Abstract
Commensal bacteria that colonize mammalian barrier surfaces are reported to influence T helper type 2 (TH2) cytokine–dependent inflammation and susceptibility to allergic disease, although the mechanisms that underlie these observations are poorly understood. In this report, we identify that deliberate alteration of commensal bacterial populations via oral antibiotic treatment resulted in elevated serum immunoglobulin E (IgE) levels, increased steady–state circulating basophil populations, and exaggerated basophil–mediated TH2 cell responses and allergic inflammation. Elevated serum IgE levels correlated with increased circulating basophil populations in mice and subjects with hyperimmunoglobulinemia E syndrome. Furthermore, B cell–intrinsic expression of MyD88 was required to limit serum IgE levels and circulating basophil populations in mice. Commensal–derived signals were found to influence basophil development by limiting proliferation of bone marrow–resident precursor populations. Collectively, these results identify a previously unrecognized pathway through which commensal–derived signals influence basophil hematopoiesis and susceptibility to TH2 cytokine–dependent inflammation and allergic disease.
Allergic diseases have reached pandemic levels1 and represent a significant source of morbidity, mortality and healthcare cost2. These chronic inflammatory diseases are characterized by interleukin (IL) –4, IL–5, IL–9 and IL–13 production by CD4+ T helper type 2 (TH2) cells, immunoglobulin E (IgE) production, and the recruitment of effector cells to sites of tissue inflammation3, 4. It is thought that susceptibility to TH2 cytokine–dependent allergic inflammation is influenced by both polymorphisms in mammalian genes5 as well as environmental factors including diet and exposure to pollutants or infectious agents6-8. However, the specific genetic and environmental stimuli that influence allergy susceptibility, and how these factors contribute to the development of allergic disease are ongoing fields of study.
The human intestine is colonized by 100 trillion microorganisms belonging to each of the three domains of life9. Of these, bacteria are the most abundant; the colon is home to trillions of commensal bacteria10 with a diversity of at least 1,000–15,000 species11. Epidemiologic studies have identified associations between alterations in the composition of commensal bacterial communities and the development of allergic disease. For example, infants who develop allergies display altered commensal populations early in life12, and children who have undergone treatment with broad–spectrum antibiotics are at an increased risk of developing allergic diseases13, 14. Studies in animal model systems have further implicated commensal–derived signals in influencing the development of TH2 cytokine–mediated allergic inflammation15-18. However, the mechanisms through which the innate immune system recognizes commensal–derived signals and regulates TH2 cytokine responses remain poorly characterized19.
Here, oral delivery of broad–spectrum antibiotics was employed to interrogate the influence of commensal bacterial–derived signals on innate cell populations that contribute to the development of TH2 cytokine–dependent allergic inflammation. Depletion or deletion of bacterial communities was associated with elevated serum IgE levels, increased circulating basophil populations, and exaggerated TH2 cell responses and allergic inflammation. Exaggerated TH2 cell responses were reduced upon depletion of basophils, implicating this cell type in contributing to the exaggerated allergic inflammation observed in antibiotic–treated mice. IgE was found to be a critical regulator of steady–state basophil responses in mice, and subjects with hyperimmunoglobulinemia E syndrome had elevated frequencies of circulating basophils compared to controls. Additionally, B cell–intrinsic expression of MyD88 was required to limit serum IgE levels and circulating basophil populations in mice. Finally, commensal–derived signals influenced circulating basophil populations by regulating the proliferative capacity of bone marrow–resident basophil progenitor populations. Together, these findings provide therapeutically relevant insights into the molecular and cellular mechanisms through which commensal bacterial–derived signals influence the development of TH2 cytokine–dependent inflammation and susceptibility to allergic diseases.
RESULTS
Higher serum IgE levels and elevated circulating basophil populations following antibiotic–mediated manipulations of commensal bacteria in mice
Oral antibiotic treatment (ABX) resulted in quantitative and qualitative alterations to commensal bacteria colonizing the murine intestine including reductions in bacteria of the Firmicutes and Bacteroidetes phyla (Supplementary Fig. 1a,b) and significant increases in serum IgE levels20, 21 (Fig. 1a). As IgE is reported to influence granulocyte homeostasis22, we investigated whether ABX–induced elevations in IgE were associated with alterations in the frequency or number of circulating mast cells, eosinophils or basophils. ABX–treatment did not alter blood mast cell (Supplementary Fig. 2a,b) or eosinophil (Supplementary Fig. 2c,d) populations. However, frequencies and numbers of basophils (identified as non–B, non–T (NBNT), CD117−, CD49b+, FcεRIα+) were significantly increased in the blood (Fig. 1b,c) and spleen (Supplementary Fig. 3a,b) of ABX–treated compared to conventionally (CNV)–reared mice. Basophils from ABX–treated mice displayed increased levels of surface–bound IgE compared to controls (Fig. 1d), while expression of other surface markers (CD69, CD123, CD200R, FcεRIα, FcγR, Gr1) were unaltered (Supplementary Fig. 3c).
Figure 1.
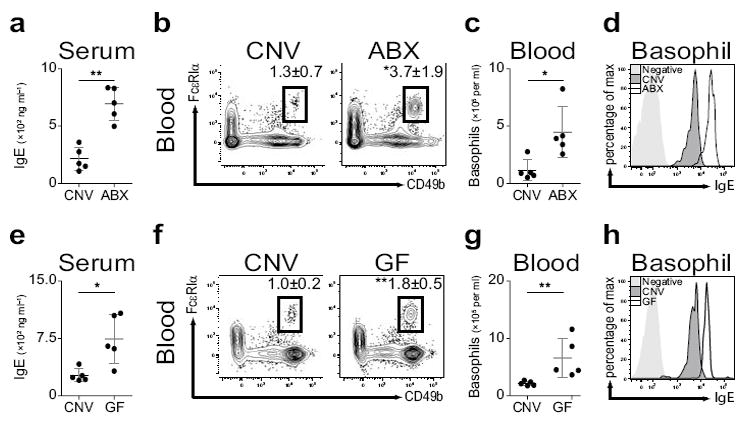
Elevated steady–state serum IgE levels and circulating basophils in antibiotic–treated or germ–free mice. (a) Serum IgE from conventionally–reared (CNV) or antibiotic–treated (ABX) mice as measured by ELISA. (b) Flow cytometric analysis of blood basophils from CNV or ABX mice. Numbers adjacent to outlined areas indicate percent cells in each gate. Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells. (c) Number of basophils per ml of blood from CNV or ABX mice. (d) Mean fluorescence intensity of surface–bound IgE on blood basophils from CNV or ABX mice as determined by flow cytometry. (e) Serum IgE from CNV or germ–free (GF) mice as measured by ELISA. (f) Flow cytometric analysis of blood basophils from CNV or GF mice. (g) Number of basophils per ml of blood from CNV or GF mice. (h) Mean fluorescence intensity of surface–bound IgE on blood basophils from CNV or GF mice as determined by flow cytometry. Data representative of three or more independent experiments, results shown as mean ± s.d., significance determined by Mann–Whitney test (CNV, n=5; ABX, n=5; GF, n=5; *, P ≤ 0.05; **, P ≤ 0.01).
ABX treatment does not eliminate all commensal bacteria23. Therefore, to investigate whether steady–state levels of IgE or basophil populations were altered in the absence of all live microbial stimuli, we employed germ–free (GF) mice24. Consistent with the effects of ABX treatment, compared to CNV–reared controls, GF mice exhibited increased serum IgE levels (Fig. 1e), increased frequencies and numbers of basophils in the blood (Fig. 1f,g) and spleen (Supplementary Fig. 3d,e), and increased basophil–surface–bound IgE levels (Fig. 1h). Consistent with commensal–derived signals being sufficient to limit serum IgE levels and circulating basophil populations, conventionalization of GF mice resulted in reductions in serum IgE levels (Supplementary Fig. 3f) and blood and spleen basophil populations (Supplementary Fig. 3g,h). Collectively, these data indicate that commensal bacterial–derived signals limit serum IgE levels and circulating basophil populations in the steady–state.
