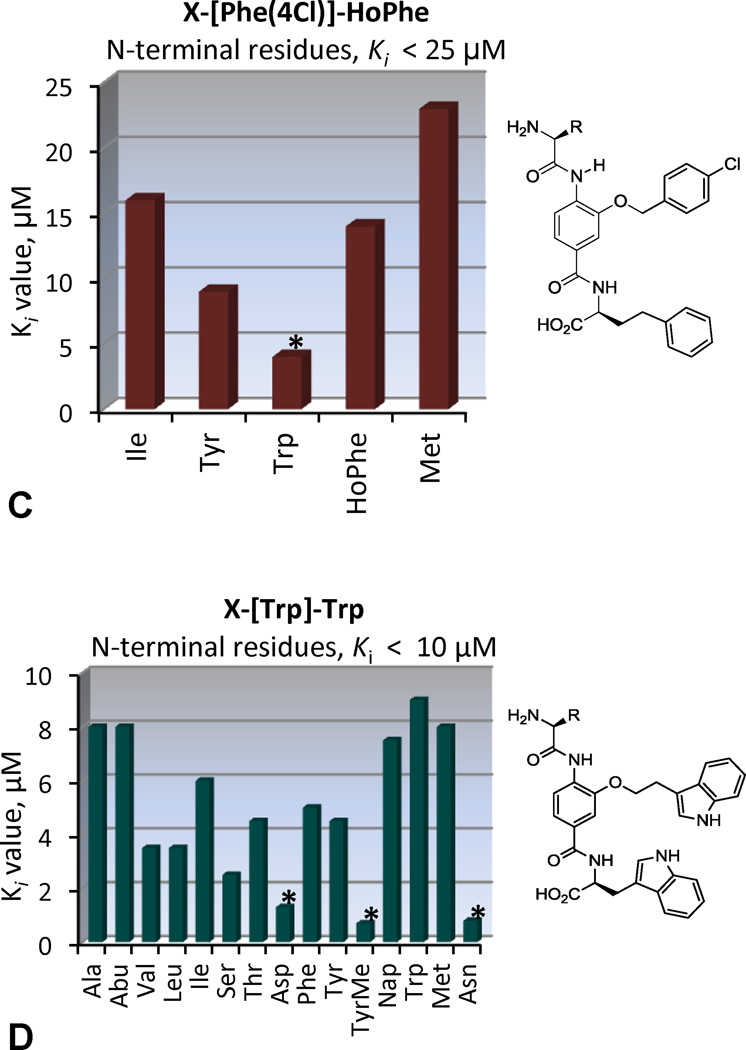
The four mixtures, XXX-[Trp]-Trp, XXX-[Trp]-Leu, XXX-[Nap]-HoPhe and XXX-[Phe(4Cl)]-HoPhe, were selected for individual compound evaluation. These mixtures were among the most active in the CCF and binding assays and evaluation of their individual components would allow further assessment of the mimicry of the CHR α-helix. These 80 compounds, prepared from archived precursor stocks using solution phase protocols and liquid–liquid acid/base extractions for isolation and purification,23–27 were examined in the binding assay, Figure 3.
Figure 3.
Ki values from the binding assay on individual compounds.
The XXX-[Nap]-HoPhe and XXX-[Phe(4Cl)]-HoPhe series display a strong N-terminal preference for Trp, mimicking of the Trp628 interaction, Figure 3B and 3C. Thus, the most active compounds in these series mimic the Trp-Trp-Ile helical face, with the central [Nap] or [Phe(4Cl)] subunits and the C-terminal HoPhe residue mimicking Trp631 and Ile635, respectively, to yield compounds with Ki’s of 2–4 µM. Presumably, this binding mode is also facilitated by an electrostatic interaction between their terminal carboxylic acid and Lys574 on the NHR pocket periphery, mimicking the endogenous interaction of Asp632.
