Abstract
Recognition of distinctly modified histones by specialized “effector” proteins constitutes a key mechanism for transducing molecular events at chromatin to biological outcomes1. Effector proteins influence DNA-templated processes, including transcription, DNA recombination, and DNA repair; however, no effector functions have yet been identified within the mammalian machinery that regulates DNA replication. Here we show that ORC1 – a component of ORC (origin of replication complex), which mediates pre-DNA replication licensing2 – contains a BAH (bromo adjacent homology) domain that specifically recognizes histone H4 dimethylated at lysine 20 (H4K20me2). Recognition of H4K20me2 is a property common to BAH domains present within diverse metazoan ORC1 proteins. Structural studies reveal that the specificity of the BAH domain for H4K20me2 is mediated by a dynamic aromatic dimethyllysine-binding cage and multiple intermolecular contacts involving the bound peptide. H4K20me2 is enriched at replication origins and abrogating ORC1 recognition of H4K20me2 in cells impairs ORC1 occupancy at origins, ORC chromatin loading, and cell-cycle progression. Mutation of the ORC1 BAH domain has been implicated in the etiology of Meier-Gorlin syndrome (MGS)3,4, a form of primordial dwarfism5, and ORC1 depletion in zebrafish results in an MGS-like phenotype4. We find that wild-type human ORC1, but not ORC1 H4K20me2-binding mutants, rescues the growth retardation of orc1 morphants. Moreover, zebrafish depleted of H4K20me2 have diminished body size, mirroring the phenotype of orc1 morphants. Together, our results identify the BAH domain as a novel methyllysine-binding module, thereby establishing the first direct link between histone methylation and the metazoan DNA replication machinery, and defining a pivotal etiologic role for the canonical H4K20me2 mark, via ORC1, in primordial dwarfism.
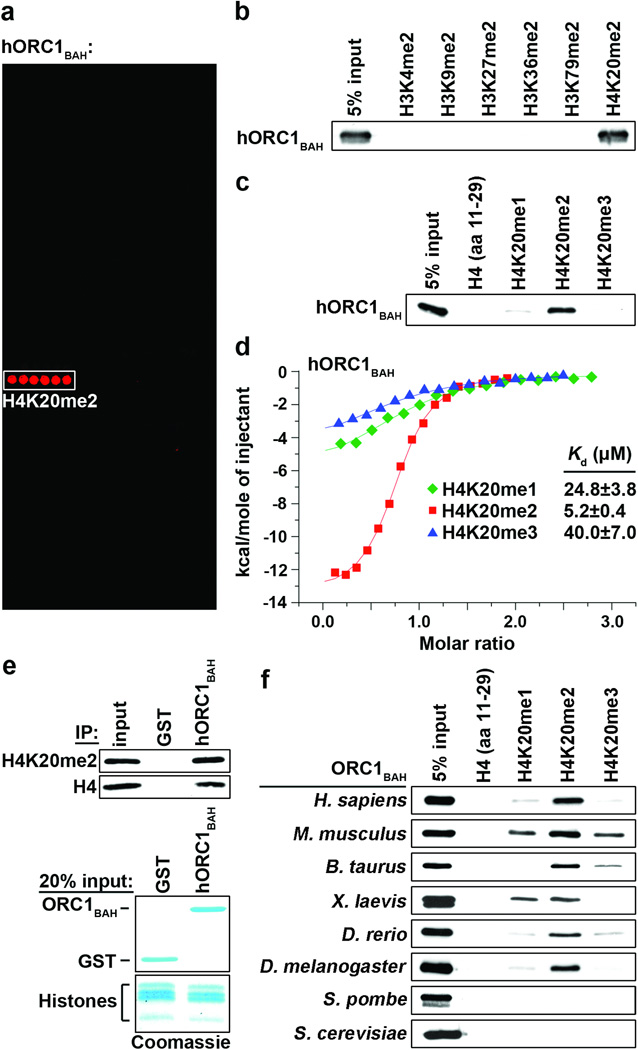
The identification of protein modules that recognize the broad spectrum of modifications present on histone proteins is critical for understanding how chromatin dynamics influence fundamental nuclear processes6. The BAH domain is an evolutionarily conserved chromatin-associated motif7. In budding yeast, the BAH domain of the silencing regulator Sir3 was shown to be a nucleosome-binding domain8–10, suggesting that other BAH domains might function as chromatin-effector modules. To test this hypothesis, we screened several BAH domains from yeast and human for binding activity on peptide microarrays containing 82 modified and unmodified histone peptides, including 54 uniquely methylated peptides (Supplementary Fig. 1)11. The BAH domain of human ORC112 (hORC1BAH: amino acids 1 – 185) bound with high specificity to H4K20me2 peptides (Fig. 1a). In peptide pull-down assays, hORC1BAH bound H4K20me2 peptides, but did not interact with several other dimethylated histone peptides (Fig. 1b). In addition, hORC1BAH did not interact with unmodified H4 peptide, and showed a strong preference for H4K20me2 over H4K20me1 or H4K20me3 (Fig. 1c). Quantitation of the interaction by isothermal titration calorimetry (ITC) demonstrated that hORC1BAH bound to H4(14-25)K20me2 peptides with a Kd of 5.2 µM, whereas monomethylation and trimethylation at K20 reduced the binding affinity five- (Kd = 24.8 µM) and eight- (Kd = 40.0 µM) fold, respectively (Fig. 1d). Finally, hORC1BAH bound to full-length H4K20me2 that is present in bulk-purified histones (Fig. 1e). Together, these in vitro results demonstrate that hORC1BAH binds with high specificity and affinity to H4K20me2.