Commensal bacteria influence basophil–mediated allergic inflammation
Given the role for commensal–derived signals in limiting circulating numbers of basophils, we hypothesized that antibiotic–treated mice might display exaggerated inflammation in models of basophil–associated allergic disease. It has previously been shown that basophils contribute to the inflammation that results from inhalation of house dust mite allergen (HDM)25. To test whether commensal–derived signals influenced the development of HDM–induced inflammation, we exposed CNV–reared or ABX–treated IL–4/eGFP reporter mice26 to HDM and examined innate cell responses, TH2 cell responses, and lung inflammation. Compared to CNV–reared mice, HDM exposure in ABX–treated mice resulted in increased circulating basophil responses in the blood (Supplementary Fig. 4a), exaggerated TH2 cell responses in the draining mediastinal lymph node (LN) (Supplementary Fig. 4b), increased LN cell–derived IL–4 and IL–13 responses (Supplementary Fig. 4c), and increased lung and bronchoalveolar lavage (BAL) eosinophil responses (Supplementary Fig. 4d). Concurrently, compared to CNV–reared controls, ABX–treated mice displayed increased HDM–elicited inflammation characterized by exaggerated alveolar infiltrates (Fig. 2a). Together, these findings indicate that commensal–derived signals limit allergic inflammation in the lung.
Figure 2.
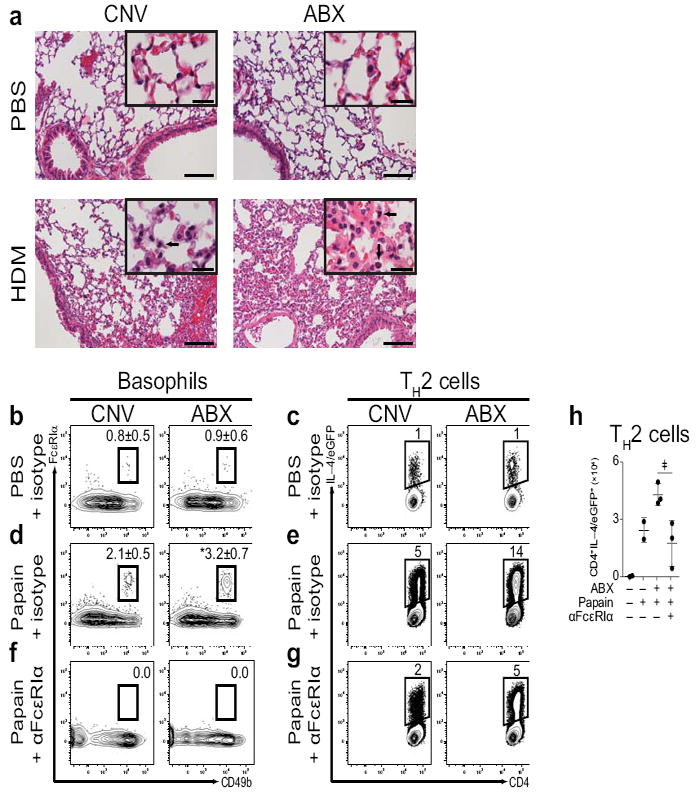
Exaggerated basophil–mediated allergic airway inflammation and TH2 cell responses in antibiotic–treated mice. (a) Histological sections of lungs from CNV or ABX mice exposed to PBS or HDM and stained with H&E (Large Bar = 100 μm; Small Bar = 20 μm; Arrows indicate eosinophilic infiltrates; CNV–PBS, n=2; ABX–PBS, n=1; CNV–HDM, n=3; ABX–HDM, n=2). (b) Flow cytometric analysis of day 3 popliteal LN (pLN) basophils from CNV or ABX mice treated with PBS and isotype control antibody. Numbers adjacent to outlined areas indicate percent cells in each gate. Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells. (c) Flow cytometric analysis of day 4 pLN IL–4/eGFP+CD4+ TH2 cells from CNV or ABX mice treated with PBS and isotype control antibody. Gated on CD8−, CD19−, CD4+ cells. (d) Flow cytometric analysis of day 3 pLN basophils from CNV or ABX mice treated with papain and isotype control antibody. (e) Flow cytometric analysis of day 4 pLN IL–4/eGFP+CD4+ TH2 cells from CNV or ABX mice treated with papain and isotype control antibody. (f) Flow cytometric analysis of day 3 pLN basophils from CNV or ABX mice treated with papain and anti–FcεRIα (αFcεRIα) antibody. (g) Flow cytometric analysis of day 4 pLN IL–4/eGFP+CD4+ TH2 cells from CNV or ABX mice treated with papain and αFcεRIα antibody (CNV–PBS–ISO, n=2–5; ABX–PBS–ISO, n=2–5; CNV–PAP–ISO, n=5; ABX–PAP–ISO, n=5; CNV–PAP–αFcεRIα, n=2; ABX–PAP–αFcεRIα, n=2; significance determined by Mann–Whitney test; *, P ≤ 0.05). (h) Number of day 4 pLN IL–4/eGFP+CD4+ TH2 cells from CNV or ABX mice treated with PBS or papain and isotype control (−) or αFcεRIα (+) antibody (means of three experiments ± s.d.; CNV–PBS–ISO, n=4; CNV–PAP–ISO, n=4; ABX–PAP–ISO, n=8; ABX–PAP–αFcεRIα, n=6; significance determined by 2 way ANOVA; ‡, P ≤ 0.06). Data representative of two or more independent experiments, results shown as mean ± s.d.
Basophils are recruited to draining LNs early in the response to infectious27 or allergic28, 29 stimuli where they may cooperate with dendritic cells (DCs) to promote optimal TH2 cell responses27, 29-31. To test whether ABX–treated mice exhibited altered allergen–induced TH2 cell responses, we exposed CNV–reared or ABX–treated IL–4/eGFP reporter mice to PBS or papain, a cysteine protease associated with occupational allergy in humans32 and TH2 cell responses in mice28. Following PBS–exposure, similar frequencies of basophils (Fig. 2b) and IL–4/eGFP+CD4+ TH2 cells (Fig. 2c) were observed in the popliteal LNs of CNV–reared or ABX–treated mice. However, following exposure to papain, ABX–treated mice exhibited increased frequencies of LN basophils (Fig. 2d) and IL–4/eGFP+CD4+ TH2 cells (Fig. 2e) compared to CNV–reared controls. Depletion of basophils by administration of anti–FcεRIα antibody28 resulted in a reduction in the frequency of LN basophils (Fig. 2f), and reduced frequencies (Fig. 2g) and numbers (Fig. 2h) of papain–elicited IL–4/eGFP+CD4+ TH2 cells, indicating that basophils contribute to the exaggerated TH2 cell responses observed in ABX–treated mice.
DCs are critically important for the development of optimal TH2 cell responses to some allergens and helminth parasites25, 33. To investigate the influence of commensal–derived signals on steady–state DC populations, we examined DCs in CNV–reared or ABX–treated mice. DC frequencies in the spleen were similar between CNV–reared or ABX–treated mice (data not shown) as was DC expression of MHC–I, MHC–II, CD40, CD80, CD86, and FcεRIα (Supplementary Fig. 5a–c), indicating that commensal–derived signals do not significantly influence splenic DC populations in the steady–state. Non–basophil FcεRIα+ cells are important for the development of inflammatory responses to some allergens25. Papain–elicited, non–basophil FcεRIα+ cell frequencies in the LN were similar between CNV–reared or ABX–treated mice (Supplementary Fig. 5d–f) indicating that non–basophil FcεRIα+ cell responses are not significantly influenced by commensal–derived signals in this setting. As FcεRIα+ cells are influenced by anti–FcεRIα antibody treatment25, we examined the effect of anti–FcεRIα antibody treatment on non–basophil FcεRIα+ cell populations. Compared to controls, anti–FcεRIα antibody treatment did not significantly influence the frequency of papain–elicited, non–basophil FcεRIα+ LN cells (Supplementary Fig. 5d–f). Together, these findings suggest that neither steady–state DC responses nor papain–elicited non–basophil FcεRIα+ cell responses are significantly dysregulated in ABX–treated mice.