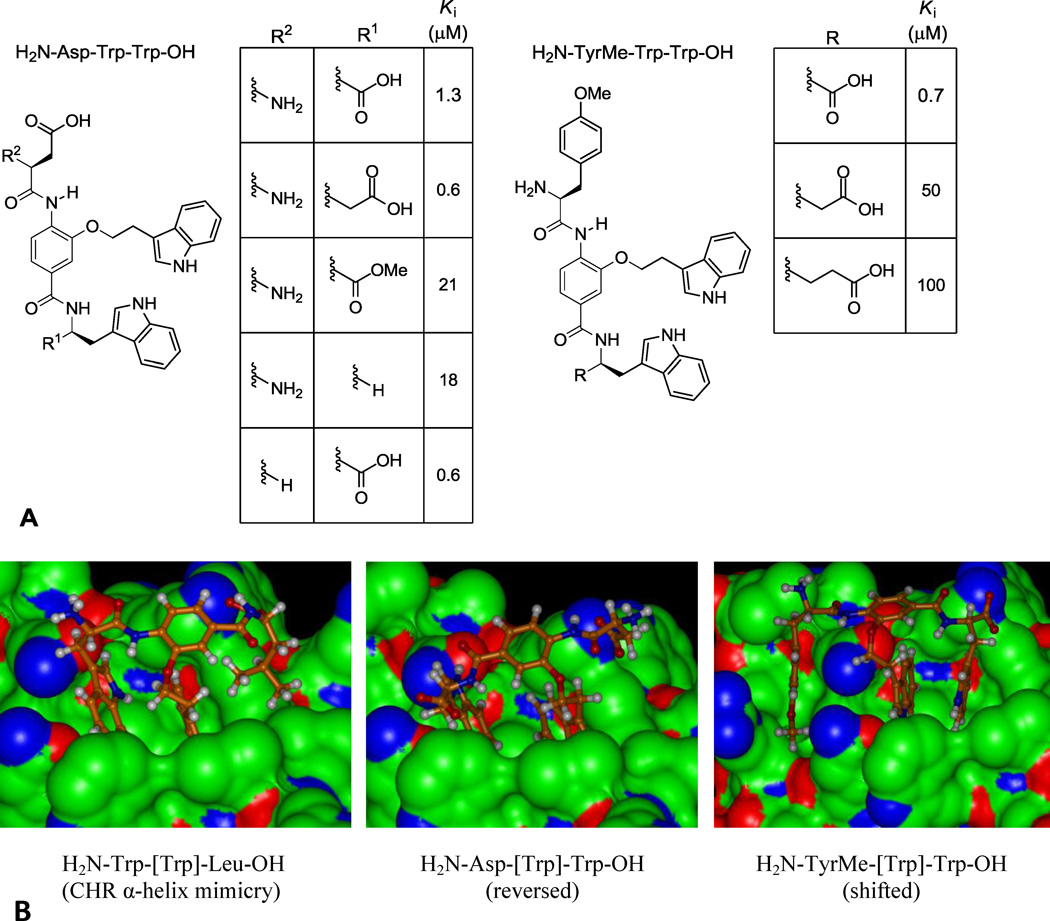
Consistent with expected mimicry of the CHR α-helix, the XXX-[Trp]-Leu series displayed an N-terminal preference for Trp with H2N-Trp-[Trp]-Leu-OH displaying a Ki = 1.3 µM (Figure 3A), being the most potent compound in the series and representing the identification of the expected binding partner from the library (Figure 4B). Also highly active were the compounds with N-terminal Tyr (Ki = 1.3 µM) and TyrMe (Ki = 3.9 µM) that are able to support a productive interaction with the Trp628 pocket as well. In a departure from these trends, the XXX-[Trp]-Trp series showed an N-terminal preference for polar side chains, with Asp or Asn giving rise to compounds with Ki = 1.3 and 0.8 µM, Figure 3D. This activity suggests that the compounds bind in the reversed orientation when Asp or Asn are present to make a favorable interaction with Lys574, leading to mimicry of the Trp628, Trp631, Asp632 sequence in the CHR α-helix, Figure 4B. The exception to this trend was the compound with N-terminal TyrMe, with a measured Ki of 0.7 µM. An alternative interaction that may be favored by H2N-XXX-[Trp]-Trp-OH (XXX = Asp, Asn, or TyrMe) is a shifted binding mode in which the C-terminal Trp mimics Trp631 and the central subunit [Trp] mimics Trp628, Figure 4B. This mode would facilitate salt bridge formation between the terminal carboxylate and Lys574 and enable a productive N-terminal interaction with Gln577 or Gln575 at the pocket periphery.
Figure 4.
(A) End group analysis of H2N-Asp-[Trp]-Trp-OH and H2N-TryMe-[Trp]-Trp-OH. (B) Proposed binding modes for indicated compounds (PDB 1IF3).