Figure 1. The ORC1 BAH domain is a novel H4K20me2-binding module.
a, hORC1BAH preferentially binds H4K20me2 peptides. Microarrays spotted with 82 distinct histone peptides as indicated in Supplementary Fig. 1 were probed with glutathione S-transferase (GST) fused to human ORC1 amino acids (aa) 1-185 (hORC1BAH). Red spots indicate positive binding. b and c, hORC1BAH specifically binds H4K20me2 peptides. Western blot analysis of histone peptide pull-downs with GST-hORC1BAH and the indicated biotinylated peptides. d, hORC1BAH binds with highest affinity to H4K20me2. ITC was used to determine the dissociation constants (Kds) for the interaction of hORC1BAH with the indicated peptides: H4K20me1 (green diamonds), H4K20me2 (red squares) and H4K20me3 (blue triangles). Standard deviation (s.d.) was derived from nonlinear fitting. e, hORC1BAH binds full-length H4K20me2. Top: Western blot of GST-hORC1BAH and GST control pull-downs from calf thymus histones (CTH) with the indicated antibodies. Bottom: Coomassie blue stain of input (20% of total). f, H4K20me2 binding is a common property of ORC1 BAH domains from diverse metazoan species. Binding assays as in (b) with GST-fused ORC1 BAH domains from the indicated species and using the indicated peptides.
ORC1 is an evolutionarily conserved protein, found in virtually all eukaryotes2,12. As shown in Figure 1f, H4K20me2 recognition is a common activity of BAH domains of ORC1 from several different metazoans. In contrast, the BAH domains of ORC1 from yeast S. cerevisiae (scORC1BAH) and S. pombe lack H4K20me2-binding activity. In both of these yeast species, it is known that ORC binds directly to DNA sequences at origins of replication, whereas in metazoans, chromatin plays a more significant role in directing ORC to replication origins2,13,14.
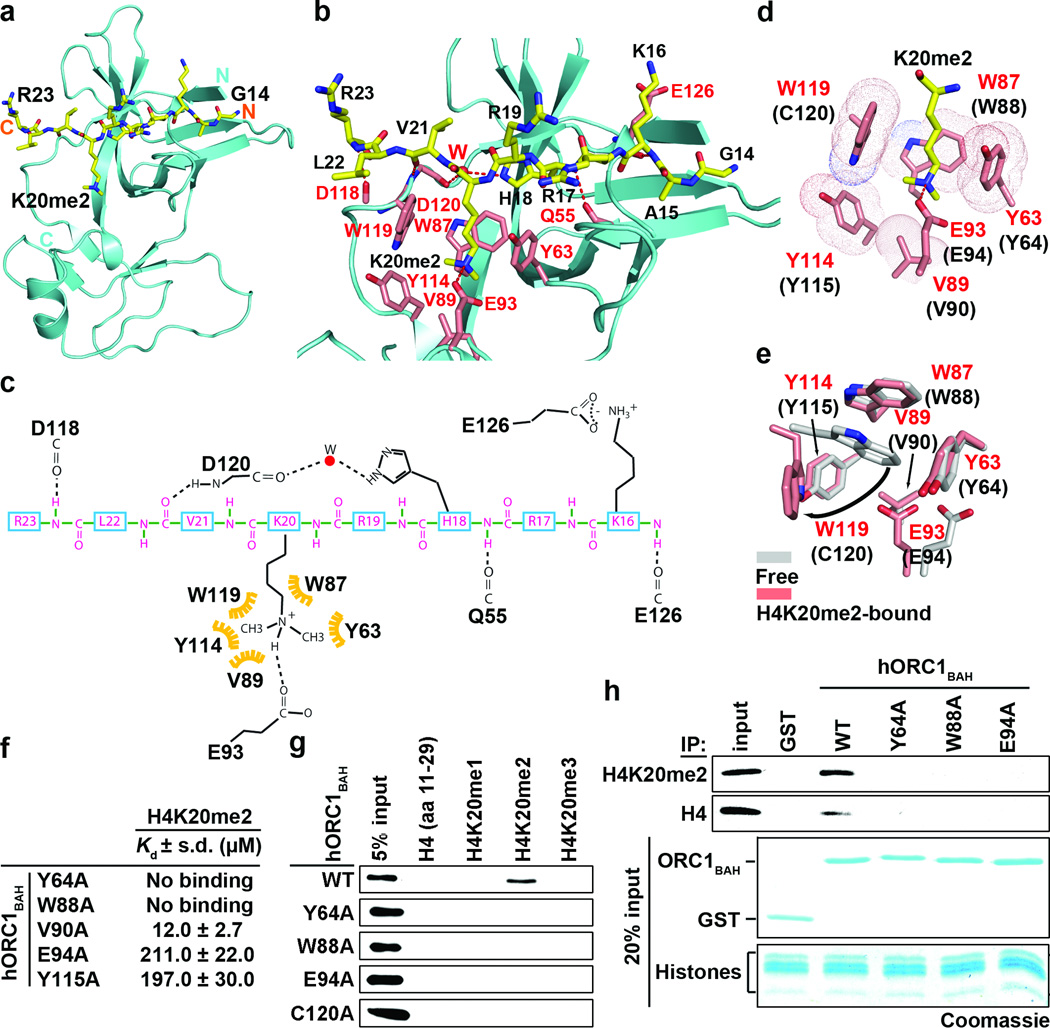
To understand the molecular basis of ORC1 BAH domain recognition of H4K20me2, the crystal structure of the complex between H4(14-25)K20me2 peptide and the BAH domain of mouse ORC1(9-170) (mORC1BAH) was solved at 1.95 Å resolution (Fig. 2a; crystallization statistics in Supplementary Table 1). mORC1BAH adopts a characteristic BAH-domain fold (Fig. 2a), as first reported for the scORC1BAH15. Complex formation between H4(14-25)K20me2 and mORC1BAH is mediated by van der Waals contacts, hydrogen-bonding