To further examine the contributions of basophils to the exaggerated TH2 cell responses observed in ABX–treated mice, we adopted a loss–of–function approach complementary to anti–FcεRIα antibody treatment by utilizing a basophil–specific diphtheria toxin (DT)–dependent depletion mouse system (BaS–TRECK)34. ABX–treatment resulted in increased frequencies of circulating basophils (Supplementary Fig. 5g) and exaggerated frequencies (Supplementary Fig. 5h) and numbers (Supplementary Fig. 5i) of papain–elicited LN TH2 cells. DT–treatment of CNV-reared or ABX–treated BaS–TRECK mice, but not littermate controls, resulted in efficient depletion of LN basophils (Supplementary Fig. 5g) and reductions in the frequency (Supplementary Fig. 5h) and number (Supplementary Fig. 5i) of TH2 cells. Together with the results of anti–FcεRIα antibody–mediated basophil depletion, DT receptor–mediated deletion indicates that basophils contribute to the exaggerated allergen–induced TH2 cell responses observed in ABX–treated mice.
Signals derived from commensal bacteria influence circulating basophil populations via an IgE–dependent mechanism
We next sought to determine the mechanisms by which commensal–derived signals regulate steady–state circulating basophil populations. The epithelial cell–derived cytokine thymic stromal lymphopoietin (TSLP) was recently found to expand circulating basophil populations34. To test whether TSLP was required for the elevated serum IgE levels or circulating basophil populations observed in ABX–treated mice, we utilized TSLP–deficient (Tslp−/−) mice. As in WT mice, antibiotic treatment of Tslp−/− mice resulted in elevated serum IgE levels (Supplementary Fig. 6a), and increased circulating basophil frequencies and numbers (Supplementary Fig. 6b,c), indicating that TSLP is not necessary for these responses to commensal alteration.
As IgE regulates mast cell homeostasis22 and has been hypothesized to have similar roles in basophils35, we examined the relationship between elevated IgE levels and increased circulating basophil populations in ABX–treated mice. Serum IgE levels significantly correlated with circulating basophil numbers in CNV–reared or ABX–treated mice (Fig. 3a), suggesting that IgE may influence basophil homeostasis. To test whether the increase in circulating basophils observed in ABX–treated mice was dependent on lymphocytes, we employed recombination activating gene 1–deficient mice (Rag1−/−)36. Consistent with a role for IgE in regulating basophil homeostasis, blood basophil frequencies and numbers were similar between CNV–reared and ABX–treated (Fig. 3b,c) or GF Rag1−/− mice (Supplementary Fig. 7a,b). As IL–4 is necessary for IgE production37, we utilized IL–4–deficient mice (Il4−/−) to test the role of IL–4 in mediating the elevated serum IgE levels and basophil responses observed in ABX–treated mice. While ABX–treatment of wild–type (WT) mice resulted in increased serum IgE levels (Supplementary Fig. 7c) and blood basophils (Supplementary Fig. 7d), IgE was not detected in Il4−/− mice (Supplementary Fig. 7c) and ABX–treatment of Il4−/− mice did not significantly influence the frequency or number of blood basophils (Supplementary Fig. 7d,e). Together, these results suggest that IL–4–induced IgE production contributes to the increased circulating basophil responses observed in ABX–treated mice.
Figure 3.
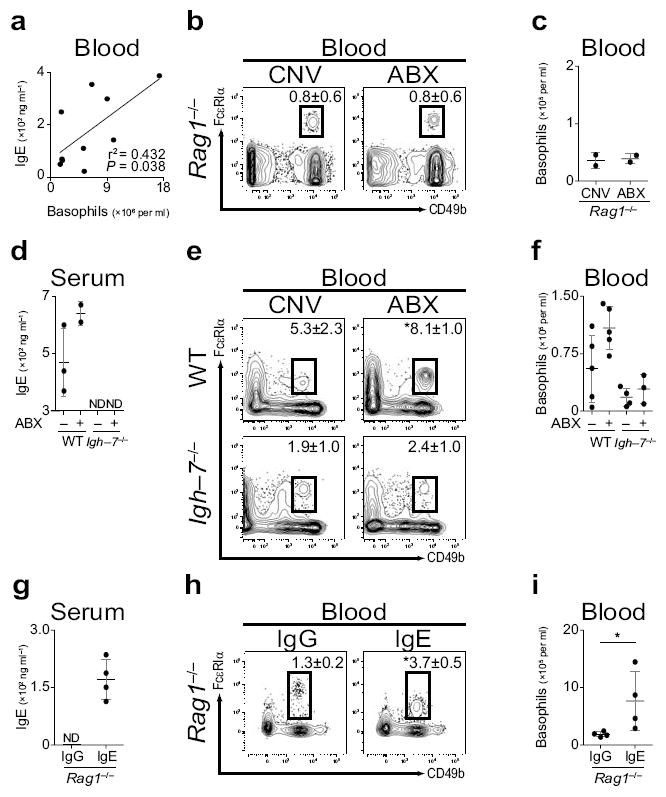
IgE correlates with circulating basophil populations in mice. (a) Statistical correlation of blood basophil number and serum IgE concentration (significance determined by linear regression analysis; r2 = 0.432; P ≤ 0.038). (b) Flow cytometric analysis of blood basophils from CNV or ABX Rag1−/− mice. Numbers adjacent to outlined areas indicate percent cells in each gate (CNV, n=4; ABX, n=5). Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells. (c) Number of basophils per ml of blood from CNV or ABX Rag1−/− mice (CNV, n=2; ABX, n=2). (d) Serum IgE from CNV or ABX wild–type (WT) or Igh–7−/− mice as measured by ELISA (ND, not detected). (e) Flow cytometric analysis of blood basophils from CNV or ABX WT or Igh–7−/− mice (CNV–WT, n=5; ABX–WT, n=5; CNV–Igh–7−/−, n=4; ABX–Igh–7−/−, n=3). (f) Number of basophils per ml of blood from CNV or ABX WT or Igh–7−/− mice (CNV–WT, n=5; ABX–WT, n=5; CNV–Igh–7−/−, n=4; ABX–Igh–7−/−, n=3). (g) Serum IgE levels from IgG or IgE treated Rag1−/− mice as measured by ELISA (IgG, n=4; IgE, n=4; ND, not detected). (h) Flow cytometric analysis of blood basophils from IgG or IgE treated Rag1−/− mice (IgG, n=4; IgE, n=4). (i) Number of basophils per ml of blood from IgG or IgE treated Rag1−/− mice (IgG, n=4; IgE, n=4). Data representative of three or more independent experiments, results shown as mean ± s.d., significance determined by Mann–Whitney test unless otherwise indicated (*, P ≤ 0.05).
To directly test the role of IgE in mediating commensal bacterial–dependent regulation of basophil responses, we utilized IgE–deficient (Igh–7−/−) mice. While ABX–treatment of WT mice resulted in increased serum IgE levels (Fig. 3d) and blood basophils (Fig. 3e,f), IgE was not detected in Igh–7−/− mice (Fig. 3d) and blood basophil populations were similar between CNV–reared or ABX–treated Igh–7−/− mice (Fig. 3e,f). Together, these results indicate that IgE is necessary to mediate the exaggerated steady–state circulating basophil responses observed in ABX–treated mice. To test whether IgE was sufficient to promote the population expansion of circulating basophils, we treated Rag1−/− mice with IgE and examined blood basophil populations. Compared to IgG, transfer of IgE into Rag1−/− mice resulted in recovery of serum IgE levels (Fig. 3g) and significantly increased frequencies and numbers of blood basophils (Fig. 3h,i). Collectively, these gain– and loss–of–function approaches identify IgE as one regulator of peripheral basophil responses in mice.