In order to clarify the binding of H2N-Asp-[Trp]-Trp-OH, we prepared four structures in which we removed or modified either the terminal amine or carboxylate (Figure 4A). The corresponding Ki’s demonstrate that the carboxylate is critical for binding, as its removal or neutralization results in a 20-fold loss in affinity. The single methylene extension of the carboxylic acid resulted in a 2-fold greater affinity (Ki 600 nM), while the removal of the amine also improved affinity 2-fold (Ki 600 nM). Extension of the analogous terminal carboxylate of H2N-TyrMe-[Trp]-Trp-OH by one or two methylenes, however, resulted in large losses in affinity (>50-fold), indicating that this compound possesses a binding mode distinct from H2N-Asp-[Trp]-Trp- OH (Figure 4A). It is likely this reflects the reversed binding mode for H2N-Asp-[Trp]-Trp-OH with mimicry of Trp628, Trp631, and Asp632 and a shifted binding mode for H2N-TyrMe-[Trp]-Trp-OH reflecting a productive additional N-terminal interaction in the pocket.
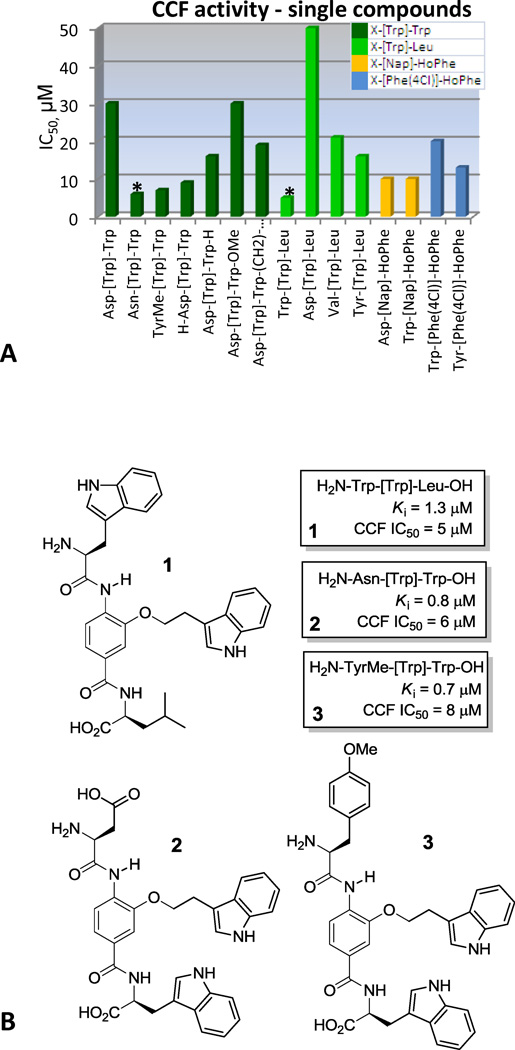
Analysis of single compounds in the functional CCF assay (Figure 5A) demonstrated that the most potent fusion inhibitor was H2N-Trp-[Trp]-Leu-OH (IC50 5 µM), whose sequence corresponds to the key CHR α-helix residues. Good fusion activity was also demonstrated by the XXX-[Trp]-Trp compounds, with N-terminal residues of Asn or TyrMe providing IC50 values of 6–8 µM, representing additional lead structures likely bound in the reversed (H2N-Asn-[Trp]-Trp-OH) or shifted (H2N-TyrMe-[Trp]-Trp-OH) orientations.
Figure 5.
(A) IC50’s of individual compounds in CCF assay. Bars are color coded according to the central and C-terminal side chains. (B) New lead compounds discovered.