and electrostatic interactions (Fig. 2b–c). The dimethylammonium group of H4K20me2 is anchored by cation-π interactions with the side chains of an aromatic cage located at the mORC1BAH surface and composed of Tyr63, Trp87, Tyr114 and Trp119, and the dimethylammonium proton is hydrogen-bonded to the side chain of Glu93 (Fig. 2d), similar to H4K20me2 bound to the 53BP1 Tudor domain16 (Supplementary Fig. 2). The aromatic cage in mORC1BAH is absent in the published structure of scORC1BAH (Supplementary Fig. 3)15, explaining why scORC1BAH does not bind H4K20me2 (Fig. 1f).
Figure 2. The molecular basis of H4K20me2 recognition by ORC1BAH.
a–c, 1.95 Å crystal structure of murine ORC1 BAH aa 9-170 (mORC1BAH) complexed with H4(14-25)K20me2 peptide. a, Ribbon representation of mORC1BAH bound to H4K20me2 peptide. The mORC1BAH (cyan) and the bound H4K20me2 peptide (yellow) are shown in ribbon and stick representations, respectively. b, Details of intermolecular contacts in the mORC1BAH-H4K20me2 complex. mORC1BAH and H4K20me2 peptide residues are colored in pink and yellow, respectively, with hydrogen bonds depicted as red dashed lines, and a water molecular as a red sphere. c, Schematic representation of intermolecular contacts in the mORC1BAH-H4K20me2 complex. The residues from the H4K20me2 peptide and mORC1BAH are colored in magenta and black, respectively. Yellow: the hydrophobic contact. d, Positioning of the K20me2 side chain within an aromatic cage of the indicated residues (red) on the surface of mORC1BAH. The equivalent cage residues in hORC1BAH are labeled in parenthesis. e, Structural overlay of the mORC1BAH K20me2-binding aromatic cage in the free (silver) and H4K20me2-bound states (salmon). Arrows indicate binding-induced structural shifts. f, Mutations in the hORC1BAH H4K20me2-binding channel impair H4K20me2 recognition. ITC analysis of the indicated hORC1BAH mutants binding to H4K20me2 peptide; s.d. derived from non-linear fitting. g–h, Mutations in the hORC1BAH dimethyllysine-binding cage abrogate H4K20me2 recognition. g, Binding assays as in (Fig. 1b) with the indicated hORC1BAH mutant proteins and biotinylated peptides. h, Top: Western analysis of CTH binding assays as in (Fig. 1e) with the indicated proteins and antibodies. Bottom: Coomassie blue stain of input GST-fusion proteins and histones (20% of total).