Serum IgE levels correlate with blood basophil frequencies in humans
As IgE was found to regulate circulating basophil populations in mice, we hypothesized that IgE would have a similar role in humans. Subjects with loss–of–function polymorphisms in the dedicator of cytokinesis 8 gene (DOCK8−) exhibit an autosomal recessive form of hyperimmunoglobulinemia E syndrome38, 39 characterized by recurrent infections, increased susceptibility to atopic eczema, and average serum IgE levels 10 times higher than those found in control subjects38, 39. Based on the identification of a role for IgE in regulating basophil responses in mice, we hypothesized that elevated serum IgE levels in DOCK8− subjects may be associated with increased circulating basophil populations. As previously reported, DOCK8− subjects displayed elevated serum IgE levels compared to control subjects38, 39 (Fig. 4a). In control subjects, human blood basophils (identified as TCRβ−, CD11c−, CD19−, CD117−, CD123+, FcεRIα+) comprised approximately 1% of the NBNT cell compartment (Fig. 4b,d). In contrast, in four of five DOCK8− subjects examined, basophils comprised approximately 4% of the NBNT compartment (Fig. 4c,d). Further, basophils isolated from DOCK8− subjects exhibited increased surface–bound IgE levels compared to those of control subjects (Fig. 4e). These data are consistent with our findings in animal models and suggest that IgE may influence peripheral basophil homeostasis and the development of basophil–mediated allergic inflammation in humans.
Figure 4.
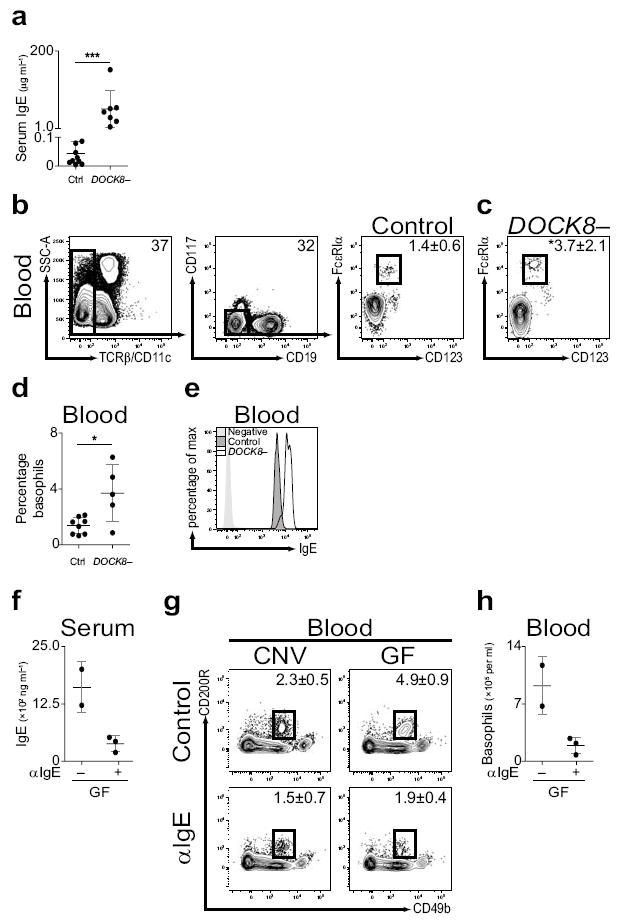
Serum IgE levels correlate with circulating basophil populations in human subjects with hyperimmunoglobulinemia E syndrome. (a) Serum IgE levels from control (Ctrl) or subjects with polymorphisms in DOCK8 (DOCK8–) (Ctrl, n=15; DOCK8–, n=7). (b) Identification of blood basophils from control subjects by flow cytometric analysis as TCRβ−, CD11c−, CD19−, CD117−, FcεRIα+, CD123+ cells. (c) Flow cytometric analysis of blood basophils from DOCK8– subjects. (d) Frequency of basophils in blood from control or DOCK8– subjects (Ctrl, n=8; DOCK8–, n=5). (e) Mean fluorescence intensity of surface–bound IgE on blood basophils from control or DOCK8– subjects as determined by flow cytometry. (f) Serum IgE from germ–free (GF) mice treated with control (−) or IgE–specific antibody (αIgE; +) antibody as measured by ELISA (control, n=2; αIgE, n=3). (g) Flow cytometric analysis of blood basophils from CNV or GF mice treated with control or αIgE antibody. Numbers adjacent to outlined areas indicate percent cells in each gate (CNV–control, n=2; GF–control, n=2; CNV–αIgE, n=3; GF–αIgE, n=3). Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells. (h) Number of basophils per ml of blood from GF mice treated with control or αIgE antibody (control, n=2; αIgE, n=3). Data representative of three or more independent experiments, results shown as mean ± s.d., significance determined by Mann–Whitney test (**, P ≤ 0.01; ***, P ≤ 0.001).
Given the potential role for IgE in regulating circulating basophil populations in mice and human subjects, we hypothesized that therapeutic depletion of serum IgE levels may reduce circulating basophil populations. Omalizumab (IgE–specific antibody) is a monoclonal, humanized mouse anti–human IgE–specific antibody and an FDA approved anti–allergy drug40, 41. To test whether IgE–specific antibody treatment influenced serum IgE levels or circulating basophil populations, we treated CNV–reared or GF mice with control or IgE–specific antibody and examined serum IgE levels and blood basophil populations. As IgE–specific antibody treatment decreases FcεRIα expression on basophils42, we identified basophils as NBNT, CD117−, CD49b+, CD200R+ cells43. IgE–specific antibody treatment of GF mice resulted in reductions in serum IgE levels (Fig. 4f), and reduced frequencies and numbers of blood basophils (Fig. 4g,h). Together, these results identify IgE–specific antibody treatment as a potential therapeutic intervention to limit IgE–mediated increases in circulating basophil populations.
MyD88–dependent signaling in B cells limits serum IgE levels and circulating basophil populations
It has been hypothesized that commensal bacterial–derived signals influence allergic responses by signaling through pattern recognition receptors (PRRs)15, 17. To interrogate the molecular mechanisms through which commensal–derived signals influence serum IgE levels and circulating basophil responses, we utilized mice deficient in myeloid differentiation primary response gene 88 (Myd88−/−) which encodes a critical adaptor molecule that regulates signaling through multiple PRRs44. Compared to littermate controls, CNV–reared Myd88−/− mice exhibited increased serum IgE levels44 (Fig. 5a), increased frequencies and numbers of blood basophils (Fig. 5b,c) and increased basophil–surface–bound IgE levels (Fig. 5d) indicating that MyD88 signaling pathways limit these responses.
Figure 5.
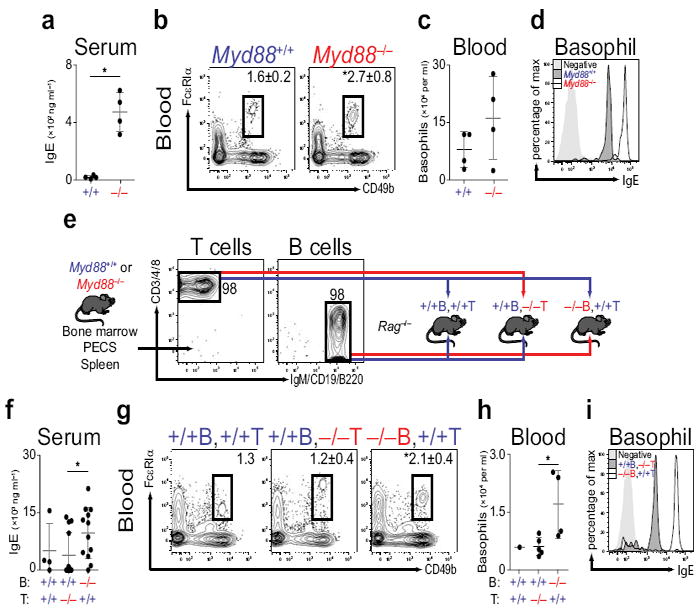
Elevated serum IgE levels and circulating basophil populations in mice lacking B cell–intrinsic MyD88 signaling. (a) Serum IgE from conventionally–reared (CNV) Myd88+/+ (+/+) or Myd88−/− (−/−) mice as measured by ELISA (Myd88+/+, n=4; Myd88−/−, n=4). (b) Flow cytometric analysis of blood basophils from CNV Myd88+/+ or Myd88−/− mice. Numbers adjacent to outlined areas indicate percent cells in each gate (Myd88+/+, n=4; Myd88−/−, n=4). Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells. (c) Number of basophils per ml of blood from CNV Myd88+/+ or Myd88−/− mice (Myd88+/+, n=4; Myd88−/−, n=4). (d) Mean fluorescence intensity of surface–bound IgE on blood basophils from CNV Myd88+/+ or Myd88−/− mice as determined by flow cytometry. (e) Experimental diagram of sort and transfer of Myd88+/+ or Myd88−/− B or T cells into Rag1−/− recipients. (f) Serum IgE from Rag1−/− mice reconstituted with Myd88+/+ or Myd88−/− B or T cells as measured by ELISA (+/+B +/+T, n=4; +/+B −/−T, n=12; −/−B +/+T, n=12). (g) Flow cytometric analysis of blood basophils from Rag1−/− mice reconstituted with Myd88+/+ or Myd88−/− B or T cells (+/+B +/+T, n=1; +/+B −/−T, n=5; −/−B +/+T, n=4). (h) Number of basophils per ml of blood from Rag1−/− mice reconstituted with Myd88+/+ or Myd88−/− B or T cells (+/+B +/+T, n=1; +/+B −/−T, n=5; −/−B +/+T, n=4). (i) Mean fluorescence intensity of surface–bound IgE on blood basophils from Rag1−/− mice reconstituted with Myd88+/+ or Myd88−/− B or T cells as determined by flow cytometry. Data representative of three or more independent experiments, results shown as mean ± s.d., significance determined by Mann–Whitney test (*, P ≤ 0.05).