The use of a comprehensive α-helix mimetic library is described, targeting the NHR hydrophobic pocket of the HIV-1 envelope glycoprotein gp41 and inhibiting binding of the CHR α-helix. The library evaluation identified the aromatic [Trp], [Phe(4Cl)], [Nap], and [Tyr] central subunits as the dominant feature in active mixtures, and Ile emerged as the best C-terminal residue. The individual compounds in the XXX-[Nap]-HoPhe, XXX-[Phe(4Cl)]-HoPhe and XXX-[Trp]-Leu series revealed that they displayed an N-terminal preference for Trp, indicating that they mimic the Trp-Trp-Ile helical face bound in an N- to C-terminal orientation. The most potent compound in the series was H2N-Trp-[Trp]-Leu-OH (Ki = 1.3 µM, CCF IC50 = 5 µM), and corresponds to the key CHR α-helix interaction residues. The XXX-[Trp]-Trp series also yielded high affinity compounds H2N-Asn-[Trp]-Trp-OH (Ki 0.8 µM) and H2N-Asp-[Trp]-Trp-OH (Ki 1.3 µM), suggesting that they may bind in the reverse orientation when a polar N-terminal residue is present, mimicking a role of Asp632 in the CHR α-helix. Modifications of H2N-Asp-[Trp]-Trp-OH to further probe the interaction provided inhibitors with Ki’s as low as 600 nM. Finally, the activity of H2N-Tyr(Me)-[Trp]-Trp-OH (Ki 700 nM) revealed an additional binding opportunity likely involving a new N-terminus interaction when bound in the expected orientation. The α-helix mimetic library therefore directly provided exciting new lead structures (Figure 5b) that match or exceed the potency of small molecules discovered over the last decade, validating its α-helix mimicry and the power it possesses for interrogating protein–protein interactions. It also directly yielded key insights into both expected as well as new binding modes that will prove valuable for lead optimization.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health [CA42056, DLB; GM87998 and NS59403, MG].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information. Supplementary data associated with this article can be found in the online version at xxxxx.
References
- 1.Tilton JC, Doms RW. Antiviral Res. 2010;85:91. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Qadir MI, Malik SA. Rev. Med. Virol. 2010;20:23. doi: 10.1002/rmv.631. [DOI] [PubMed] [Google Scholar]
- 3.Hertje M, Zhou M, Dietrich U. ChemMedChem. 2010;5:1825. doi: 10.1002/cmdc.201000292. [DOI] [PubMed] [Google Scholar]
- 4.Eggink D, Berkhout B, Sanders RW. Curr. Pharm. Des. 2010;16:3716. doi: 10.2174/138161210794079218. [DOI] [PubMed] [Google Scholar]
- 5.Chan DC, Chutkowski CT, Kim PS. Proc. Natl. Acad. Sci. USA. 1998;95:15613. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou G, Wu D, Snyder B, Ptak RG, Kaur H, Gochin M. J. Med. Chem. 2011;54:7220. doi: 10.1021/jm200791z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch BD, Francis JN, Redman JS, Paul S, Weinstock MT, Reeves JD, Lie YS, Whitby FG, Eckert DM, Hill CP, Root MJ, Kay MS. J. Virol. 2010;84:11235. doi: 10.1128/JVI.01339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai L, Jiang S. ChemMedChem. 2010;5:1813. doi: 10.1002/cmdc.201000289. [DOI] [PubMed] [Google Scholar]
- 9.Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Proc. Natl. Acad. Sci. USA. 2007;104:16828. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joly V, Jidar K, Tatay M, Yeni P. Expert Opin. Pharmacol. 2010;11:2701. doi: 10.1517/14656566.2010.522178. [DOI] [PubMed] [Google Scholar]
- 11.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Proc. Natl. Acad. Sci. USA. 2009;106:14751. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bautista AD, Stephens OM, Wang L, Domaoal RA, Anderson KS, Schepartz A. Bioorg. Med. Chem. Lett . 2009;19:3736. doi: 10.1016/j.bmcl.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J. Am. Chem. Soc. 2005;127:13126. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst JT, Kutzki O, Debnath AK, Jiang S, Lu H, Hamilton AD. Angew. Chem. Int. Ed. 2002;41:278. doi: 10.1002/1521-3773(20020118)41:2<278::aid-anie278>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 15.Tsou LK, Dutschman GE, Gullen EA, Telpoukhovskaia M, Cheng YC, Hamilton AD. Bioorg. Med. Chem. Lett. 2010;20:2137. doi: 10.1016/j.bmcl.