The preference of hORC1BAH for H4K20me2 over its me1/3 counterparts (see Fig. 1c) likely reflects the contribution from the hydrogen bond formed between the dimethylammonium proton and the side chain carboxylate of Glu93 (Fig. 2c). Indeed, previous studies demonstrated that a single Tyr-to-Glu substitution within an otherwise aromatic cage of the BPTF PHD finger reverses the binding preference from trimethyl- to dimethyllysine17. Engineering a binding preference for H4K20me1 likely would require incorporation of a second carboxylate group to line the aromatic cage of hORC1BAH and facilitate hydrogen bond formation with both monomethylammonium protons. The high degree of specificity of mORC1BAH for the sequence surrounding H4K20 is conferred by a series of intermolecular hydrogen-bonding interactions involving Lys16, His18, Val21, and Arg23 of the bound peptide and residues lining the peptidebinding channel of mORC1BAH (Fig. 2b–c). While backbone-backbone interactions represent the majority of contacts, a water-mediated hydrogen bond involving the imidazole ring of His18, as well as a salt bridge between Lys16 of the H4 peptide and Glu126 of mORC1BAH are also observed (Fig 2b–c). In this regard, acetylation at K16 slightly reduced the binding affinity of hORC1BAH and mORC1BAH for the H4K16acK20me2 peptide (Supplementary Fig. 4).
To investigate the structural dynamics of H4K20me2 binding, we determined the 1.70 Å crystal structure of mORC1BAH in the free state (crystallization statistics in Supplementary Table 1). Superimposition of the mORC1BAH structures in the free and peptide-bound states demonstrated that while the interaction with H4K20me2 does not alter the conformation for the majority of the protein, the K20me2-binding pocket of mORC1BAH undergoes a significant rearrangement upon complex formation (Fig. 2e; Supplementary Fig. 5). Specifically, formation of the mORC1BAH-H4K20me2 complex prompts the indole ring of Trp119, which in the free state stacks with the indole ring of Trp87, to swing aside and accommodate insertion of H4K20me2 (Fig. 2e). In addition, the interaction elicits reorientation of Glu93 to allow its side chain to hydrogen bond with the methylammonium proton of H4K20me2 (Fig. 2e).
ITC studies establish that mORC1BAH binds to H4K20me2 peptide with a Kd of 9.6 µM, with the binding affinity towards H4K20me1 and H4K20me3 weaker by a factor of 2 to 3 (Kd = 32.7 µM and 17.4 µM, respectively (Supplementary Fig. 6a)). The mORC1BAH domain shows moderate sequence conservation through evolution, with relatively high conservation of residues lining the H4K20me2-binding pocket (Supplementary Fig. 6b); hORC1BAH retains all the aromatic cage residues that recognize H4K20me2, except for Trp119 of mORC1BAH, which is substituted by Cys120 in hORC1BAH. Replacing mORC1BAHTrp119 with a cysteine (human counterpart) resulted in increased discrimination of H4K20me2 over H4K20me3 from two-fold to three-fold, approaching the eight-fold discrimination observed for hORC1BAH (Supplementary Fig. 6a). However, given that this mutant could not fully recapitulate the H4K20me2-binding specificity of hORC1BAH (Supplementary Fig. 6a), other residues likely contribute to H4K20me2 discrimination. Indeed a C120A substitution in hORC1BAH decreases binding to all three methylation states of H4K20me, with retention of H4K20me2-specificity (Supplementary Fig. 6a).
ITC analysis also demonstrated that alanine substitutions of the remaining residues lining the H4K20me2-binding pocket of hORC1BAH (Y64A, W88A, E94A, and Y115A) abolished or largely diminished H4K20me2 recognition (Fig. 2f). Similar results were observed with hORC1BAH and mORC1BAH cage mutants in peptide-binding assays (Fig. 2g; Supplementary Fig. 6c) and in histone-binding assays (Fig. 2h). These results further support the structural analysis and establish the molecular basis for the H4K20me2-ORC1BAH interaction.