Nucleotide–binding oligomerization domain–containing protein 1 (NOD1) is a MyD88–independent intracellular PRR that mediates innate and adaptive immunity by recognizing commensal bacterial–derived signals45. To test whether NOD1 influenced steady–state circulating basophil populations, we examined serum IgE levels and circulating basophil populations in control or NOD1–deficient (Nod1−/−) mice. Neither serum IgE levels (Supplementary Fig. 8a) nor blood basophils populations were altered in Nod1−/− compared to control mice (Supplementary Fig. 8b,c), indicating that NOD1–dependent signaling does not significantly influence steady–state serum IgE levels or circulating basophil responses in this setting. To examine whether MyD88–dependent bacterial–derived signals were sufficient to limit serum IgE levels or circulating basophil populations, we treated CNV–reared or ABX–treated mice with CpG15, 46 and examined serum IgE levels and circulating basophil populations. CpG treatment of ABX–treated mice resulted in reductions in serum IgE levels (Supplementary Fig. 8d) and reduced frequencies and numbers of blood basophil populations (Supplementary Fig. 8e,f). Together, these findings identify MyD88–dependent signaling pathways as important regulators of steady–state serum IgE levels and circulating basophil populations in mice.
MyD88–dependent signaling in B cells can inhibit IgE class switching in vitro47. To test whether B cell–intrinsic expression of MyD88 influenced serum IgE levels or basophil homeostasis in vivo, we generated chimeras by sorting and adoptively transferring B or T cells from Myd88+/+ or Myd88−/− mice into Rag1−/− recipients (Fig. 5e). Compared to controls, mice that received Myd88−/− B cells with Myd88+/+ T cells displayed elevated serum IgE levels (Fig. 5f), increased frequencies (Fig. 5g) and numbers (Fig. 5h) of blood basophils, and increased basophil–surface–bound IgE levels (Fig. 5i). Together, these results indicate that B cell–intrinsic MyD88 expression limits steady–state serum IgE levels and circulating basophil populations in vivo.
Commensal bacteria influence basophil development by limiting IgE–mediated proliferation of a bone marrow–resident precursor population
We hypothesized that commensal–derived signals may regulate circulating basophil populations by influencing basophil survival. To test this, we sort–purified basophils from CNV–reared or ABX–treated mice and cultured them in the presence of IL–3. Basophils isolated from CNV–reared or ABX–treated mice exhibited similar survival (Supplementary Fig. 9a), suggesting that commensal–derived signals do not readily influence basophil survival in this setting. As IgE was found to regulate commensal–dependent alterations in circulating basophil populations, we sought to investigate whether IgE influenced basophil survival in this assay. Basophils isolated from control or Igh–7−/− mice exhibited comparable survival rates (Supplementary Fig. 9b), suggesting that IgE does not readily influence basophil survival in this setting.
Rather than influencing survival, we hypothesized that commensal–derived signals may influence basophil development from the bone marrow. To test whether proliferation of basophils from bone marrow cells was influenced by commensal–derived signals, we CFSE labeled bone marrow from CNV–reared or ABX–treated mice, cultured it in the presence of IL–3, and examined mature basophil populations48. Compared to basophils derived from bone marrow of CNV–reared mice, a greater proportion of basophils derived from bone marrow of GF mice had diluted CFSE fluorescence and expressed the cell proliferation marker Ki67 (Fig. 6a) indicating that they originated from precursors that had undergone more rounds of cell division. Consistent with this, cultures of bone marrow cells from GF (Fig. 6b) or ABX–treated (Supplementary Fig. 9c) mice exhibited a greater population expansion of basophils compared to equal numbers of bone marrow cells isolated from CNV–reared mice. Together, these results indicate that commensal–derived signals limit the proliferative capacity of bone marrow–resident basophil precursor populations.
Figure 6.
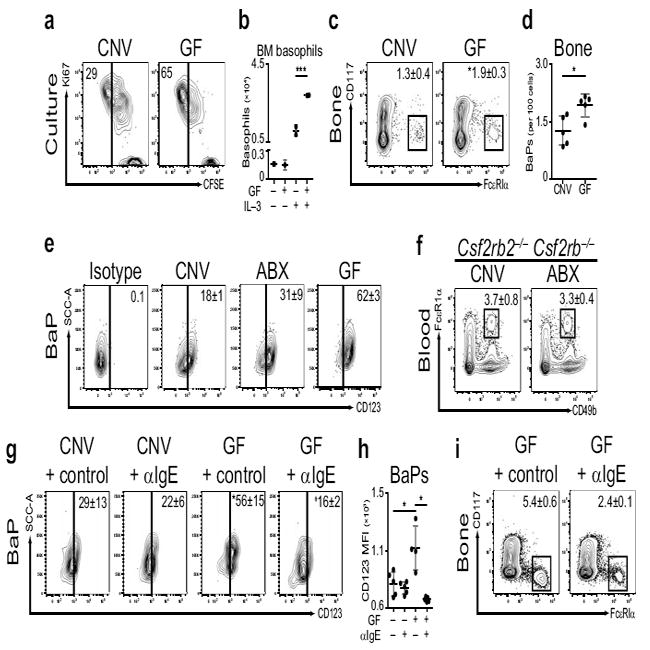
Dysregulated basophil development in germ–free or antibiotic–treated mice. (a) Bone marrow of conventionally–reared (CNV) or germ–free (GF) mice was CFSE labeled, cultured in the presence of IL–3, and basophil populations were examined by flow cytometry. Numbers adjacent to outlined areas indicate percent cells in each gate. Gated on CD3−, CD4−, CD8−, CD19−, CD117−, CD49b+, FcεR1α+ cells. (b) Bone marrow of CNV (−) or GF (+) mice was cultured in the absence (−) or presence (+) of IL–3 and basophil numbers were determined (means of two experiments ± s.d.; CNV–media, n=5; GF–media, n=3; CNV–IL–3, n=10; GF–IL–3, n=8; significance determined by 2 way ANOVA; ***, P ≤ 0.001). (c) Flow cytometric analysis of BaPs in bone marrow of CNV or GF mice. Gated on CD3−, CD4−, CD8−, CD19−, CD34+ cells (CNV, n=5; GF, n=5). (d) Number of BaPs in bone marrow of CNV or GF mice (CNV, n=5; GF, n=5). (e) Frequency of CD123+ BaPs in bone marrow of CNV, antibiotic–treated (ABX) or GF mice as determined by flow cytometry. Gated on CD3−, CD4−, CD8−, CD19−, CD117−, CD34+, FcεR1α+ cells (CNV, n=2; ABX, n=2; GF, n=2). (f) Flow cytometric analysis of blood basophils from CNV or ABX Csf2rb−/−Csf2rb−/− mice. Gated on CD3−, CD4−, CD8−, CD19−, CD117− cells (CNV, n=3; ABX, n=3). (g) Frequency of CD123+ BaPs in bone marrow of CNV or GF mice treated with control or IgE–specific antibody (αIgE) (CNV–control, n=5; CNV–αIgE, n=5; GF–control, n=4; GF–αIgE, n=5; CNV–control vs. GF–control, *, P ≤ 0.05; GF–control vs. GF–αIgE, ‡, P ≤ 0.05). (h) Mean fluorescence intensity of CD123 on BaPs in bone marrow of CNV or GF mice treated with control or αIgE antibody (CNV–control, n=5; CNV–αIgE, n=5; GF–control, n=4; GF–αIgE, n=5; *, P ≤ 0.05). (i) Flow cytometric analysis of BaPs in bone marrow of GF mice treated with control or αIgE antibody (GF–control, n=2; GF–αIgE, n=3). Data representative of two or more independent experiments, results shown as mean ± s.d., significance determined by Mann–Whitney test unless otherwise indicated (*, P ≤ 0.05).