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitby LR, Ando Y, Setola V, Vogt PK, Roth BL, Boger DL. J. Am. Chem. Soc. 2011;133:10184. doi: 10.1021/ja201878v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaginian A, Whitby LR, Hong S, Hwang I, Farooqi B, Searcey M, Chen JC, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:5564. doi: 10.1021/ja810025g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambrus G, Whitby LR, Singer EL, Trott O, Choi E, Olson AJ, Boger DL, Gerace L. Bioorg. Med. Chem. 2010;18:7611. doi: 10.1016/j.bmc.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst JT, Becerril J, Park HS, Yin H, Hamilton AD. Angew. Chem. Int. Ed. 2003;42:535. doi: 10.1002/anie.200390154. [DOI] [PubMed] [Google Scholar]
- 20.Cai L, Gochin M. Antimicrob. Agents Chemother. 2007;51:2388. doi: 10.1128/AAC.00150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gochin M, Savage R, Hinckley S, Cai L. Biol. Chem. 2006;387:477. doi: 10.1515/BC.2006.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gochin M, Kiplin Guy R, Case MA. Angew. Chem. Int. Ed. 2003;42:5325. doi: 10.1002/anie.200352006. [DOI] [PubMed] [Google Scholar]
- 23.Boger DL, Desharnais J, Capps K. Angew. Chem. Int. Ed. 2003;42:4138. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- 24.Cheng S, Tarby CM, Comer DD, Williams JP, Caporale LH, Myers PL, Boger DL. Bioorg. Med. Chem. 1996;4:727. doi: 10.1016/0968-0896(96)00069-7. [DOI] [PubMed] [Google Scholar]
- 25.Cheng S, Comer DD, Williams JP, Myers PL, Boger DL. J. Am. Chem. Soc. 1996;118:2567. doi: 10.1016/0968-0896(96)00069-7. [DOI] [PubMed] [Google Scholar]
- 26.Boger DL, Tarby CM, Myers PL, Caporale LH. J. Am. Chem. Soc. 1996;118:2109. [Google Scholar]
- 27.For library applications see: Chang MW, Giffin MJ, Muller R, Savage J, Lin YC, Hong S, Jin W, Whitby LR, Elder JH, Boger DL, Torbett BE. Biochem. J. 2010;429:527. doi: 10.1042/BJ20091645. Lee AM, Rojek JM, Spiropoulou CF, Gundersen AT, Jin W, Shaginian A, York J, Nunberg JH, Boger DL, Oldstone MBA, Kunz S. J. Biol. Chem. 2008;283:18734. doi: 10.1074/jbc.M802089200. Whitby LR, Lee AM, Kunz S, Oldstone MBA, Boger DL. Bioorg. Med. Chem. Lett. 2009;19:3771. doi: 10.1016/j.bmcl.2009.04.098. Eubanks LM, Hixon MS, Jin W, Hong S, Clancy CM, Tepp WH, Malizio CJ, Goodnough MC, Barbieri JT, Johnson EA, Boger DL, Dickerson TJ, Janda KD. Proc. Natl. Acad. Sci. USA. 2007;104:2602. doi: 10.1073/pnas.0611213104. Capps KJ, Humiston J, Dominique R, Hwang I, Boger DL. Bioorg. Med. Chem. Lett. 2005;15:2840. doi: 10.1016/j.bmcl.2005.03.094. Ambroise Y, Yuspan B, Ginsberg MH, Boger DL. Chem. Biol. 2002;9:1219. doi: 10.1016/s1074-5521(02)00246-6. Goldberg J, Jin Q, Ambroise Y, Satoh S, Desharnais J, Capps K, Boger DL. J. Am. Chem. Soc. 2002;124:544. doi: 10.1021/ja0118789. Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Proc. Natl. Acad. Sci. USA. 2002;99:3830. doi: 10.1073/pnas.062036999. Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Bioorg. Med. Chem. Lett. 2009;19:6038. doi: 10.1016/j.bmcl.2009.09.044. Silletti S, Kessler T, Goldberg J, Boger DL, Cheresh DA. Proc. Natl. Acad. Sci. USA. 2001;98:119. doi: 10.1073/pnas.011343298. Boger DL, Goldberg J, Silletti S, Kessler T, Cheresh DA. J. Am. Chem. Soc. 2001;123:1280. doi: 10.1021/ja003579+. Boger DL, Goldberg J, Satoh S, Ambroise Y, Cohen SB, Vogt PK. Helv. Chim. Acta. 2000;83:1825. For related DNA binding library applications, see: Stover JS, Shi J, Jin W, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:3342. doi: 10.1021/ja809083d. Boger DL, Fink BE, Hedrick MP. J. Am. Chem. Soc. 2000;122:6382. Boger DL, Fink BF, Brunette SR, Tse W, Hedrick MP. J. Am. Chem. Soc. 2001;123:5878. doi: 10.1021/ja010041a. Tse W, Boger DL. Acc. Chem. Res. 2004;37:61. doi: 10.1021/ar030113y. Hauschild KE, Stover JS, Boger DL, Ansari AZ. Bioorg. Med. Chem. Lett. 2009;19:3779. doi: 10.1016/j.bmcl.2009.04.097.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.