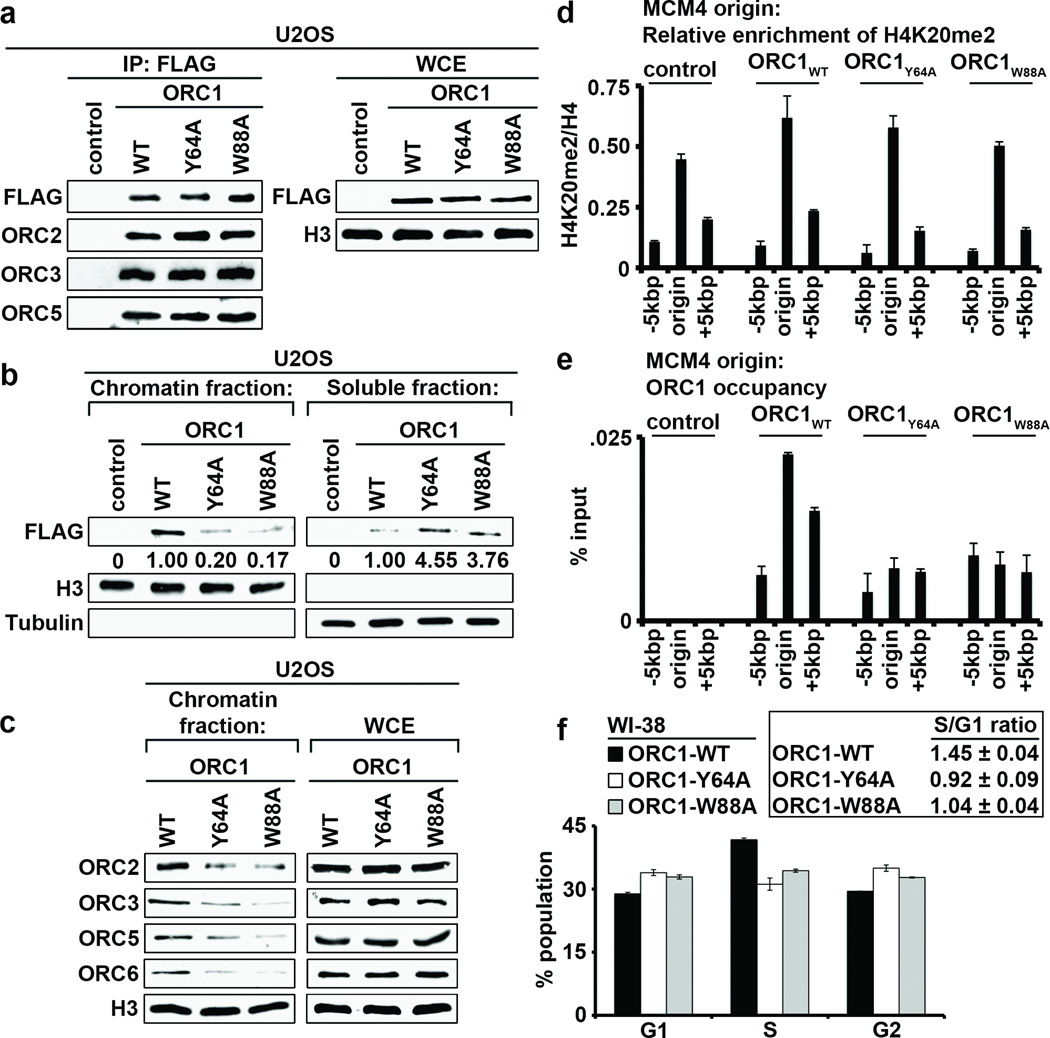
H4K20me2 is an abundant H4 modification18, and the BAH domain of ORC1, while dispensable for ORC complex assembly, has been shown to be important for loading of the complex onto chromatin in humans cells19. We therefore postulated that the interaction between ORC1 and H4K20me2 might regulate ORC stabilization at chromatin. First, ORC components (ORC2, ORC3 and ORC5) affinity purified with two structure-guided H4K20me2-binding mutants (hORC1Y64A and hORC1W88A) with an efficiency equal to that observed with the wild-type protein in both U2OS (Fig. 3a) and HT1080 (Supplementary Fig. 7a) cell lines, suggesting that H4K20me2 binding by ORC1 is dispensable for ORC complex assembly. Next, analysis of lysates biochemically separated into chromatin-enriched and soluble fractions from cells stably expressing hORC1, hORC1Y64A, or hORC1W88A demonstrated that hORC1Y64A and hORC1W88A enrichment at chromatin was considerably reduced in comparison to hORC1 (Fig. 3b; Supplementary Fig. 7b). Moreover, chromatin association of the ORC components ORC2, ORC3, ORC5 and ORC6 was compromised in cells expressing the ORC1 H4K20me2-binding pocket mutants (Fig. 3c; Supplementary Fig. 7c). Thus, hORC1BAH binding to H4K20me2 is required for efficient stabilization of ORC1 and other ORC components at chromatin.