Bone marrow–resident basophils precursors (BaPs) are characterized by expression of FcεRIα and the hematopoietic progenitor adhesion molecule CD3449. To determine whether BaPs were influenced by alterations in commensal–derived signals, we isolated bone marrow from CNV–reared or GF mice and examined BaP populations. Frequencies of BaPs (identified as NBNT, CD117−, FcεRIα+, CD34+) were significantly increased in the bone marrow of GF compared to CNV–reared mice (Fig. 6c,d). Having identified a role for IgE in mediating the influence of commensal–derived signals on circulating basophil populations, we hypothesized that IgE might mediate commensal–dependent effects on bone marrow BaP populations. To test this, we isolated bone marrow from CNV–reared or ABX–treated Igh–7−/− mice and examined BaP populations. Frequencies of BaPs were similar between CNV–reared or ABX–treated Igh–7−/− mice (Supplementary Fig. 9d) suggesting that IgE contributes to the increased numbers of bone marrow BaP cells observed following ABX–treatment.
Commensal bacteria limit BaP surface expression of CD123 and responsiveness to IL–3
As the development of basophils from bone marrow–resident precursors is regulated in part by IL–3–IL–3 receptor (IL–3R) signaling50, we examined whether commensal–derived signals influenced BaP expression of the IL–3R subunit CD123. Compared to CNV–reared controls, BaPs in the bone marrow of ABX–treated or GF mice displayed increased surface–bound CD123 (Fig. 6e). Based on elevated surface-bound CD123, we hypothesized that BaPs from ABX–treated mice may be more responsive to IL–3 compared to BaPs from CNV–reared mice. To test this, we sort–purified BaPs from CNV–reared or ABX–treated mice and cultured them in equal numbers in the presence or absence of IL–3 (Supplementary Fig. 9e). In the presence of IL–3, BaPs from both CNV–reared or ABX–treated mice developed into mature basophil populations as indicated by reduced expression of CD34 (Supplementary Fig. 9f), expression of CD49b and FcεRIα (Supplementary Fig. 9g), and mature cellular morphology (Supplementary Fig. 9h,i). However, culture of BaPs isolated from ABX–treated mice resulted in the development of a greater number of mature basophils compared to culture of an equal number of BaPs isolated from CNV–reared mice (Supplementary Fig. 9j). Together, these results indicate that commensal–derived signals limit BaP populations in the bone marrow, limit surface–bound CD123 on BaPs, and regulate responsiveness of BaPs to IL–3.
Due to the increased surface–bound CD123 on BaPs of ABX–treated mice, and the increased responsiveness of these BaPs to IL–3, we hypothesized that the elevations in circulating basophils observed upon ABX–treatment of mice would be dependent on IL–3R–dependent signaling. To test this, we examined basophil responses in CNV–reared or ABX–treated mice deficient in both IL–3Rβil3 and IL–3Rβc (Csf2rb−/−Csf2rb−/−). ABX–treatment of Csf2rb−/−Csf2rb−/− mice did not significantly alter the frequency of blood basophil populations (Fig. 6f), suggesting that IL–3R signaling is necessary for ABX–induced elevations in circulating basophil populations. Therefore, the effects of commensal–derived signals on basophils are dependent in part on both IgE and IL–3R signaling.
IgE mediates increased surface–bound CD123 on BaP cells isolated from germ–free mice
Based on our findings that ABX–induced elevations in circulating basophils were dependent on IgE and IL–3R signaling, and correlated with bone marrow BaP surface–bound CD123, we hypothesized that IgE may regulate the increased CD123 surface expression on BaPs of GF mice. To test this hypothesis, we treated CNV–reared or GF mice with control or IgE–specific antibody and examined both surface–bound CD123 on BaPs and total bone marrow BaP frequencies. Compared to BaPs in the bone marrow of CNV–reared mice, BaPs in the bone marrow of GF mice exhibited significantly elevated expression of CD123 (Fig. 6g,h). Treatment of GF mice with IgE–specific antibody, but not control antibody, significantly reduced surface–bound CD123 on GF bone marrow BaPs (Fig. 6g,h) and reduced frequencies of BaPs in the bone marrow of GF mice (Fig. 6i). Collectively, these results indicate that commensal–derived signals influence bone marrow BaP populations in part by regulating IgE–mediated expression of CD123.
Discussion
In this study, oral delivery of broad–spectrum antibiotics was employed to interrogate the influence of commensal bacterial–derived signals on innate immune cell populations known to contribute to the development of TH2 cytokine–dependent allergic inflammation. Antibiotic treatment resulted in elevated serum IgE levels, increased circulating basophil populations, and exaggerated basophil–associated TH2 cell responses and allergic inflammation. IgE was identified as a critical regulator of steady–state circulating basophil populations in mice. These findings are consistent with studies that indicate that IgE can influence mast cell survival and function and suggest a broader immunoregulatory role for IgE in settings of infection and allergy 22. Additionally, serum IgE levels were found to correlate with increased circulating basophil frequencies in subjects with hyperimmunoglobulinemia E syndrome implicating IgE–mediated regulation of basophil populations in contributing to the increased susceptibility to allergies observed in these and other subjects with genetic or pathogenic elevations in circulating IgE levels38, 39.
Previous reports that utilized genomic deletions in Myd88 identified MyD88–dependent signaling as an important regulator of TH2 cytokine–associated inflammation44, 51. Here, B cell–intrinsic expression of MyD88 was identified as one regulatory pathway that limits steady–state serum IgE levels and circulating basophil populations in mice. This finding is supported by in vitro studies that indicate that CpG can inhibit IgE class switching by B cells47. Consistent with this, human subjects with deficiencies in MyD88 or IRAK–4 signaling pathways exhibit elevated serum IgE levels52. Given these associations, further examination of circulating basophil populations or susceptibility to allergic inflammation in these subject populations may be of value.
Commensal bacterial–derived signals were found to regulate basophil development from bone marrow–resident precursor populations by influencing the responsiveness of these precursors to IL–3. These findings suggest that in addition to regulating immune cells in the periphery, commensal–derived signals can alter mammalian hematopoietic programs in the bone marrow to promote or protect against the development of allergic responses. The identification of a role for commensal–derived signals in regulating hematopoiesis could have implications beyond allergy to other chronic inflammatory disease states that are associated with alterations in commensal bacteria including cancer, infection or autoimmunity.
While the focus of the present report has been on the influence of commensal–bacterial derived signals on steady–state basophil populations and basophil–associated allergic inflammation, these findings do not exclude a potential role for DCs or regulatory T cells (Tregs) in influencing aspects of commensal regulation of allergic responses25, 53. Additionally, the findings described here do not exclude a synergistic role for other mediators of allergic inflammation such as thymic stromal lymphopoietin, a cytokine that is regulated by IgE and elevated in the intestines of germ-free mice54, 55. Further analysis of the effects of selective manipulation of commensal bacteria on DCs and Tregs (including inducible LAP+ cell populations56) in murine models and human subjects is therefore warranted.