Figure 3. ORC1-H4K20me2 interaction regulates ORC chromatin association and cell cycle progression.
a, Western blot analysis with the indicated antibodies of wild-type (WT) and H4K20me2-binding pocket mutants (Y64A and W88A) affinity-purified FLAG-tagged ORC1 complexes from U2OS cells. Control, empty vector control IP. WCE, whole cell extract. b, The ORC1BAH-H4K20me2 interaction is required for efficient ORC1 chromatin association. Western blot analysis of lysates biochemically separated into chromatin-enriched and soluble fractions from U2OS cells stably expressing the indicated ORC1 protein. Quantitation of FLAG-ORC1 levels is shown. Control, empty vector control lysates. Tubulin and H3 levels are shown as control for the integrity of fractionation. c, Disruption of ORC1 binding to H4K20me2 destabilizes ORC chromatin association. Western blot analysis of biochemically purified chromatin from U2OS cells as in (b) with the indicated antibodies. Total ORC protein levels in WCE are shown. d, H4K20me2 is enriched at DNA replication origins. H4K20me2 signal normalized to total H4 at the MCM4 origin and indicated flanking regions in G1 phase synchronized U2OS cells stably expressing the indicated ORC protein; y-axis: H4K20me2 ChIP/H4 ChIP. e, An intact BAH domain is required for ORC1 occupancy at replication origins. Occupancy of FLAG tagged hORC1, hORC1Y64A, hORC1W88A, or control was determined by ChIP analysis (y-axs: % input) as in (d). Error bars in (d, and e,) indicate s.e.m. from three experiments. f, The ORC1-H4K20me2 interaction is required for efficient cell cycle progression. The cell-cycle profile of WI38 cells transiently expressing GFP and the indicated ORC1 protein was determined by flow cytometry. GFP-positive cells were used to ensure that only transfected cells were analyzed. (Left) Percentage of cells in the indicated cell-cycle phase is shown. (Right) The S/G1 ratio for the indicated transfections is shown. Error bars indicate s.d. from two experiments.
Local chromatin structure is thought to play a role in the mechanism that determines metazoan origins of replication13. In this context, chromatin immunoprecipitation (ChIP) assays with a highly specific H4K20me2 antibody in G1 synchronized U2OS cells (Supplementary Figs. 8–9) demonstrated enrichment of H4K20me2 signal at two defined human replication origins20–23 relative to adjacent sequences (Fig. 3d; Supplementary Fig. 10). Moreover, like the pattern of H4K20me2, hORC1 occupancy peaked at origins relative to adjacent sequences (Fig. 3e; Supplementary Fig. 10). In contrast, hORC1Y64A and hORC1W88A enrichment at origins was not observed, even though the H4K20me2 peak at origins is present in these cell lines (Figs. 3de; Supplementary Fig. 10). These data suggest that H4K20me2 may play a role at human origins by stabilizing ORC1 at chromatin.
ORC marks genomic origins of replication during the G1 phase of the cell cycle and serves to nucleate assembly of the pre-DNA replication complex (pre-RC) to coordinate DNA replication during S phase14. As a core component of this licensing machinery, ORC1 is required for efficient cell-cycle progression and the transition from G1 to S phase4,19. In this context, cell-cycle analysis demonstrated a decreased S phase population and a lower S/G1 ratio in WI38 primary fibroblast transiently expressing the H4K20me2-binding mutants hORC1Y64A and hORC1W88A relative to wild-type hORC1-overexpressing cells (Fig. 3f). Notably, mouse embryonic fibroblast isolated from mice lacking the enzymes that generate H4K20me2 (Suv4-20h1 and Suv4-20h2) show DNA replication defects, including delayed S-phase entry18. Thus, ORC1 recognition of the canonical H4K20me2 mark appears to facilitate ORC chromatin loading to promote DNA replication and cell-cycle progression.
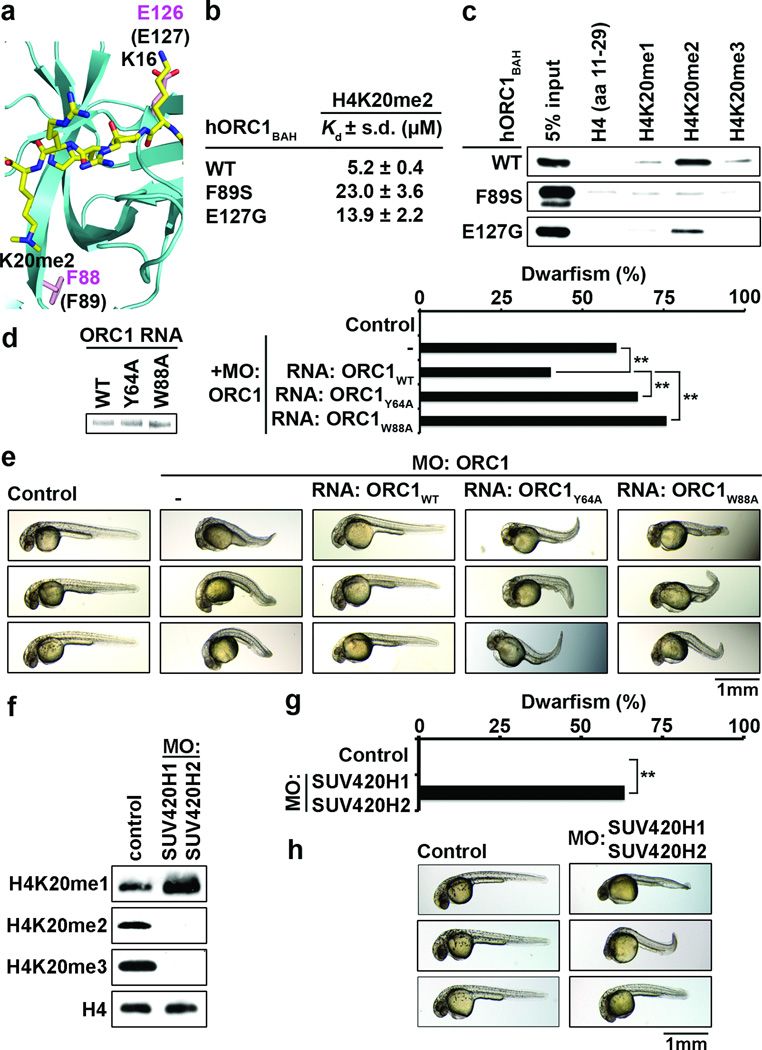
Mutations in components of the pre-DNA replication machinery, including ORC1, have recently been identified in individuals with MGS, and in a zebrafish model, orc1 morphants display an MGS-like proportionate dwarfism phenotype3,4,24. In MGS individuals, the majority of ORC1 mutations fall within the protein’s BAH domain3,4, including two mutants (F89S and E127G) that are in regions that impinge upon H4K20me2 recognition (Fig. 4a) and decrease binding of hORC1BAH to H4K20me2 peptides (Figs. 4b–c). Furthermore, mice deficient in H4K20me2 synthesis are born significantly smaller than control littermates18. Together, these observations implicate disruption of the ORC1-H4K20me2 interaction in MGS pathogenesis. To test this hypothesis, an orc1 zebrafish morphant reconstitution system was established. Injection of two independent orc1-targeting morpholino oligonucleotides (MO) resulted in growth retardation, recapitulating published results4 (Fig. 4d–e; data not shown). Co-injection of hORC1 mRNA with zebrafish orc1-targeting MOs attenuated the dwarf phenotype observed in the orc1 morphant alone (Fig. 4d–e). In contrast, co-injection of hORC1Y64A and hORC1W88A mRNA failed to rescue orc1 morphants, moderately aggravating the growth retardation phenotype (Fig. 4d–e).
Figure 4. Disruption of the ORC1BAH-H4K20me2 interaction leads to dwarfism in zebrafish.
a–c, MGS-associated mutations F89S and E127G impair H4K20me2 binding by hORC1. a, Close-up view of mORC1BAH bound to H4K20me2, with residues F88 and E126 (equivalent to F89 and E127 in hORC1, respectively) shown in stick representation. b, ITC analysis as in (Fig. 1d) of F89S and E127G hORC1 BAH mutants binding to H4K20me2 peptides. *ITC data for wild-type hORC1BAH from Fig. 1d. c, Binding assays as in (Fig. 1b) with the indicated hORC1BAH mutant proteins and biotinylated peptides. d–e, The H4K20me2-recognition activity of hORC1 is required to rescue the dwarfism phenotype of orc1 morphants. d, Quantification of dwarf phenotype in zebrafish injected with morphilino oligos (MO) targeting the orc1 translation start site (tss) alone or MO co-injected with the indicated hORC1 mRNAs. Insert on left shows electrophoresis analysis of the indicated hORC1 mRNAs used for reconstitution. Control, uninjected embryos. Dwarfism was defined as a reduction of body length of ≥ 3 s.d. relative to the average size of the control zebrafish. Zebrafish analyzed: control: 26; orc1 MO: 86; MO+hORC1WT: 67; MO+hORC1Y64A: 85; MO+hORC1W88A: 83. P values are calculated with a two-tailed unpaired Student’s t-test. ** P < 0.01. e, Representative images of zebrafish in (b) 1 day post-fertilization (dpf). f, Depletion of H4K20me2/3 in suv4-20h1/suv4-20h2 morphants. Western blot analysis using the indicated antibodies of whole animal extracts 1 dpf from control zebrafish or zebrafish injected with MOs targeting the tss’s of suv4-20h1 and suv4-20h2. Control, uninjected embryos. g–h, Dwarfism in suv4-20h1/suv4-20h2 morphants. g, Quantification of dwarfism in suv4-20h1/suv4-20h2 morphants relative to controls as described in (d). Zebrafish analyzed: control: 38; MO: 60. ** P<0.01. h, representative images of zebrafish in (e) 1 dpf.