In summary, the data presented here indicate that commensal bacterial–derived signals limit steady–state serum IgE levels, circulating basophil populations, and basophil–associated allergic inflammation. We propose that in addition to influencing mature basophils in the periphery, IgE binds to bone marrow–resident basophil precursors increasing their responsiveness to IL–3 and resulting in both the population expansion of mature circulating basophils in the steady–state and exaggerated allergen–induced inflammation (Supplementary Fig. 10). These mechanistic insights into the influence of the commensal–IgE–basophil axis on the development of allergic inflammation may have utility in the design of new strategies to prevent or treat allergic diseases in humans.
METHODS
Mice
BALB/c, Swiss–Webster, C57BL/6, Rag1−/−, BaS–TRECK, Csf2rb−/−Csf2rb−/−, Igh–7−/−, IL–4/eGFP reporter, Il4−/−, Myd88−/−, Nod1−/− or Tslp−/− mice (8–24 weeks of age) were bred at the University of Pennsylvania, purchased from Jackson, Taconic or Charles River Laboratories, or provided by Amgen. Germ–free mice were provided by the Penn Gnotobiotic Mouse Facility. Conventional animals were housed in specific pathogen–free conditions. Experiments were performed under Institutional Animal Care and Use Committee (IACUC) approved protocols and in accordance with the guidelines of the IACUC of the University of Pennsylvania.
Reagents and treatments
We fed mice autoclaved water +/− ampicillin (0.5 mg ml−1), gentamicin (0.5 mg ml−1), metronidazole (0.5 mg ml−1), neomycin (0.5 mg ml−1), and vancomycin (0.25 mg ml−1) continuously via water bottle for four weeks. In the event of poor animal hydration, we supplemented control and antibiotic water with artificial sweetener. We conventionalized germ–free mice by housing with conventionally–reared mice for 4–8 weeks. We inoculated mice intranasally on D–17, D–14, D–7 with 50 μl of PBS +/− 100 μg of D. pteronyssinus extract (Greer)25. We injected mice subcutaneously in contralateral footpads with 50 μl of PBS +/− 50 μg of papain (Calbiochem)28 and sacrificed on D3 or D4 post injection for basophil or TH2 cell analyses. We treated mice by intraperitoneal injection (i.p.) with 10 μg of MAR1 or isotype antibody (eBioscience) on D–3, –2, –1. We treated littermate control or BaS–TRECK mice i.p with 750 ng DT on D–2 and D0. We treated Rag1−/− mice i.p. with 50 μg of IgG or IgE antibody (BD Bioscience) daily for seven days. We treated mice i.p. with 200 μg of control IgG or Omalizumab (IgE–specific) antibody (Genentech) daily for 7–9 days. We treated mice i.p. weekly with 100 μl of PBS containing 100 μg of GpC–Phos (5’-ZOOFZEFEOZZOOZEFEZOZT-3’) or CpG–Phos (5’-ZOOFZEFOEZZOOZEFOEZZT-3’).
Flow cytometry, cell sorting and adoptive transfer
We collected blood, spleen, bone marrow, lung, bronchoalveolar lavage or lymph node tissues, digested with collagenase and dispase (lung), homogenized, purified of red blood cells by histopaque (Sigma–Aldrich) or RBC lysis, and stained with mouse fluorochrome–conjugated monoclonal antibodies against B220, CD3ε, CD4, CD8, CD11c, CD19, CD34, CD40, CD49b, CD69, CD80, CD86, CD117, CD123, CD200R, FcεRIα, FcγR, Gr–1, IgE, IgM, IL–4, Ki67, MHC–I, MHC–II or Siglec F (BD Bioscience, BioLegend, eBioscience). We obtained samples from Hyperimmunoglobulinemia E or control subjects with participant consent, assent, and/or parental consent under protocols approved by the Children’s Hospital of Philadelphia and the Ludwig Maximilians University Institutional Review Boards. We isolated PBMCs by Ficoll (GE) gradient, stained with antibodies against human CD11c, CD19, CD117, CD123, FcεRIα, IgE or TCRβ (BD Bioscience, eBioscience) and fixed with 4% PFA. We acquired/sorted cells with a FACSCanto II, LSR II or FACSAria with DiVa software (BD Bioscience) and analyzed with FlowJo software (version 8.7.1; Tree Star). We transferred purified B or T cells i.p. in equal ratios and allowed to reconstitute for 8–10 weeks.
Culture, ELISA and histology
We cultured single–cell LNs suspensions for 2–4 days at 200–250,000 cells in 200 μl of complete media +/− anti–CD3 and anti–CD28 (0.5–1.0 μg ml−1), stimulated for 4 hours with 50 ng ml−1 phorbol 12–myristate 13–acetate, 750 ng ml−1 ionomycin, 10 μg ml−1 Brefeldin A (Sigma–Aldrich) and treated with Fix/Perm (eBioscience). We CFSE labeled and cultured bone marrow cells for 4–5 days at 2–2.5×106 cells per ml +/− 10 ng ml−1 of IL–3 (R&D Systems). We cultured sort–purified BaPs for 4–5 days at 50,000 cells per ml +/− 10 ng ml−1 of IL–3 (R&D Systems). We assayed IgE, IL–4 or IL–13 levels by sandwich ELISA (BD Bioscience) or chart review. We fixed lung tissue sections in 4% paraformaldehyde, embedded in paraffin, cut 5 μm sections and stained with hematoxylin and eosin.
16S rDNA sample acquisition and quantification of 16S rDNA
We collected stool samples and extracted DNA with the QIAamp DNA Stool Mini Kit (Qiagen) as described previously23. We quantified 16S rDNA by real–time RT–PCR with degenerate bacterial 16S rDNA–specific primers (5′-AGAGTTTGATCCTGGCTCAG-3′; forward), (5′-CTGCTGCCTYCCGTA-3′; reverse), (5′-FAM-TA+ACA+CATG+CA+AGTC+GA-BHQ1-3′; probe; + precedes position of LNA base).
DNA manipulations, sequencing and bioinformatic analysis
We obtained 16S rRNA gene fragments as described previously23. We carried out PCR reactions (50 μl) with the AmpliTaq System (Applied Biosystems). We gel purified PCR products with the QIAquick Gel extraction kit (Qiagen). We pooled and pyrosequenced amplicons (100 ng). We assessed sequence quality, and samples with <100 sequences were discarded, and sequences were inserted into a 16S rRNA gene tree using parsimony insertion implemented in ARB. We obtained taxonomic assignments using RDP Classifier.
Statistical analysis
Results are shown as mean ± standard deviation for individual animals. We determined significance by non–parametric two–tailed Mann–Whitney test, unweighted means analysis two–way ANOVA, or linear regression analysis.
Supplementary Material
Acknowledgments
We thank members of the Artis lab for helpful discussions; M.R. Comeau (Tslp−/− mice, Amgen, Inc), L. Shawver, R. Sinha, R. Custers–Allen, H. Oettgen (Igh–7−/− mice, Harvard), J. Weiser (Nod1−/− mice, UPenn) for reagents and experimental advice; J. Sawalle–Beloradsky and B. Hagl for performing DOCK8 sequencing; A. Jansson, G. Notheis, B.H. Belohradsky, M. Albert and the referring physicians for patient care; H. Pletcher, A. Bantly, R. Wychowanec and the UPenn FCCSRL for FACS support; the UPenn Gnotobiotic Mouse Facility for germ–free mice. This work was supported by the National Institutes of Health (T.K., AI067946 to J.O., UH2DK083981 to F.D.B). Research in the Artis lab is supported by the National Institutes of Health (AI061570, AI087990, AI074878, AI095608, AI083480, AI095466, and U01095608 to D.A., T32–AI060516 to D.A.H., F32–AI085828 to M.S., T32–AI05528 to M.C.A.), the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D.A.), the Penn Genome Frontiers Institute (D.A. and F.D.B.), and pilot grants from the NIDDK Center for the Molecular Studies in Digestive and Liver Diseases Molecular Pathology and Imaging Core (DK50306) and the University of Pennsylvania Veterinary Center of Infectious Diseases (D.A.).
Acronyms/Abbreviations
- IL
Interleukin
- TH2
T Helper Type 2
- IgE
Immunoglobulin E
- CNV
Conventional
- ABX
Antibiotic
- GF
Germ–Free
- NBNT
Non–B Non–T
- LN
Lymph Node
- WT
Wild–Type
- PRR
pattern recognition receptor
- DOCK8
Dedicator of Cytokinesis 8
Footnotes
AUTHOR CONTRIBUTIONS D.A.H., M.C.S., M.C.A., B.S.K., D.K. and D.A. designed and performed the research, D.F.L, E.D.R, J.S.O., M.K, T.K. and F.D.B provided reagents, D.A.H., M.C.S., M.C.A., B.S.K., and D.A analyzed the data, D.A.H., M.C.S. and D.A. wrote the manuscript.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Bahadori K, et al. Economic burden of asthma: a systematic review. BMC Pulm Med. 2009;9:24. doi: 10.1186/1471-2466-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mowen KA, Glimcher LH. Signaling pathways in Th2 development. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 4.Holgate ST. Pathogenesis of asthma. Clin Exp Allergy. 2008;38:872–897. doi: 10.1111/j.1365-2222.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- 5.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 6.Zeiger RS. Food allergen avoidance in the prevention of food allergy in infants and children. Pediatrics. 2003;111:1662–1671. [PubMed] [Google Scholar]
- 7.Gilliland FD. Outdoor air pollution, genetic susceptibility, and asthma management: opportunities for intervention to reduce the burden of asthma. Pediatrics. 2009;123(Suppl 3):S168–73. doi: 10.1542/peds.2008-2233G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ege MJ, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 9.Eckburg PB, Lepp PW, Relman DA. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Kalliomaki M, et al. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 13.Kummeling I, et al. Early life exposure to antibiotics and the subsequent development of eczema, wheeze, and allergic sensitization in the first 2 years of life: the KOALA Birth Cohort Study. Pediatrics. 2007;119:e225–31. doi: 10.1542/peds.2006-0896. [DOI] [PubMed] [Google Scholar]
- 14.Marra F, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009;123:1003–1010. doi: 10.1542/peds.2008-1146. [DOI] [PubMed] [Google Scholar]
- 15.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 16.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbst T, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 19.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy KD, et al. Natural IgE production in the absence of MHC Class II cognate help. Immunity. 2006;24:329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Sudo N, et al. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–1745. [PubMed] [Google Scholar]
- 22.Kitaura J, et al. Evidence that IgE molecules mediate a spectrum of effects on mast cell survival and activation via aggregation of the FcepsilonRI. Proc Natl Acad Sci U S A. 2003;100:12911–12916. doi: 10.1073/pnas.1735525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Hammad H, et al. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp Med. 2010;207:2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 2005;23:419–429. doi: 10.1016/j.immuni.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrigoue JG, et al. MHC class II-dependent basophil-CD4+ T cell interactions promote T(H)2 cytokine-dependent immunity. Nat Immunol. 2009;10:697–705. doi: 10.1038/ni.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimoto T, et al. Basophils contribute to T(H)2-IgE responses in vivo via IL-4 production and presentation of peptide-MHC class II complexes to CD4+ T cells. Nat Immunol. 2009;10:706–712. doi: 10.1038/ni.1737. [DOI] [PubMed] [Google Scholar]
- 30.Sokol CL, et al. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat Immunol. 2009;10:713–720. doi: 10.1038/ni.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan BM, et al. Genetic analysis of basophil function in vivo. Nat Immunol. 2011;12:527–535. doi: 10.1038/ni.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novey HS, Marchioli LE, Sokol WN, Wells ID. Papain-induced asthma--physiological and immunological features. J Allergy Clin Immunol. 1979;63:98–103. doi: 10.1016/0091-6749(79)90198-2. [DOI] [PubMed] [Google Scholar]
- 33.Phythian-Adams AT, et al. CD11c depletion severely disrupts Th2 induction and development in vivo. J Exp Med. 2010;207:2089–2096. doi: 10.1084/jem.20100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siracusa MC, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–233. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang Z, Moller C, Nilsson G. IgE-receptor activation induces survival and Bfl-1 expression in human mast cells but not basophils. Allergy. 2006;61:1040–1046. doi: 10.1111/j.1398-9995.2006.01024.x. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 37.Delphin S, Stavnezer J. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline epsilon promoter: regulation by NF-IL-4, a C/EBP family member and NF-kappa B/p50. J Exp Med. 1995;181:181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelhardt KR, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124:1289–302.e4. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361:2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holgate S, et al. The use of omalizumab in the treatment of severe allergic asthma: A clinical experience update. Respir Med. 2009;103:1098–1113. doi: 10.1016/j.rmed.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Pace E, et al. Clinical benefits of 7 years of treatment with omalizumab in severe uncontrolled asthmatics. J Asthma. 2011;48:387–392. doi: 10.3109/02770903.2011.561512. [DOI] [PubMed] [Google Scholar]
- 42.Lin H, et al. Omalizumab rapidly decreases nasal allergic response and FcepsilonRI on basophils. J Allergy Clin Immunol. 2004;113:297–302. doi: 10.1016/j.jaci.2003.11.044. [DOI] [PubMed] [Google Scholar]
- 43.Shiratori I, et al. Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200. J Immunol. 2005;175:4441–4449. doi: 10.4049/jimmunol.175.7.4441. [DOI] [PubMed] [Google Scholar]
- 44.Schnare M, et al. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 45.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu N, Ohnishi N, Ni L, Akira S, Bacon KB. CpG directly induces T-bet expression and inhibits IgG1 and IgE switching in B cells. Nat Immunol. 2003;4:687–693. doi: 10.1038/ni941. [DOI] [PubMed] [Google Scholar]
- 48.Gessner A, Mohrs K, Mohrs M. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J Immunol. 2005;174:1063–1072. doi: 10.4049/jimmunol.174.2.1063. [DOI] [PubMed] [Google Scholar]
- 49.Siracusa MC, Perrigoue JG, Comeau MR, Artis D. New paradigms in basophil development, regulation and function. Immunol Cell Biol. 2010;88:275–284. doi: 10.1038/icb.2010.1. [DOI] [PubMed] [Google Scholar]
- 50.Ohmori K, et al. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. 2009;182:2835–2841. doi: 10.4049/jimmunol.0802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layland LE, Wagner H, da Costa CU. Lack of antigen-specific Th1 response alters granuloma formation and composition in Schistosoma mansoni-infected MyD88-/- mice. Eur J Immunol. 2005;35:3248–3257. doi: 10.1002/eji.200526273. [DOI] [PubMed] [Google Scholar]
- 52.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim BS, et al. Conversion of Th2 memory cells into Foxp3+ regulatory T cells suppressing Th2-mediated allergic asthma. Proc Natl Acad Sci U S A. 2010;107:8742–8747. doi: 10.1073/pnas.0911756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fink LN, et al. Establishment of tolerance to commensal bacteria requires a complex microbiota and is accompanied by decreased intestinal chemokine expression. Am J Physiol Gastrointest Liver Physiol. 2011 doi: 10.1152/ajpgi.00428.2010. [DOI] [PubMed] [Google Scholar]
- 55.Redhu NS, et al. IgE induces transcriptional regulation of thymic stromal lymphopoietin in human airway smooth muscle cells. J Allergy Clin Immunol. 2011;128:892–896.e2. doi: 10.1016/j.jaci.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 56.Chen ML, Yan BS, Bando Y, Kuchroo VK, Weiner HL. Latency-associated peptide identifies a novel CD4+CD25+ regulatory T cell subset with TGFbeta-mediated function and enhanced suppression of experimental autoimmune encephalomyelitis. J Immunol. 2008;180:7327–7337. doi: 10.4049/jimmunol.180.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