The H4K20me2/3 lysine methyltransferases (KMTs) Suv4-20h1 and Suv4-20h2 are conserved in zebrafish25. As shown in Figure 4f, injection of MOs targeting Danio rario suv4-20h1 and suv4-20h2 resulted in global depletion of H4K20me2 and H4K20me3, and an increase in H4K20me1; these changes in H4K20 methylation are similar to those observed in cells derived from Suv4-20h1/20h2 double knockout mice18. Analysis of body size in the suv4-20h1/20h2 morphants demonstrated that H4K20me2 depletion results in a significant dwarfism phenotype compared to controls (Fig. 4g–h). Finally, orc1/suv4-20h1/20h2 morphants did not display significantly more dwarfisms than orc1 or suv4-20h1/20h2 morphants (Supplementary Fig. 11). Together, these data argue that the specific recognition of H4K20me2 by the BAH domain of ORC1 plays a crucial role in determining organism body size.
In summary, we have identified the BAH domain as a novel methyllysine-binding module, demonstrating that H4K20me2 recognition is an evolutionarily conserved function for BAH domains from diverse metazoan ORC1 proteins. These results provide the first direct link between histone methylation and the metazoan DNA replication machinery. The mechanism that determines metazoan origins of replication is thought to be dependent upon both the information encoded in the DNA sequence at origins, as well as the local chromatin architecture13,14. Several histone modifications have been detected at origins20–22,26,27, including H3K27me1, which regulates replication of DNA at heterochromatin in Arabidopsis28. The mark H4K20me1 is present in early G1 at human origins and likely serves as the chromatin template for H4K20me2 catalysis18,29,30. Since H4K20me1 is generated in a highly cell-cycle regulated manner by the KMT PR-Set7/SET8, this modification may regulate licensing by governing the temporal and spatial availability of H4K20me2 at origins29. We postulate that the recognition of H4K20me2 by ORC1 cooperates with other ORC chromatin-loading mechanisms in marking replication origins. Based on this model, during developmental phases requiring rapid cell division, inefficient pre-RC formation due to abrogation of the ORC1-H4K20me2 interaction would result in delayed cell-cycle progression and insufficient cellular proliferation, a characteristic of proportional “hypocellular” dwarfism disorders like MGS5. Together, our findings reveal a new function for histone methylation signaling at chromatin in the regulation of DNA replication and organismal growth.
METHODS
Materials, Peptide microarray experiments, binding assays, structure analysis and crystallization conditions, and zebrafish experiments and data analysis are described in detail in Supplementary Information. For the zebrafish experiments, wild-type AB zebrafish (Danio rerio) was maintained and raised using standard protocols. All zebrafish were treated in accordance with AAALAC approved guidelines at Stanford University (protocol number: 10511).
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Tennen for critical reading of the manuscript. This work was supported in part by grants to O.G. (R01 GM079641), D.J.P (Abby Rockefeller Mauze, STARR and Maloris Foundations) and J.KC. (DP1 OD003792), and a predoctoral fellowship to A.J.K. (Genentech Foundation). O.G. is a recipient of an Ellison Senior Scholar in Aging Award.
Footnotes
AUTHOR CONTRIBUTIONS
A.J.K and P.C. performed the molecular biology, cellular, and zebrafish studies; J.S. performed structural and binding affinity studies; S.Y. and J.K.C. advised on zebrafish experiments; S.I.M. assisted in protein production and crystallization. A.J.K., P.C., J.S., D.J.P., and O.G. designed studies, analyzed data, and wrote the paper. All authors discussed and commented on the manuscript.
Atomic coordinates have been deposited with the Protein Data Bank under the accession codes for the free (accession no: 4DOV) and H4K20me2-bound mORC1BAH (accession no: 4DOW) for the reported crystal structures. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
REFERENCES
- 1.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bicknell LS, et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bicknell LS, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43:350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 5.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–2024. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 8.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Sampath V, et al. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol Cell Biol. 2009;29:2532–2545. doi: 10.1128/MCB.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bua DJ, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 13.Dorn ES, Cook JG. Nucleosomes in the neighborhood: new roles for chromatin modifications in replication origin control. Epigenetics. 2011;6:552–559. doi: 10.4161/epi.6.5.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 2002;21:4600–4611. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tardat M, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 21.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human betaglobin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 24.Guernsey DL, et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43:360–364. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 25.Sun XJ, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costas C, et al. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol. 2011;18:395–400. doi: 10.1038/nsmb.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob Y, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature. 2010;466:987–991. doi: 10.1038/nature09290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brustel J, Tardat M, Kirsh O, Grimaud C, Julien E. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 2011;21:452–460. doi: 10.1016/j.tcb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.